Abstract
The nuclear progesterone receptor (nPR) mediates many of the physiological effects of progesterone by regulating the expression of genes, however, progesterone also exerts non-transcriptional (non-genomic) effects that have been proposed to rely on a receptor that is distinct from nPR. Several members of the Progestin and AdipoQ Receptor (PAQR) family were recently identified as potential mediators of these non-genomic effects. Membranes from cells expressing these proteins, called mPRα, mPRβ and mPRγ, were shown to specifically bind progesterone and have G-protein coupled receptor (GPCR) characteristics, although other studies dispute these findings. To clarify the role of these mPRs in non-genomic progesterone signaling, we established an assay for PAQR functional evaluation using heterologous expression in Saccharomyces cerevisiae. Using this assay, we demonstrate unequivocally that mPRα, mPRβ and mPRγ can sense and respond to progesterone with EC50 values that are physiologically relevant. Agonist profiles also show that mPRα, mPRβ and mPRγ are activated by ligands, such as 17α-hydroxyprogesterone, that are known to activate non-genomic pathways but not nPR. These results strongly suggest that these receptors may indeed function as the long-sought-after membrane progesterone receptors. Additionally, we show that two uncharacterized PAQRs, PAQR6 and PAQR9, are also capable of responding to progesterone. These mPR-like PAQRs have been renamed mPRδ (PAQR6) and mPRε (PAQR9). Additional characterization of mPRγ and mPRα indicate that their progesterone-dependent signaling in yeast does not require heterotrimeric G-proteins, thus calling into question the characterization of the mPRs as a novel class of G-protein coupled receptor.
Keywords: membrane progestin receptors, progesterone, yeast, adiponectin, osmotin
Introduction
The classic paradigm for progesterone-mediated signal transduction involves diffusion of the steroid hormone into cells and binding to soluble intracellular progesterone receptors.[1] This type of signal transduction is often referred to as genomic signaling because this receptor, also called the nuclear progesterone receptor (nPR), doubles as a DNA binding transcription factor and, as such, modulates gene transcription in response to progesterone. It has long been recognized, however, that many of the physiological effects of progesterone occur far too rapidly to require gene transcription and often occur in cells that are transcriptionally silent, such as sperm.[2] Moreover, the receptor(s) responsible for mediating the non-genomic effects of progesterone have a markedly different agonist profile from nPR,[3] suggesting that it is a distinct receptor. These facts have led researchers to propose the existence of an integral membrane progesterone receptor (mPR) that binds progesterone at the cell surface and rapidly generates intracellular second messengers.[4] This mechanism for progesterone response has been called non-genomic signaling because it does not absolutely require transcription to proceed, although there is no reason to suspect that non-genomic mechanisms do not also regulate transcription.[5] The existence of mPR is a matter of intense debate. Indeed, it has been argued that there is no need to invoke the existence of a distinct mPR because the nPR itself is capable of mediating rapid non-genomic effects by binding to and regulating the activity of other signaling proteins.[6–9] Nevertheless, several candidate mPRs have emerged and, while it is clear that some of these candidates bind progesterone, it is not clear how, or even if, they function as receptors to mediate the non-genomic effects of progesterone.
Recently, a series of seminal papers were published identifying three integral membrane proteins from fish that not only bind progesterone, but also seem to mediate many of its rapid non-genomic effects.[10, 11] These three proteins, renamed mPRα, mPRβ and mPRγ, are conserved in vertebrates and belong to a newly characterized family of proteins that includes the yeast osmotin receptor (Izh2p) [12] and human adiponectin receptors (AdipoR).[13] This family is known as the Progestin- and AdipoQ-Receptor family (PAQR).[14]
A series of follow-up studies produced compelling evidence that mPRα, mPRβ and mPRγ function as membrane progesterone receptors, however, as a recent review in this journal demonstrates [15], the field remains embroiled in controversy. The primary reason for this is the fact a recent study was unable to reproduce the original results.[16, 17] Another significant source of contention is the assertion that these receptors function as a new class of G-protein coupled receptor (GPCR) [18, 19, 10, 11] despite the fact that no other class of PAQR seems to couple to G-proteins and that members of the PAQR family of receptors bear only superficial similarity with GPCRs.
Further investigation is needed before mPRα, mPRβ and mPRγ can be universally accepted as membrane progesterone receptors. One side of this equation involves the study of the physiological roles of these proteins in vivo. To date, no studies have demonstrated an unequivocal role for these proteins in the physiology of progesterone. Part of the reason for this is the overabundance of progesterone binding proteins and putative progesterone receptors in vertebrates that can potentially confound data analysis. Not only do vertebrate cells possess classical nuclear progesterone receptors, they also possess the other members of the nuclear hormone receptor family for which progesterone may function as either an agonist or antagonist.[20, 21] Vertebrate cells also contain a variety of other progesterone-binding proteins that have been proposed to function as mPRs.[22, 23] Progesterone has even been shown to function as an allosteric regulator of a variety of enzymes and receptors.[24–26] Thus, it is difficult to attribute progesterone-dependent effects solely to one protein. This line of research is beyond the scope of this study and, ultimately, the discovery of the true physiological roles of mPRα, mPRβ and mPRγ may require the development of mouse knockout strains.
