Abstract
Using an acetylcholine-derivatized affinity column, we have purified human α4β2 neuronal nicotinic acetylcholine receptors (nAChR) from a stably transfected HEK-293 cell line. Both the quantity and the quality of the purified receptor are suitable for applying biochemical methods to directly study the structure of the α4β2 nAChR. In this first study, the lipid-protein interface of purified and lipid reconstituted α4β2 nAChRs was directly examined using photoaffinity labeling with the hydrophobic probe 3-trifluoromethyl-3-(m-[125I]iodophenyl) diazirine ([125I]TID). [125I]TID photoincorporated into both α4 and β2 subunits, and for each subunit the labeling was initially mapped to fragments containing the M4 and M1-M3 transmembrane segments. For both the α4 and β2 subunits, ~ 60% of the total labeling was localized within fragments that contain the M4 segment which suggests that the M4 segment has the greatest exposure to lipid. Within M4 segments, [125I]TID labeled homologous amino acids α4-Cys582/β2-Cys445 which are also homologous to the [125I]TID-labeled residues α1-Cys418 and β1-Cys447 in the lipid exposed face of Torpedo nAChR α1M4 and β1M4, respectively. Within the α4M1 segment, [125I]TID labeled residues Cys226 and Cys231 which correspond to the [125I]TID-labeled residues Cys222 and Phe227 at the lipid exposed face of the Torpedo α1M1 segment. In β2M1, [125I]TID labeled β2-Cys220 which is homologous to α4-Cys226. We conclude from these studies that the α4β2 nAChR cam be purified from stably transfected HEK-293 cells in sufficient quantity and purity for structural studies and that the lipid-protein interface of the neuronal α4β2 nAChR and the Torpedo nAChR display a high degree of structural homology.
Neuronal nicotinic acetylcholine receptors (nAChR1) are members of the Cys-loop superfamily of ligand gated ion channels that mediate the actions of the neurotransmitter acetylcholine (1, 2). Neuronal nAChRs are widely distributed in the nervous system and play a role in many physiological functions including arousal, sleep, attention, memory, mood, emotion, pain perception, food intake and cognition and are implicated in numerous pathophysiological conditions including epilepsy, schizophrenia, Alzheimer’s and Parkinson’s diseases, anxiety, and nicotine addiction (reviewed in 3–5). To date twelve mammalian neuronal nAChR subunit genes have been cloned; nine neuronal alpha subunits (α2-α10), and three neuronal beta subunits (β2-β4). Based on the three dimensional structure of the Torpedo nAChR (6, 7) and the available structural information regarding neuronal nAChRs (reviewed in 8, 9), the α4β2 neuronal nAChR is a pentameric membrane protein that is formed by the assembly of two α4 and three β2 subunits. Each nAChR subunit contains a large extracellular N-terminus and a bundle of four transmembrane alpha helices (M1–M4). The five M2 helices are arranged about a central axis orthogonal to the membrane forming the channel lumen and the M1, M3, and M4 helices form an outer ring that shield M2 from the lipid bilayer.
Due to the abundance and purity of muscle-type nAChR in preparations from the electric organ of the Torpedo electric ray, it is the most studied and understood member of the Cys-loop superfamily of ligand-gated ion channels, and many of the structural/functional features of the Torpedo nAChR may be generalized to other member of the superfamily including neuronal nAChRs. This approach is largely justified by the high sequence identity/homology between Torpedo and neuronal nAChR subunits (e.g. Torpedo α1 and neuronal α4 subunits display 53% sequence identity). At the same time, expression of neuronal nAChR in heterologous expression systems, combined with site-directed mutagenesis and electrophysiological analysis, have associated particular amino acid residues with agonist binding, channel gating, ion conductance and desensitization (2, 9, 10). Nevertheless, direct structural information regarding neuronal nAChRs is currently lacking. This is in large part due to the low expression level of individual neuronal nAChRs, the high level of diversity among neuronal nAChR subunits in a given brain area, and the lack of subunit-specific ligands.
Purification of individual neuronal nAChRs is an important step in the effort to directly study the structure of neuronal nAChRs and to reveal the structural differences among neuronal nAChR subtypes. We report here a purification strategy that provides highly purified neuronal nAChR in a lipid environment and in a quantity that is suitable for direct structural studies (e.g. photoaffinity labeling, Fourier transform infrared spectroscopy (FTIR), etc). Membrane preparations from HEK-293 cells stably transfected with human α4β2 nAChRs were detergent solubilized and the receptors were purified on an acetylcholine-derivatized affinity column and reconstituted into lipid. To begin direct structural studies of the α4β2 nAChR, we chose to examine the structure of the lipid-protein interface of the receptor and to determine amino acid residues of α4 and β2 subunits that are in contact with membrane lipid. For this we employed the hydrophobic photoreactive probe 3-trifluoromethyl-3-(m-[125I] iodophenyl) diazirine ([125I]TID). In earlier studies, [125I]TID was used to define the lipid-protein interface of the Torpedo nAChR (11, 12). [125I]TID photoincorporated into the M4 and M1 segments of both the α4 and β2 subunits with ~60% of the total subunit labeling localized in the M4 segment. The labeled amino acids within the α4M4/β2M4 and α4M1/β2M1 segments correspond to residues labeled in the Torpedo nAChR M4/M1 segments, consistent with the existence of a high degree of structural homology between the transmembrane domain of the neuronal α4β2 nAChR and the Torpedo nAChR at the lipid-protein interface.
EXPERIMENTAL PROCEDURES
Materials
3-trifluoromethyl-3-(m-[125I]iodophenyl) diazirine ([125I]TID; ~10 Ci/mmol) was obtained from Amersham Biosciences (Piscataway, NJ) and stored in ethanol:water 3:1 at −4°C. [3H]nicotine (L-(-)-[N-methyl-3H]nicotine; ~ 70 Ci/mmol) was obtained from Perkin Elmer Life Sciences, Inc. (Boston, MA) and stored in 95% ethanol at 4°C. Carbamylcholine chloride and bromoacetylcholine bromide were purchased from Sigma-Aldrich (St. Louis, MO); S. aureus glutamyl endopeptidase (V8 protease) from MP Biochemicals; Trypsin (TPCK-treated) from Worthington; o-Phthalaldehyde (OPA) and Trifluoroacetic acid (TFA) from Pierce; Sodium cholate and CHAPS from USB Corporation (Cleveland, OH); Affi-Gel 10 from Bio Rad; protease inhibitor cocktail III and Genapol C-100 from Calbiochem. Prestained low range molecular weight standards were purchased from Life Technologies, Inc. Dulbecco’s Modification of Eagle’s Medium/Ham’s F-12 50/50 mix (DMEM/Ham’s F-12) was purchased from Mediatech, Inc (Herndon, VA). Natural and synthetic lipids were from Avanti Polar lipids, Inc. (Alabaster, AL).
Cell Culture and Membrane Preparation
HEK-293 cells stably transfected with human α4β2 nAChRs (HEK-hα4β2 hereafter) were obtained from Dr. Joseph H. Steinbach (Department of Anesthesiology, Washington University School of Medicine, Saint Louis, MO; 13). Cells were grown at 37°C in a humidified incubator at 5% CO2, in 140 mm tissue culture dishes and were maintained in DMEM/Ham’s F-12 (Mediatech, Inc.), supplemented with 10% fetal bovine serum, 100 units/mL penicillin G, 100 µg/mL streptomycin and 450 µg/mL geneticin (G418) as a selection agent. The use of 140 mm dishes for culturing these adherent cells (HEK-hα4β2) was determined to be the most cost-effective method for large-scale production of receptor protein. However, we are currently exploring culturing HEK-hα4β2 cells in spinner flasks (5–15 L) with the addition of glass microcarriers (Cytodex-3) in order to scale-up receptor protein production. In most cases, 100 µM nicotine was added to the media 24 h prior to harvesting to enhance the expression of α4β2 nAChRs (10). Cells were harvested by gentle scraping in 5 mL of growth media in the presence of protease inhibitor cocktail III (Calbiochem; 0.2 µL/mL), pelleted by centrifugation (210g for 4 min), resuspended in small volume of vesicle dialysis buffer (VDB: 100mM NaCl, 0.1mM EDTA, 0.02% NaN3, 10mM MOPS, pH 7.5) and then pelleted by centrifugation. The final cell pellet was stored at −80° C.
For membrane preparation, HEK-hα4β2 cells were thawed and homogenized in VDB in the presence of protease inhibitor cocktail III (Calbiochem; 1 µL/mL) using a glass homogenizer. Membrane fractions were pelleted by centrifugation (39,000g for 1 h), then the membrane pellets were resuspended in VDB (~ 0.5 ml/140 mm dish) and the protein concentration determined by Lowry protein assay (Lowry et al. 1951).
[3H]Nicotine Binding Assay
Equilibrium binding of [3H]nicotine (~ 70 Ci/mmol; Perkin Elmer Life Sciences) to HEK-hα4β2 membranes was determined using a centrifugation assay. HEK-hα4β2 membranes were suspended in VDB (final concentration 0.33 mg protein/mL, final volume 150 uL) and incubated for 1 h at room temperature with increasing concentrations of [3H]nicotine (final concentrations 2–60 nM). Bound [3H]nicotine was separated from free by centrifugation (39,000g for 1 h). Free [3H]nicotine was determined by counting 50 µL of the supernatant in a liquid scintillation counter. Bound [3H]nicotine was determined by suspending the pellet in 200µl of 10 % SDS for liquid scintillation counting. Non-specific binding was determined in the presence of 10 µM nicotine. Total, non-specific and specific 3H cpm were converted to fmol bound [3H]nicotine/mg protein and free 3H cpm was converted to nM [3H]nicotine. Curve fitting and parameter estimation were performed using Graph pad Prism v4.0 software (San Diego, CA).
Solubilization, Purification and Reconstitution
Human α4β2 nAChRs were affinity-purified on a bromoacetylcholine bromide-derivatized Affi-Gel 10 column (Biorad) originally developed for purification of the Torpedo nAChR from electric organ tissue (15, 16) but with some modifications. Briefly, the affinity column matrix was prepared by amino-coupling cystamine to Affi-Gel 10, reduction with dithiothreitol and final modification with bromoacetylcholine bromide. HEK-hα4β2 membranes were solubilized by adding an equal volume of 2% CHAPS or 2% cholate in VDB (final concentration 2 mg/mL protein; 1% detergent; final volume 800 mL), stirred for 5 h at 4° C and then centrifuged (91,500g for 1 h) to pellet insoluble material. The solubilized material was dialyzed for 5 h against 1% cholate in VDB (solubilized material to dialysis buffer ratio 1:10). This dialysis step is a critical aspect of the purification strategy; in its absence nAChRs are not retained on the ACh-affinity column. The dialyzed solubilized material was then treated with diisopropyl fluorophosphates (0.1 mM), slowly applied to the affinity column (0.3 mL/min, ~24 h, at 4°C), and the column then washed extensively with a defined lipid solution (e.g. total lipid extract from porcine brain, or dioleoylphosphatidylcholine: dioleoylphosphatic acid: cholesterol 3:1:1; 0.2 mg/mL lipid) in 1% cholate in VDB (15 column volumes; >15 h). This extensive wash ensures complete exchange of endogenous lipids for the defined lipid mixture and removal of nonspecific protein (16, 17). Receptors were eluted from the column using the defined lipid solution containing 10 mM carbamylcholine. Fractions of 2.5 mL were collected, and the protein concentration determined (A280 * 0.6; 15). Peak protein fractions were pooled and dialyzed against 2 L of VDB (4 d with buffer change once a day) to remove carbamylcholine and detergent, thereby reconstituting nAChRs into membrane vesicles containing a defined lipid mixture. The purified nAChRs were stored at −80°C.
[125I]TID Photolabeling
For analytical labelings, 50 µg of affinity-purified and lipid-reconstituted α4β2 nAChRs were incubated with ~0.4 µM [125I]TID (~10 Ci/mmol; Amersham Biosciences) in the absence or presence of 400 µM carbamylcholine in 1 ml VDB. For preparative labelings, 1 mg of affinity-purified and lipid- reconstituted α4β2 nAChRs in 4 mL of VDB were incubated with ~ 8 µM [125I]TID. After 1 h incubation at room temperature under reduced light conditions, the samples were irradiated with a 365 nm hand-held UV lamp (Spectroline EN-280L) for 7 min (analytical labeling) or 20 min (preparative labeling) at a distance of less than 1 cm and then pelleted by centrifugation (39,000g for 1 h). Pellets were solubilized in electrophoresis sample buffer (12.5 mM Tris-HCL, 2% SDS, 8% sucrose, 1% glycerol, 0.01% bromophenol blue, pH 6.8) and the polypeptides were resolved by SDS-PAGE.
SDS-Polyacrylamide Gel Electrophoresis
SDS-PAGE was performed according to Laemmli (18) with the 1.0 mm thick separating gel comprised of 8% polyacrylamide/0.33 bisacrylamide. Gels were stained for 1 h with Coomassie Blue R-250 (0.25% (w/v) in 45% methanol, 10% acetic acid, 45% H2O), and destained (25% methanol, 10% acetic acid, 65% H2O) to visualize bands. Gels were then dried and exposed to Kodak X-OMAT LS film with an intensifying screen at −80°C (12–48 h exposure). After autoradiography, bands that correspond to [125I]TID-labeled α4 and β2 subunits were excised, soaked in overlay buffer (5% sucrose, 125 mM Tris-HCl, 0.1% SDS, pH 6.8) for 30 min and transferred to the wells of a 15% acrylamide mapping gel (19). Each gel slice was overlaid with 2 µg (analytical labeling) or 100 µg (preparative labeling) of V8 protease in overlay buffer. After electrophoresis, the gels were stained for 2 h with Coomassie Blue R-250, destained and then either prepared for autoradiography (analytical labeling) or soaked in distilled water overnight (preparative labeling). The bands corresponding to labeled subunit proteolytic fragments, (α4V8-8, α4V8-14, α4V8-16, β2V8-8, β2V8-13 and β2V8-21) were excised from preparative gels, and the labeled peptides were retrieved by passive diffusion into 25 mL of elution buffer (0.1M NH4HCO3, 0.1% (w/v) SDS, 1% β-mercaptoethanol, pH 7.8) for 4 d at room temperature with gentle mixing. Gel pieces were removed by filtration (Whatman No. 1 paper), and the peptides were concentrated using Centriprep-10 concentrators (10 KDa cutoff, Amicon; final volume < 150 uL). Samples were then either directly purified using reversed phase HPLC (α4V8-16, and β2V8-8) or acetone precipitated (>85% acetone at −20°C overnight) to remove excess SDS and then subjected to additional proteolytic digestion (α4V8-14, β2V8-13, and β2V8-21).
Proteolytic Digestions
For digestion with trypsin, acetone-precipitated subunit fragments (α4V8-14, β2V8-13, and β2V8-21) were suspended in 60 µL 0.1M NH4HCO3, 0.1% SDS, pH 7.8, then the SDS content was diluted by addition of 225 µL 0.1M NH4HCO3 and 35 µL Genapol C-100 (final concentrations: 0.02% (w/v) SDS, 0.5% Genapol C-100, pH 7.8). Trypsin was added at a 200% (w/w) enzyme to substrate ratio, and the digestion was allowed to proceed for 4 d at room temperature.
Tricine SDS-PAGE
The tryptic digestion products of [125I]TID-labeled fragment β2V8-21 were resolved on a 1.0 mm thick small pore (16.5%T, 6%C) Tricine SDS-PAGE gel (12, 20). After electrophoresis, Tricine gels were processed for autoradiography as described above for 8% gels. Tricine gel bands containing [125I]TID-labeled peptide fragments β2T6K, β2T10K, and β2T12K were excised and processed for HPLC purification.
Reversed-Phase HPLC Purification
All of the [125I]TID-labeled peptides were purified using reversed-phase HPLC prior to sequence analysis. HPLC was performed on a Shimadzu LC-10A binary HPLC system, using a Brownlee Aquapore C4 column (100 × 2.1mm). Solvent A was comprised of 0.08% trifluoroacetic acid (TFA) in water and Solvent B, 0.05% TFA in 60% acetonitrile/ 40% 2- propanol. A nonlinear elution gradient at 0.2 mL/min was employed (25% to 100% Solvent B in 100 min, shown as dotted line in figures) and fractions were collected every 2.5 min (42 fractions/run). The elution of peptides was monitored by the absorbance at 210 nm, and the amount of 125I associated with each fraction was determined by γ-counting.
Sequence Analysis
Amino terminal sequence analysis of nAChR subunits and some subunit fragments was performed on a Beckman Instruments (Porton) 20/20 automated protein sequencer using gas phase cycles (Texas Tech Biotechnology Core Facility). Pooled HPLC fractions were dried by vacuum centrifugation, resuspended in 20 µL 0.1% SDS and immobilized on chemically modified glass fiber disks (Beckman Instruments). Peptides were subjected to at least 10 sequencing cycles.
Sequencing of purified [125I]TID-labeled subunit fragments was performed on an Applied Biosystems PROCISE™ 492 protein sequencer configured to utilize 1/6 of each cycle of Edman Degradation for amino acid identification / quantification and collect the other 5/6 for 125I counting. HPLC fractions of interest were diluted 3-fold with 0.1 % trifluoroacetic acid and loaded onto PVDF filters using Prosorb® Sample Preparation Cartridges (Applied Biosystems #401959). Before sequencing, filters were processed as recommended by the manufacturer. To determine the amount of the sequenced peptide, the pmol of each amino acid in a detected sequence was quantified by peak height and fit to the equation f(x) = I0Rx, where I0 was the initial amount of the peptide sequenced (in pmol), R was the repetitive yield, and f(x) was the pmol detected in cycle x. Ser, His, Trp, and Cys were not included in the fits due to known problems with their accurate detection / quantification. The fit was calculated in SigmaPlot 2001 (SPSS) using a non-linear least-squares method and Figures containing 125I release profiles include this fit as a dotted line. Some sequencing samples were treated with o-phthalaldehyde (OPA) prior to a cycle known to contain a proline (21). OPA reacts with all N-terminal amino acids (but not with the imino acid proline) and blocks further Edman degradation (22). Thus release of 125I in a cycle after an OPA treatment establishes that the 125I release originates from a peptide with a proline in the OPA-treated cycle. Quantification of 125I incorporated into a specific residue was calculated by (cpmx-cpm(x-1))/5IoRx, then the value was decay corrected to the date of labeling.
Molecular Modeling
A homology model of the human α4β2 nAChR was built on the Torpedo marmorata nAChR structure (PDB # 2BG9) using the homology module within the Insight II molecular modeling package (Accelrys). Two α4 sequences were aligned with and substituted into the two α1 subunits in the model, and β2 sequences were aligned with and substituted into each of the remaining subunits (β1, γ, and δ) to create a receptor with a stochiometry of 2α4:3β2.
RESULTS
Affinity-purification of α4β2 nAChR
In the absence of a natural rich source of α4β2 nAChRs, one alternative is a cell line that stably expresses α4β2 nAChRs. HEK-293 cells stably or transiently transfected with α4β2 nAChRs express functional receptors and have been used by several groups as an in vitro system to study structural and functional aspects of the neuronal nAChR including the pharmacology of ligand binding (13, 23–25), agonist-induced upregulation (10, 26), subunit stoichiometry (27) and the effect of steroids (28, 29).
The expression level of hα4β2 nAChRs in HEK-hα4β2 is typically ~5 pmol receptor/mg protein with a [3H]nicotine binding affinity (Kd) of ~ 6 nM (data not shown), consistent with the equilibrium binding affinity for nicotine measured using cell homogenates expressing rat or human α4β2 nAChRs (30). Treatment of HEK-α4β2 cells with 100 µM nicotine 24 h prior to harvesting enhanced the level of hα4β2 nAChRs expression by 3-fold (~15 pmol receptor/mg protein; data not shown).
When receptor solubilization in various detergents including CHAPS, sodium cholate, and Triton X100 was tested, we found similar levels of solubilization (40–60 % of total protein), with no preferential nAChR solubilization. We decided to use cholate for solubilization because: i) it can be readily removed by dialysis following purification allowing reconstitution of the receptor into a lipid environment, and ii) Torpedo nAChRs solubilized in cholate are stabilized in the resting (closed) state and retain the ability to undergo agonist-induced conformational transitions (31). [3H]nicotine binding affinity following detergent solubilization in cholate and dialysis was identical to that measured with HEK-α4β2 membranes.
Human α4β2 nAChRs were affinity-purified using a bromoacetylcholine bromide-derivatized Affi-Gel 10 column (see Experimental Procedures). This ACh-affinity column has been used to purify Torpedo nAChRs (32, 15), but its application to the purification of neuronal nAChRs has been very limited (33, 34). A number of factors, including the low abundance of neuronal nAChRs in native tissues (<1 pmol/mg) as well as the heterogeneity of nAChR subtypes, have all contributed to the scarcity of reports involving neuronal nAChR purification. For each column purification, HEK-α4β2 membranes from ~1000 culture dishes (20×140 mm) were collected over a six week period and solubilized in 1% cholate in VDB. The solubilized material was dialyzed for 5 h against 1% cholate in VDB and applied slowly to the affinity column. The dialysis step was a critical aspect of the purification strategy; in its absence nAChRs were not retained on the ACh-affinity column. The column was then washed extensively with a defined lipid/1% cholate solution, and bound nAChRs then eluted from the column using the same solution containing 10 mM carbamylcholine. Fractions were collected, the protein concentration determined, (see supplementary Figure 1) and peak protein fractions were pooled and dialyzed against VDB to remove carbamylcholine and detergent (reconstitution). For a typical column run (for HEK cells grown in the absence of nicotine), we started with ~3 grams of HEK-hα4β2 membrane protein containing ~10 nmol of nicotine binding sites (~4 mg of α4β2 nAChR protein), and the yield following detergent solubilization and affinity purification was ~50% (2.2 mg of receptor at ~4 nmol [3H]nicotine binding sites /mg protein).
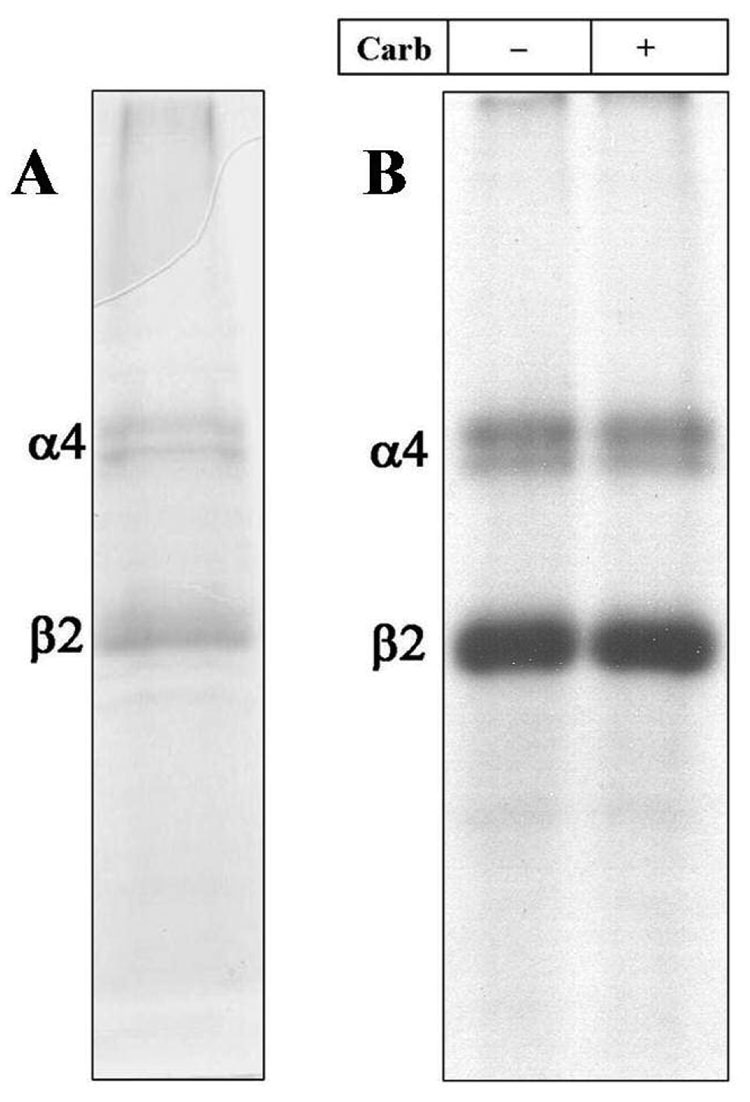
When an aliquot (~50 µg) of affinity-purified α4β2 nAChRs was resolved on a 1 mm thick 8% polyacrylamide gel, two primary Commassie blue stained bands were visible with apparent molecular mass of 70 KDa and 48 KDa (Figure 1A). The 70 KDa and 48 KDa bands correspond to the expected electrophoretic mobility of the α4 and β2 nAChR subunits respectively (35). This conclusion was confirmed by N-terminal sequencing and by LC/MS (Taplin Biological Mass Spectrometry Facility, Boston, MA). Based on densitometric scans of the Commassie blue stained gel (Supplementary Figure 2A) and saturation [3H]nicotine binding (~4 nmol/mg protein), we estimated that the α4β2 nAChR preparation was greater than 50% pure. The relative intensities of the α4 and β2 subunit bands in the stained 8% gel (Supplementary Figure 2A) were consistent with a subunit stoichiometry of (α4)2(β2)3 (36). Both the percent yield and the purity based on SDS-PAGE were reproducible in three successive purifications starting with ~1000 culture dishes.
Figure 1. Photoincorporation of [125I]TID into Purified α4β2 nAChR.
An aliquot of affinity-purified α4β2 receptor (50 µg) was equilibrated for 1 h with [125I]TID (0.4 µM; 10 µCi), in the absence (− lanes) and in the presence (+ lanes) of 400 µM carbamylcholine (Carb) then irradiated at 365 nm for 7 min. The protein was pelleted by centrifugation, resuspended in electrophoresis sample buffer and fractionated by SDS-PAGE. Following electrophoresis, the mapping gel was stained with Coomassie Blue R-250, destained, dried and exposed to X-ray film with an intensifying screen. A, Coomassie stained gel. B, Autoradiograph, 15 h exposure. The electrophoretic mobility of α4β2 receptor subunits are indicated on the left.
[125I]TID Photolabeling of the α4β2 nAChR
The hydrophobic photoreactive probe [125I]TID was used to study the structure of the lipid-protein interface of the α4β2 nAChR and to identify individual amino acid residues in the α4 and β2 subunits that are in contact with membrane lipid. [125I]TID is a small hydrophobic photoreactive compound that partitions efficiently (>95%) into the lipid bilayer and upon activation with UV light (365 nm) covalently tags amino acid residues that are in contact with the lipid bilayer (12). Purified α4β2 nAChRs were equilibrated for 1 h with [125I]TID in the absence or presence of 400 µM carb, irradiated at 365 nm for 7 min, and then the polypeptides were resolved by SDS-PAGE. After electrophoresis, the gel was stained, destained, dried and exposed to X-ray film. As shown in Figure 1B, [125I]TID photoincorporated into the α4 and β2 subunits, with the amount of subunit incorporation the same in the absence or presence of agonist. The ratio of [125I]TID photoincorporation into the α4 vs. β2 subunit (α4/β2 labeling ratio: 0.6; Supplementary Figure 2B) indicates that [125I]TID was incorporated into both subunits with equal efficiency, consistent with labeling at the lipid-protein interface.
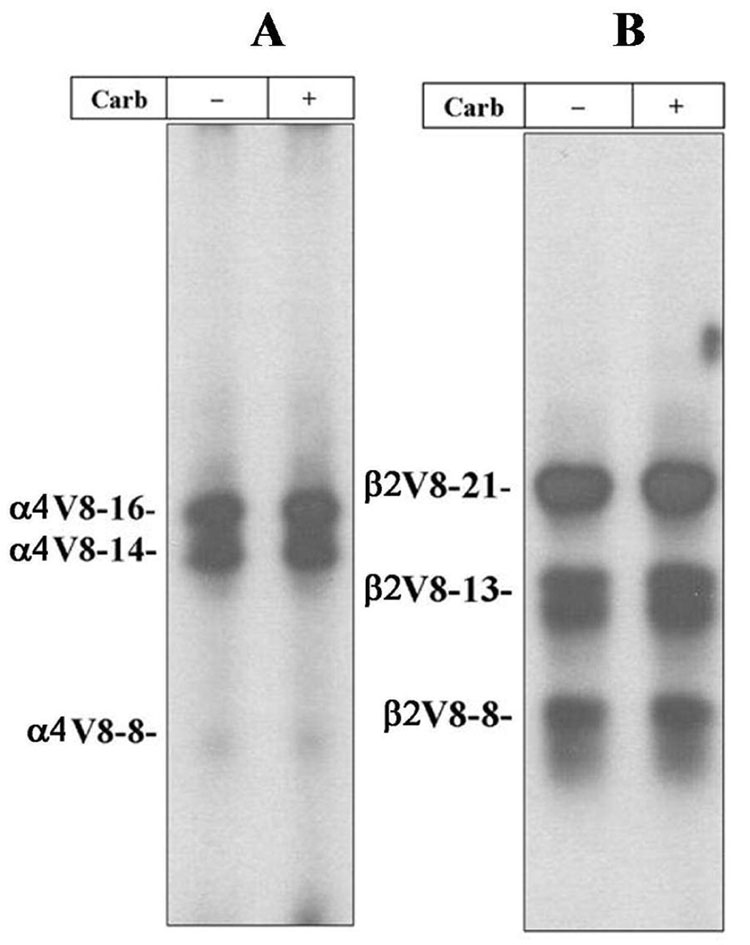
To generate subunit fragments containing incorporated [125I]TID, the α4 and β2 subunit bands were excised from the stained 8% polyacrylamide gel, transferred to the wells of 15% acrylamide mapping gels and subjected to “in-gel-digestion” with S. aureus V8 protease as described in the Experimental Procedures. Based upon autoradiography of the dried mapping gel (Figure 2A), [125I]TID photoincorporation within the α4 subunit was detected primarily in two proteolytic fragments with apparent molecular masses of 16 KDa (α4V8-16) and 14 KDa (α4V8-14), with minor labeling in an 8 KDa fragment (α4V8-8). Within the β2 subunit (Figure 2B), [125I]TID photoincorporation was detected in three fragments with apparent molecular masses of 21 KDa (β2V8-21), 13 KDa (β2V8-13) and 8 KDa (β2V8-8). Addition of agonist had no significant effect on the extent of [125I]TID labeling for any of these subunit fragments.
Figure 2. Proteolytic mapping of the [125I]TID-labeled α4 and β2 subunits.
The α4 and β2 subunit bands isolated from an 8% SDS-PAGE gel containing [125I]TID-labeled α4β2 nAChRs (Figure 1B) were transferred to the well of a 15% acrylamide mapping gel and subjected to in-the-gel digestion with V8 protease. Following electrophoresis, the mapping gel was stained, destained, dried and exposed to X-ray film with an intensifying screen. A, Autoradiograph of a 15% acrylamide mapping gel showing the photoincorporation of [125I]TID into three α4 subunit proteolytic fragments with apparent molecular masses of 16 KDa (α4V8-16), 14 KDa (α4V8-14) and 8 KDa (α4V8-8). B, Autoradiograph of a 15 % acrylamide mapping gel showing the photoincorporation of [125I]TID into three β2 subunit proteolytic fragments with apparent molecular masses of 21 KDa (β2V8-21), 13 KDa (β2V8-13) and 8 KDa (β2V8-8).
Protein microsequencing established that the N-termini of α4V8-8 and α4V8-14 were at α4-Asp562 and α4-Ala474, respectively. Based on the apparent molecular mass of each fragment, they are predicted to contain the M4 segment (α4-Ile573-Pro592) and to extend to the C-terminus of the subunit, α4-Ile599. The amino acid sequence for α4V8-16 began at α4-Ile201. Based on its apparent molecular mass (16 KDa) and the likely cleavage site of V8 protease, α4V8-16 is predicted to include the M1 (α4-Pro214-Pro240), M2 (α4-Ile247-Ile264) and M3 (α4-Leu282-Arg305) segments. The amino acid sequences for β2V8-8 and β2V8-13 began at β2-Asp425 and β2-Gly347, respectively, and are predicted to contain the β2M4 (β2-Leu436-β2-Leu445) segment. The amino acid sequence for β2V8-21 began at β2-Val166 and is predicted to contain the M1 (β2-Pro209-Pro234), M2 (β2-Met241-Ile258) and M3 (β2-Ile276-Arg299) segments.
Amino acids in the M4 segments of α4 and β2 nAChR subunits photolabeled by [125I]TID
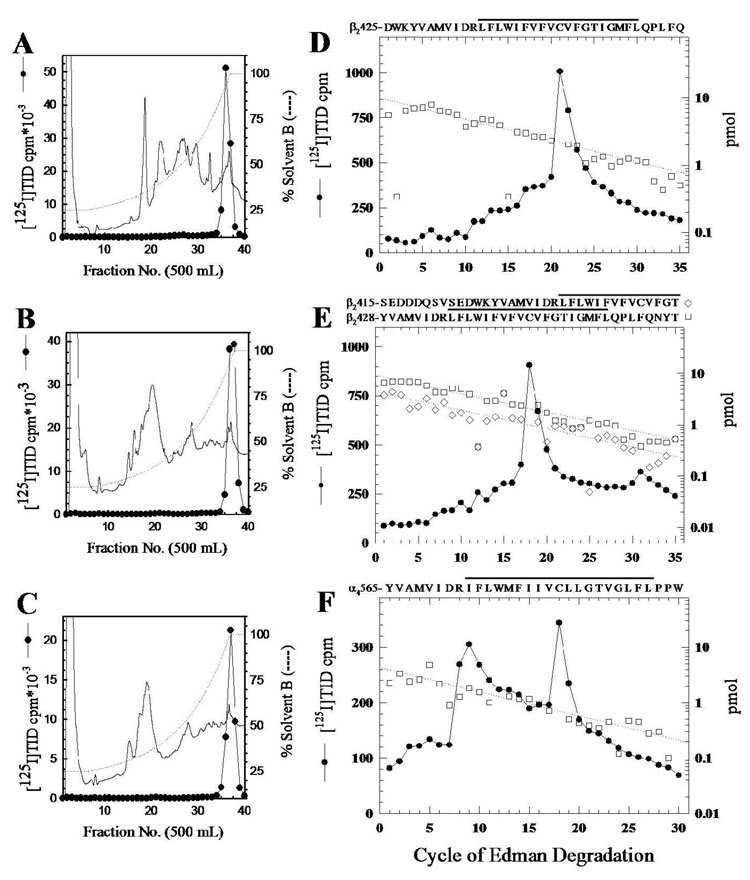
To identify the individual amino acid residue(s) labeled by [125I]TID within the M4 segments, [125I]TID-labeled β2V8-8, with an N-terminus just 11 amino acids before the beginning of M4, was isolated from a preparative labeling (1 mg affinity-purified hα4β2) and purified by reversed-phase HPLC (Figure 3A). The [125I]TID-labeled fragments, β2V8-13 and α4V8-14, with N-termini 64 and 99 amino acids, respectively, before the beginning of M4, were digested with trypsin and the digests were fractionated by reversed-phase HPLC (Figure 3B and 3C). Peak 125I HPLC fractions were pooled, loaded onto PVDF supports and sequenced.
Figure 3. Reversed-phase HPLC purification and sequence analysis of [125I]TID-labeled fragments containing the M4 segments.
Reversed-phase HPLC purification of β2V8-8 (A) and the tryptic digest of β2V8-13 (B) and α4V8-14 (C). The elution of peptides was monitored by absorbance at 210 nm (solid line) and elution of 125I (closed circles) by γ-counting. D–F, 125I (●) and PTH-amino acids released (□, ◇) during sequencing of HPLC-purified β2V8-8 (D) and fractions containing the peak of 125I from the HPLC purification of the tryptic digest of β2V8-13 (E) and α4V8-14 (F). For β2V8-8, a single amino acid sequence begining at β2-Asp425 (□, Io, 9.9 pmol; R, 93%) was detected (44,250 cpm loaded in the filter and 22,000 cpm left after 35 cycles), with 125I release in cycle 21 corresponding to labeling of β2-Cys445 (590 cpm). E, For the 125I peak fraction from the tryptic digest of β2V8-13, there were amino acid sequences beginning at β2-Tyr428 (□, Io, 9 pmol; R, 92%) and at β2-Ser415 (◇, Io, 4.5 pmol; R, 92%), (46,200 cpm loaded in the filter and 26,000 cpm left after 35 cycles). Peaks of 125I release in cycles 18 (507 cpm) and 31 (59 cpm) correspond to labeling of β2-Cys445 in the fragments beginning at β2-Tyr428 and β2-Ser415, respectively. F, For the 125I peak fraction from the tryptic digest of α4V8-14, the primary amino acid sequence begin at α4-Tyr565 (□, Io, 4.3 pmol; R, 90%; 23,000 cpm loaded in the filter and 6188 cpm left after 30 cycles). The 125I release in cycle 18 (72 cpm) corresponds to labeling of α4-Cys582. The amino acid sequences of the detected fragment(s) are shown above each panel, with a line indicating the M4 region.
When HPLC-purified β2V8-8 was sequenced (Figure 3D), a single amino acid sequence was detected beginning at β2-Asp425 (10 pmol). There was a peak of 125I release in cycle 21 (590 cpm) corresponding to the labeling of β2-Cys445 (70 cpm/pmol), the only cysteine in the α4M4 segment and homologous to one of the amino acids in the α1M4 segment of the Torpedo nAChR (12) labeled by [125I]TID. Sequence analysis of the 125I peak from the HPLC fractionation of the β2V8-13 tryptic digest (Figure 3E) revealed the presence of two amino acid sequences, one beginning at β2-Tyr428 (9 pmol), corresponding to the trypsin cleavage site β2-Lys427, and the other beginning at β2-Ser415 (5 pmol), corresponding to the trypsin cleavage site at β2-Arg414. The major peak of 125I release was in cycle 18 (510 cpm) with a minor peak in cycle 31 (60 cpm). The 125I release in cycle 18 is consistent with labeling of β2-Cys445 (70 cpm/pmol) in the fragment beginning at β2-Tyr428, and the release in cycle 31 is also consistent with labeling of β2-Cys445 but in the fragment beginning at β2-Ser415. Sequence analysis of the 125I peak from the HPLC purification of the tryptic digest of α4V8-14 (Figure 3F) revealed an amino acid sequence beginning at α4-Tyr565 (4 pmol). The largest 125I release was in cycle 18 (150 cpm), corresponding to labeling of α4-Cys582 (70 cpm/pmol). While there was a clear increase in 125I release in cycle 8, the progressive decline of released radioactivity in cycles 11–17 was unusual. Further studies are required to determine whether the release in cycle 8 resulted from labeling of α4-Arg572 or from enhanced wash-off of the peptide from the PVDF support after cleaving at Arg572, which is the last charged residue in the peptide.
Amino acids in the M1 segments of α4 and β2 nAChR subunits photolabeled by [125I]TID
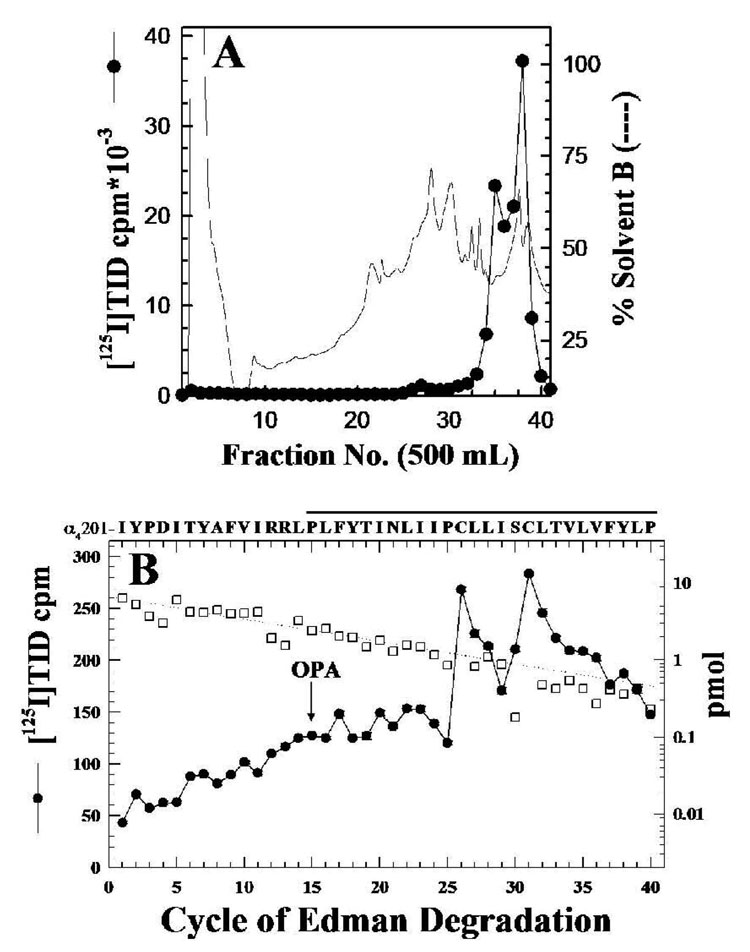
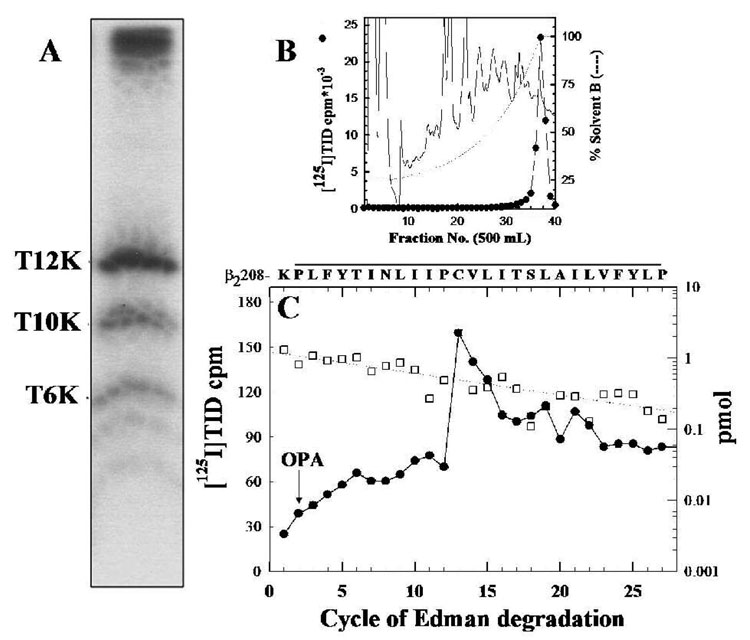
To identify the amino acids photolabeled within the α4M1 segment, the labeled fragment α4V8-16 was recovered from the mapping gel, HPLC-purified (Figure 4A) and sequenced. The amino acid sequence detected (Figure 4B) began at α4-Ile201 (6 pmol), shortly before the N-terminus of the M1 segment (α4-Pro214-Pro240). Peaks of 125I release were seen in cycle 26 (150 cpm) and cycle 31 (74 cpm), corresponding to the labeling of α4-Cys226 and α4-Cys231 at 46 and 35 cpm/pmol, respectively. The presence of the 125I release in cycles 26 and 31, along with the persistence of the amino acid sequence of α4M1 after treatment of the sequencing filter at cycle 15 with o-phthalaldehyde (OPA, which reacts with all non-proline amino-termini and blocks their further Edman Degradation (21, 22), established that [125I]TID labeled α4-Cys226 and α4-Cys231. The [125I]TID-labeled amino acid within the β2V8-21 fragment, which begins at β2-Val166 and includes M1, M2, and M3, was identified when material from the β2V8-21 band was digested with trypsin and fractionated by Tricine SDS-PAGE. An autoradiograph (Figure 5A) of the dried Tricine gel revealed a primary radioactive band migrating with an apparent molecular mass of 12 KDa (β2T12K), as well as secondary bands of 10 KDa (β2T10K) and 6 KDa (β2T6K). When the labeled bands were recovered from the gel and purified by reversed-phase HPLC, 125I was recovered in broad hydrophobic peaks, as seen in Figure 5B for β2T12K. N-terminal sequence analysis for β2T12K and β2T10K established that each band contained a fragment beginning at β2-Lys208, the N-terminal of β2M1. Since the labeled fragment began at β2-Lys208, the peak of 125I from the HPLC purification of β2T12K was sequenced with OPA treatment prior to cycle 2, corresponding to β2-Pro209. The amino acid sequence of the peptide beginning at β2-Lys208 (1 pmol; Figure 5C) continued after treatment with OPA, and there was 125I release in cycle 13 (90 cpm) corresponding to labeling of β2-Cys220 (70 cpm/pmol).
Figure 4. [125I]TID labels α4-Cys226 and α4-Cys231 in α4M1.
A, α4V8-16, produced by in-gel digestion of [125I]TID-labeled α4 subunit with V8 protease (Figure 3A), was eluted and purified by reversed-phase HPLC. B, 125I (●) and PTH-amino acids released (□) when pooled fractions 37–40 were sequenced with sequencing interrupted for OPA treatment before cycle 15. The primary amino acid sequence detected began at α4-Ile201 (Io, 6 pmol; R, 94%; 45,560 cpm loaded in the filter and 26,850 cpm left after 40 cycles). The peak of 125I release in cycle 26 (148 cpm) and 31 (74 cpm) correspond to labeling of α4-Cys226 and α4-Cys231, respectively. The amino acid sequence of the detected fragment is shown, with a line indicating the M1 region.
Figure 5. [125I]TID labels β2-Cys220 in β2M1.
The β2V8-21 fragment, produced by in-gel digestion of [125I]TID-labeled β2 subunit with V8 protease (Figure 2B), was digested with trypsin for 5 days and the digest was fractionated on Tricine SDS-PAGE. A, Autoradiograph of a Tricine gel showing [125I]TID photoincorporation into three peptide fragments with apparent molecular masses of 12 KDa (β2T12K), 10 KDa (β2T10K) and 6 KDa (β2T6K). B, Reversed-phase HPLC purification of [125I]TID-labeled β2T12K. C, 125I (●) and PTH-amino acids released (□) during sequencing of β2T12K HPLC fractions 36–38, with sequencing interrupted for OPA treatment before cycle 2. The primary amino acid sequence began at β2-Lys208 ( Io, 1.2 pmol; R, 93 %; 18,630 cpm loaded in the filter and 11,909 cpm left after 30 cycles) and the peak of 125I release was in cycle 13 (90 cpm) correspond to labeling of β2-Cys220. The amino acid sequence detected is shown above the panel, with a line indicating the M1 region.
DISCUSSION
The crucial role of neuronal nAChRs in several neuronal diseases makes them an important drug target. Development of agonist, antagonist and allosteric modulators of α4β2 nAChRs provide a potential hope for a better understanding of and improved treatment for pathophysiological conditions that include Alzheimer’s disease and nicotine addiction (37, 38). Among other factors, a more refined understanding of the molecular structure of the α4β2 nAChR is a prerequisite for the development of such ligands. In this study, we sought to obtain large quantities of highly purified human α4β2 nAChRs and to begin direct structural studies of the purified receptor. Starting with membrane preparations from a HEK-hα4β2 stable cell line, we were able to purify α4β2 nAChRs using an acetylcholine-derivatized affinity column. The yields (2–3 mg) and levels of purity (>50%), which were reproduced in three different purifications, are sufficient to apply a variety of structural techniques, including photoaffinity labeling, to characterize the structure of the α4β2 nAChR. Our purification results have several advantages compared with previous purifications of neuronal nAChRs from rat brain using monoclonal antibody affinity chromatography (39) or an acetylcholine affinity column alone (33) or in combination with other columns (34): i) We used a human cell line (HEK-293) that expresses a single population of functional nAChRs (hα4β2 nAChRs). This avoids co-purification of other neuronal nAChRs, maintains a fixed subunit stoichiometry, and preserves receptor posttranslational modifications; ii) Although earlier studies reported high levels of receptor enrichment (e.g. 13,000-fold enrichment (34); 7,000–13,000-fold enrichment (33)), the specific activity of the purified receptor was at best 0.4 nmol [3H]ACh binding site/mg protein, and the reported protein yields were either at the microgram level (20 µg, 34) or below the level of detection (33). Furthermore, the yields from rat brain were low and variable (4–20%), suggesting that different populations of neuronal nAChR subtypes were purified depending on the type of column used (33); iii) The use of an acetylcholine-derivatized affinity chromatography in a single step purification minimizes the time of the purification procedure and, most importantly, minimizes the time in which the receptor is complexed with detergent; iv) In the present study, the receptor was solubilized using 1% cholate, a relatively mild detergent, which has been shown to retain the resting state conformation of the Torpedo nAChR (31), and can be removed efficiently by dialysis for reconstitution of the purified receptor into lipid (bilayer containing) vesicles. This one-step purification and lipid-reconstitution protocol increases the likelihood that the purified receptor will retain its native three dimensional structure in a membrane environment that is supportive of functionality (i.e agonist induced - conformational transitions).
To begin structural characterization of purified α4β2 nAChRs, we employed [125I]TID as a probe of the receptor lipid-protein interface. [125I]TID is a small hydrophobic photoreactive compound that has been used extensively to study the structure of Torpedo nAChRs (11, 12, 40–42). When affinity-purified and lipid-reconstituted α4β2 nAChRs were labeled with [125I]TID, both α4 and β2 subunits incorporated equal amounts of the probe. The α4/β2 labeling ratio (0.6; Supplementary Figure 2B) was reflective of the relative amounts of α4 versus β2 subunit (2α4:3β2; Supplementary Figure 2B) that other approaches have shown likely represents the predominate subunit stoichiometry (36). Identical results were obtained for α4β2 nAChRs purified from HEK-α4β2 cells that had been exposed to 100 µM nicotine 24 h prior to harvesting, indicating that nicotine exposure did not significantly alter the subunit stoichiometry for this stably transfected HEK cell line (but see ref. 27).
Unlike [125I]TID labeling of the Torpedo nAChR, addition of agonist did not alter the extent nor the pattern of [125I]TID labeling within the α4 or β2 subunits (Figure 1B). There are two primary components of [125I]TID incorporation into the Torpedo AChR: i) For Torpedo nAChRs in the absence of agonist (resting state), [125I]TID labels amino acids in the channel lumen (M2 helix), and this component of labeling is inhibited by >90% for nAChRs in the desensitized state (in the presence of agonist) or by the addition of an excess of nonradioactive TID (specific, agonist-sensitive component; 40, 41); ii) [125I]TID labels amino acids at the lipid-protein interface (M1, M3 and M4 segments; 11, 12), and this labeling is neither affected by the addition of agonist nor by an excess of nonradioactive TID (nonspecific, lipid-protein interface component). Therefore, the absence of agonist-sensitivity for [125I]TID labeling of the α4β2 nAChR indicates that either the affinity-purified α4β2 nAChRs are stabilized in a desensitized state (i.e. unable to undergo agonist-induced conformational changes) or that [125I]TID labeling of the α4β2 has no agonist-sensitive component. Additional studies are needed to distinguish between these two possibilities.
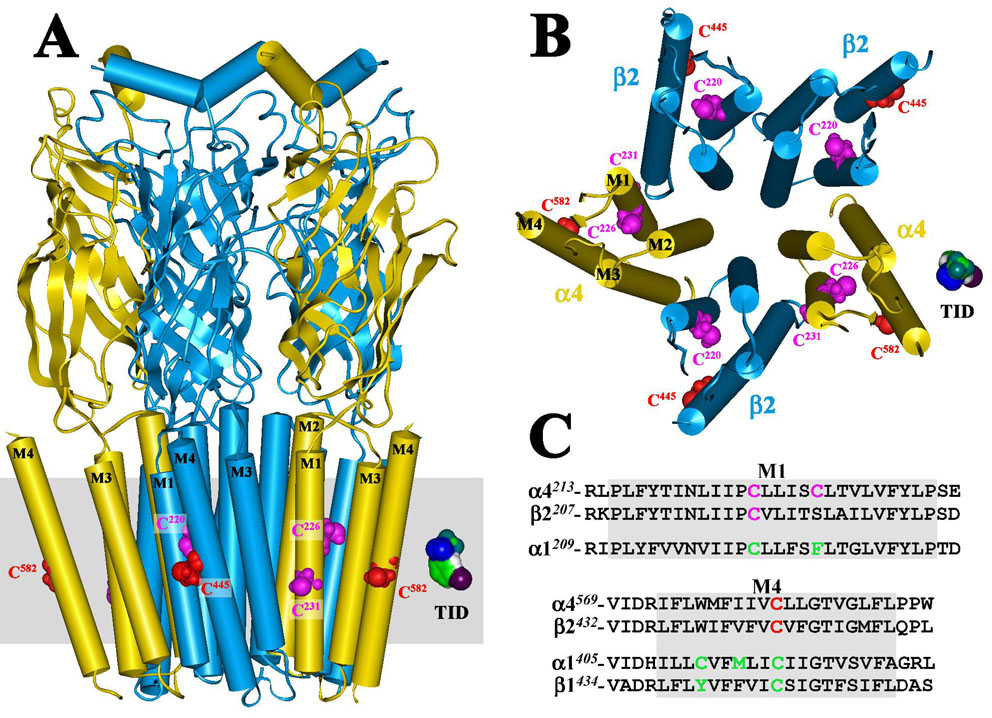
[125I]TID labeling was mapped to amino acids in the M4 and M1 segments of both the α4 and β2 subunits. For both α4 and β2 subunits ~ 60% of the total labeling was localized within fragments that contain the M4 segment, which suggests that the M4 helix has the greatest exposure to lipid, consistent with the published structure of the Torpedo nAChR (7). Amino acid sequence analysis of the [125I]TID-labeled subunit fragment β2V8-8 and the 125I HPLC peaks from the tryptic digests of β2V8-13 and α4V8-14 revealed labeling within β2M4 and α4M4 of the homologous amino acids β2-Cys445 and α4-Cys582 (Figures 4D cycle 21; Figures 4E cycles 18 and 31; Figures 4F cycle 18). Amino acid sequence alignment of the M4 segments of human α4, human β2 and Torpedo α1 nAChR subunits (Figure 6C) shows that the labeled residues β2-Cys445 and α4-Cys582 correspond to α1-Cys418 and β1-Cys447 which are labeled by [125I]TID in the Torpedo nAChR α1/β1M4 segment (12). N-terminal sequencing of the [125I]TID-labeled fragments α4V8-16 (Figure 4B) and β2T12K (Figure 5C) established [125I]TID labeling of Cys226 and Cys231 within the α4M1 segment and Cys220 within the β2M1 segment. α4-Cys226/β2-Cys220 and α4-Cys231 are homologous to α1-Cys222 and α1-Phe227 (Figure 6C), the two amino acids labeled by [125I]TID in the Torpedo α1M1 segment (12). A homology model of the hα4β2 nAChR was constructed using the three dimensional structure of the Torpedo nAChR as a template (Figure 6A and 6B) and shows that the [125I]TID-labeled residues (α4-Cys582, β2-Cys445, α4-Cys226, α4-Cys231 and β2-Cys220) are all located at the lipid-protein interface of the α4β2 nAChR and are situated near the middle of the lipid bilayer. These results provide the first experimental evidence that support the existence of a high degree of structural homology between the lipid-protein interfaces of the neuronal α4β2 nAChR and the Torpedo nAChR.
Figure 6. Homology model of the human α4β2 nAChR displaying [125I]TID labeled residues situated at the lipid-protein interface.
A homology model of a human α4β2 nAChR was constructed from the published structure of the Torpedo marmorata nAChR (PDB # 2BG9) as described in the Experimental Procedures. A) A side view of the extracellular and transmembrane domains of the α4 (yellow) β2 (blue) nAChR and B) a view of the transmembrane domain looking through the channel from the synaptic side. The polypeptide chains are traced with regions of α-helix (cylinders) or β-sheet (ribbons) denoted. Residues labeled by [125I]TID are shown in CPK representation within the M1 (magenta) and M4 (red) helices. An approximation of the membrane is included in A (gray) and a Connelly surface model of TID is included for scale. C) An alignment of the M1 and M4 transmembrane helices from human α4 and β2 and Torpedo α1 and β1 (M4 only) nAChRs, with [125I]TID labeled residues highlighted in green (α1M1/M4 and β1M1; ref 12), magenta (M1 of α4 and β2) or red (M4 of α4 and β2).
The predominant incorporation of [125I]TID into cysteine residues undoubtedly reflects the high intrinsic reactivity of the cysteine side chain compared to the side chains of other amino acids (43). [125I]TID labeled each of the cysteines in the M4 segments of the Torpedo nAChR α1, β1, and γ subunits, while in δM4, which lacks cysteines, δSer457 was labeled (12). Within α1M4, Cys412 and αCys418 were labeled at ~3-fold higher efficiency than Met415, while in β1M4, β1-Tyr441, homologous to α1-Cys412, was labeled at 2-fold higher efficiency than β1-Cys447. No aliphatic side chains were labeled in the Torpedo M4 segments, but there were unlabeled cysteines within γM3 and δM3 (non lipid-exposed helical face) as well as labeled aliphatic side chains. The pattern of labeling within the Torpedo αM4 and βM4 segments also predicts labeling of α4-Trp576 and β2-Trp439 (Figure 6C), especially since tryptophan, at least as a free amino acid, also has high intrinisic reactivity for [125I]TID (43). However, despite the extensive characterization of the Torpedo nAChR amino acids photolabeled by [125I]TID, no labeled tryptophan has been identified, which suggests that tryptophans in proteins may be less reactive than as a free amino acid or that the adduct is unstable under the conditions used to identify labeled amino acids by Edman degradation. However, since the selective labeling of cysteines in α4β2 nAChR M1 and M4 segments was somewhat unexpected, we also examined the pattern of [125I]TID labeling of the Torpedo nAChR after purification and reconstitution into lipid by the same protocol used for the α4β2 nAChR. [125I]TID labeled both Cys222 and Phe227 within α1-M1 (data not shown), consistent with the published data for native Torpedo nAChR-rich membranes (12). Collectively these results support the conclusion that the position of the amino acid, that is the degree of exposure to lipid, is the primary determinant of labeling by [125I]TID, with side-chain reactivity then providing a secondary factor. The combination of these two factors then determines the overall labeling pattern.
To identify potential sites of [125I]TID labeling in the M3 and/or M2 segments of α4 and β2 subunits, [125I]TID-labeled α4V8-16 (Figure 2A) and β2V8-21 (Figure 2B) fragments were exhaustively digested with trypsin (> 1:1 w/w ratio of protease/substrate). When the tryptic digest of α4V8-16 was fractionated by Tricine SDS-PAGE, two labeled bands with apparent molecular masses of 10 KDa (α4T10K) and 6 KDa (α4T6K) were evident (data not shown). While the N-terminal sequencing of HPLC-purified α4T6K gave no clear sequence, N-terminal sequencing of the HPLC-purified α4T10K revealed a peptide beginning at α4-Leu214, one residue before the N-terminus of α4M1 segment. Digestion of β2V8-21 with trypsin (Figure 5) did not result in the isolation of either the M2 or M3 segment. We conclude from these results that site(s) for tryptic cleavage that are present at the N-terminus of M2 and M3 are very resistant to proteolysis. Additional work, including the use of alternative proteases, will be necessary to determine whether or not [125I]TID photolabels amino acids in the M3 and/or M2 segments.
Supplementary Material
A typical elution profile of the hα4β2 neuronal nAChR from an acetylcholine-derivatized affinity column; densiometric analysis of Coomassie Blue R-250 stained gel (Figure 1A); and quantification of [125I]TID incorporation into α4 and β2 nAChR subunits (Figure 1B). This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
We thank Dr. Joseph H. Steinbach (Washington University School of Medicine) for kindly providing us with the stable α4β2 nAChR HEK-293 cell line and Dr. Jose-Luis Redondo (Department of Pharmacology and Neuroscience, TTUHSC) for his cell culture assistance.
Footnotes
This research was supported in part by an Intramural Grant from Texas Tech University Health Sciences Center School of Medicine (M.P.B), by American Heart Association Texas Affiliate Grant-In-Aid 0755029Y (M.P.B.), and by grant GM-58448 from the National Institute of General Medical Sciences (J.B.C.)
Abbreviations: nAChR, nicotinic acetylcholine receptor; HEK-hα4β2, human embryonic kidney cells stably expressing human α4β2 nAChR; HPLC, high-performance liquid chromatography; OPA, o-Phthalaldehyde; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TFA, trifluoroacetic acid; PTH, phenylthiohydantoin; [125I]TID, 3-trifluoromethyl-3-(m-[125I] iodophenyl) diazirine; Tricine, N-tris(hydroxymethyl)methylglycine; VDB, vesicle dialysis buffer; V8 protease, S. aureus glutamyl endopeptidase.
REFERENCES
- 1.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature Rev. Neurosci. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 2.Gotti C, Clementi F. Neuronal nicotinic receptors: From structure to pathology. Progress in Neurobiology. 2004;74:363–396. doi: 10.1016/j.pneurobio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Leonard S, Bertrand D. Neuronal nicotinic receptors: From structure to function. Nicotine & tobacco Research. 2001;3:203–223. doi: 10.1080/14622200110050213. [DOI] [PubMed] [Google Scholar]
- 4.Hogg RC, Raggenbass M, Bertrand D. Nicotinic acetylcholine receptors: From structure to brain function. Rev Physiol Biochem Pharmacol. 2003;147:1–46. doi: 10.1007/s10254-003-0005-1. [DOI] [PubMed] [Google Scholar]
- 5.Jensen A, Jensen, Frolund B, Liljefors T, Krogsgaard-Larsen P. Neuronal Nicotinic Acetylcholine Receptors: Structural Revelations, Target Identifications, and Therapeutic Inspirations. J. Med. Chem. 2005;48:4705–4745. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- 6.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 7.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4Å resolution. J. Mol. Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends in Pharmacological Sciences. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Sine SM, Engel AG. Recent advances in Cys-loop receptor structure and function. Nature. 2006;440:448–455. doi: 10.1038/nature04708. [DOI] [PubMed] [Google Scholar]
- 10.Xiao Y, Kellar KJ. The comparative pharmacology and up-regulation of rat neuronal nicotinic receptor subtype binding sites stably expressed in transfected mammalian cells. J. Pharm & Exp. Therap. 2004;310:98–107. doi: 10.1124/jpet.104.066787. [DOI] [PubMed] [Google Scholar]
- 11.Blanton MP, Cohen JB. Mapping the lipid-exposed regions in the Torpedo californica nicotinic acetylcholine receptor. Biochemistry. 1992;31:3738–3750. doi: 10.1021/bi00130a003. [DOI] [PubMed] [Google Scholar]
- 12.Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Steinbach JH. Cytisine binds with similar affinity to nicotinic α4β2 receptors. Brain Research. 2002;959:98–102. doi: 10.1016/s0006-8993(02)03733-2. [DOI] [PubMed] [Google Scholar]
- 14.Lowry OH, Rosebrough NJ, Farr L, Randall RJ. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 15.Bhushan A, McNamee MG. Differential scanning calorimetry and fourier transform infrared analysis of lipid-protein interactions involving the nicotinic acetylcholine receptor. Biochim. Biophys. Acta. 1990;1027:93–101. doi: 10.1016/0005-2736(90)90053-q. [DOI] [PubMed] [Google Scholar]
- 16.Hamouda AK, Sanghvi M, Sauls D, Machu TK, Blanton MP. Assessing the lipid requirements of the Torpedo Californica nicotinic acetylcholine receptor. Biochemistry. 2006;45(13):4327–4337. doi: 10.1021/bi052281z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.daCosta CJB, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE. Lipid-protein interactions at the nicotinic acetylcholine receptor. J. Biol. Chem. 2002;227:201–208. doi: 10.1074/jbc.M108341200. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 20.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.Middleton RE, Cohen JB. Mapping of the acetylcholine binding site of the nicotinic acetylcholine receptor: [3H]nicotine as an agonist photoaffinity label. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 22.Brauer AW, Oman CL, Margolies MN. Use of o-phthalaldehyde to reduce background during automated Edman degradation. Anal. Biochem. 1984;137:134–142. doi: 10.1016/0003-2697(84)90359-2. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Y, Meyer EL, Thompson J, Surin A, Wroblewski J, Kellar KJ. Rats α3β4 subtype of neuronal nicotinic acetylcholine receptor stably expressed in a transfected cell line: pharmacology of ligand binding and function. Mol Pharmacol. 1998;54:322–333. doi: 10.1124/mol.54.2.322. [DOI] [PubMed] [Google Scholar]
- 24.Xiao Y, Baydyuk M, Wang H, Davis HE, Kellar KJ. Pharmacology of the agonist binding sites of rat neuronal nicotinic receptor subtypes expressed in HEK 293 cells. Bioorganic and medicinal chemistry lett. 2004;14:1845–1848. doi: 10.1016/j.bmcl.2003.09.105. [DOI] [PubMed] [Google Scholar]
- 25.Meyer LE, Xiao Y, Kellar KJ. Agonist regulation of rat α3β4 nicotinic acetylcholine receptors stably expressed in human embryonic kidney 293 cells. Mol Pharmacol. 2001;60:568–576. [PubMed] [Google Scholar]
- 26.Wang F, Nelson ME, Kuryatov A, Olale F, Cooper J, Keyser K, Lindstrom J. Chronic nicotine treatment up-regulates human alpha3 beta2 but not alpha3 beta4 acetylcholine receptors stably transfected in human embryonic kidneycells. J. Biol. Chem. 1998;273:28721–28732. doi: 10.1074/jbc.273.44.28721. [DOI] [PubMed] [Google Scholar]
- 27.Nelson ME, Kuryakov A, Chol CH, Zhou Y, Lindstrom J. Alternate stoichiometries of α4β2 nicotinic acetylcholine receptors. Mol Pharmacol. 2003;63:332–341. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- 28.Paradiso K, Sabey K, Evers A, Zorumski CF, Covey DF, Steinbach JH. Steroid inhibition of rat neuronal nicotinic α4β2 receptors expressed in HEK-293 cells. Mol Pharmacol. 2000;58:341–351. doi: 10.1124/mol.58.2.341. [DOI] [PubMed] [Google Scholar]
- 29.Paradioso K, Zhang J, Steinbach JH. The C Terminus of the Human Nicotinic α4β2 Receptor Forms a Binding Site Required for Potentiation by an Estrogenic Steroid. J. Neuroscience. 2001;21:6561–6568. doi: 10.1523/JNEUROSCI.21-17-06561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabey K, Paradiso K, Zhang J, Steinbach jH. Ligand binding and activation of rat nicotinic α4β2 receptors stably expressed in HEK293 cells. Mol. Pharm. 1999;55:58–66. [PubMed] [Google Scholar]
- 31.McCarthy MP, Moore MA. Effect of lipids and detergents on the conformation of the nicotinic acetylcholine receptor from Torpedo californica. J. Biol. Chem. 1992;267:7655–7663. [PubMed] [Google Scholar]
- 32.Huganir RL, Racker E. Properties of proteoliposomes reconstituted with acetylcholine receptor from Torpedo californica. J Biol Chem. 1982;257:9372–9378. [PubMed] [Google Scholar]
- 33.Dwork AJ, Desmond JT. Purification of a nicotinic acetylcholine receptor from rat brain by affinity chromatography directed at the acetylcholine binding site. Brain Res. 1991;21:119–123. doi: 10.1016/0006-8993(91)90668-l. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama H, Shirase M, Nakashima T, Kurogochi Y, Lindstrom JM. Affinity purification of nicotinic acetylcholine receptor from rat brain. Brain Res Mol Brain Res. 1990;7:221–226. doi: 10.1016/0169-328x(90)90031-8. [DOI] [PubMed] [Google Scholar]
- 35.Arroyo-Jimnez MM, Bourgeois JP, Marubio LM, LeSourd AM, Ottersen OP, Rinvik E, Fairen A, Changeux JP. Ultrastructural localization of the alpha4-subunit of the neuronal acetylcholine nicotinic receptor in the rat substantia nigra. J Neurosci. 1999;19:6475–6487. doi: 10.1523/JNEUROSCI.19-15-06475.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper E, Couturier S, Ballivet M. Pentameric structure and subunit stoichiometry of a neuronal nicotinic acetylcholine receptor. Nature. 1991;350:235–238. doi: 10.1038/350235a0. [DOI] [PubMed] [Google Scholar]
- 37.Kalamida D, Poulas K, Avramopoulou V, Fostieri E, Lagoumintzis G, Lazaridis K, Sideri A, Zouridakis M, Tzartos SJ. Muscle and neuronal nicotinic acetylcholine receptors. Structure, function and pathogenicity. FEBS J. 2007;274:3799–3845. doi: 10.1111/j.1742-4658.2007.05935.x. [DOI] [PubMed] [Google Scholar]
- 38.Romanelli MN, Gratteri P, Guandalini L, Martini E, Bonaccini C, Gualtieri F. Central Nicotinic Receptors: Structure, Function, Ligands, and Therapeutic Potential. ChemMedChem. 2007;2:746–767. doi: 10.1002/cmdc.200600207. [DOI] [PubMed] [Google Scholar]
- 39.Whiting PJ, Lindstrom JM. Purification and characterization of a nicotinic acetylcholine receptor from rat brain. PNAS. 1986;84:595–599. doi: 10.1073/pnas.84.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White BH, Howard S, Cohen S, Cohen JB. The hydrophobic photoreagent 3-(trifluoromethyl)-3-m-([125I]iodophenyl) diazirine is a novel noncompetitive antagonist of the nicotinic acetylcholine receptor. J. Biol. Chem. 1991;266:21595–21607. [PubMed] [Google Scholar]
- 41.White BH, Cohen JB. Agonist-induced changes in the structure of the acetylcholine receptor M2 regions revealed by photoincorporation of an uncharged nicotinic noncompetitive antagonist. J. Biol. Chem. 1992;267:15770–15783. [PubMed] [Google Scholar]
- 42.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor's δ-subunit. A time-resolved photolabeling study. J. Biol. Chem. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 43.Sigrist H, Muhlemann M, Dolder M. Philicity of amino acid side-chains for photoreactive carbenes. J. Photochem. Photobiol. 1990;B 7:277–287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A typical elution profile of the hα4β2 neuronal nAChR from an acetylcholine-derivatized affinity column; densiometric analysis of Coomassie Blue R-250 stained gel (Figure 1A); and quantification of [125I]TID incorporation into α4 and β2 nAChR subunits (Figure 1B). This material is available free of charge via the Internet at http://pubs.acs.org.