Herein, we address another important aspect of characterizing mPRα, mPRβ and mPRγ - their biochemistry. To date, the biochemical characterization of mPRα, mPRβ and mPRγ has involved their expression in either vertebrate cell culture or E. coli.[19, 10, 11] Expression in vertebrate cell culture has been fruitful, however, conflicting results were obtained by different groups.[17, 19] Moreover, this approach is limited by the aforementioned overabundance of progesterone binding proteins in vertebrate cells that make data interpretation difficult. On the other hand, E. coli do not contain known progesterone receptors and heterologous expression in this organism has been successfully used to confirm progesterone binding to isolated mPRs embedded in intact E. coli membranes.[10, 11] However, while expression in E. coli is certainly useful for studying progesterone binding to these receptors, it cannot yet be used to investigate functionality since E. coli it is not known if PAQR receptors function the same in this organism as they do in higher eukaryotes.
Thus, a resolution to the debate about whether mPRα, mPRβ and mPRγ can sense and respond to progesterone requires a system with two fundamental properties. First, such a system must be devoid of known progesterone binding/sensing proteins so that the mPRs can be studied in isolation. Second, this system must have an intact signaling apparatus that allows for monitoring signal transduction in response to progesterone. Herein we report the development of such a system that entails the heterologous expression of mPRα, mPRβ and mPRγ in the yeast Saccharomyces cerevisiae. This choice of model systems is advantageous for several reasons. First, yeast is a eukaryotic system for which simple yet powerful genetic tools exist. Second, yeast possess receptors in the PAQR family suggesting that the machinery required to read second messengers produced by these proteins is present.[27] Third, S. cerevisiae neither makes nor uses progesterone. In fact, a recent publication showed that massive doses of progesterone (1 mM) did little more than weakly induce the general stress response.[28] The low background biological activity of progesterone in this system makes it an ideal model for the study of individual progesterone receptors in a living cell. Indeed, S. cerevisiae has already been successfully adapted for the biochemical characterization of nuclear progesterone receptors.[29]
We recently published a study showing that the yeast osmotin receptor (Izh2p) controls a signaling pathway in S. cerevisiae that negatively regulates the expression of a gene called FET3.[30] The physiological importance of this regulation is not yet clear - nor is it actually important in the context of this investigation. What is important is that we also showed that activation of human adiponectin receptors, when heterologously expressed in yeast, had the same effect on FET3 expression. The regulation of the same pathway by fungal and human PAQRs suggests that PAQRs from diverse sources generate similar second messengers when expressed in yeast. The identity of this second messenger and the mechanism of signal transduction for PAQRs in yeast are still under investigation and these topics are beyond the scope of this study.
Herein, we demonstrate that the expression of the FET3 gene can be used as a general reporter of the functionality of human PAQR receptors. Using this reporter system to study the mPRs, we made several important findings. First and foremost, we demonstrated mPRα, mPRβ and mPRγ repress FET3 in response to progesterone while other PAQR receptors do not. This confirms beyond reasonable doubt that these proteins can sense and respond to progesterone. Moreover, the ED50 values for the activation of the mPRs by progesterone demonstrate that the mPRs are most responsive to progesterone at physiologically relevant concentrations. Second, we show that this assay can be used to probe the agonist profiles for these receptors. Not only do these receptors have specificities that are distinct from nPR, they are activated by ligands, such as 17α-hydroxyprogesterone, that are known to activate non-genomic pathways but not nPR.[3] We also demonstrate that mifepristone, an important antagonist of nuclear progesterone receptors, actually serves as a weak agonist of the mPRs, fact that is consistent with what is known about non-genomic progesterone signaling.[31] Third, we show that human PAQR6 and PAQR9, both uncharacterized members of the PAQR family likely function as additional vertebrate mPRs. Accordingly, we renamed them mPRδ and mPRε, respectively. Finally, we demonstrate that the ability of these proteins to sense and respond to progesterone requires neither human nor yeast Gα-proteins making it unlikely that the mPRs function as GPCRs in the classical sense.
Materials and Methods
Yeast strains
Wild type BY4742 (Mat α), BY4741 (Mat a) and gpa2Δ (Mat α, BY4742 background) mutant yeast strains were obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/). gpa1Δ mutants are inviable due to constitutive growth arrest caused by the hyperactivation of the Ste4p/Ste118p Gβγ subunit in the absence of the Gpa1p Gα subunit. Viability of gpa1Δ mutant strains can be restored by concomitant deletion of the Ste7p MAP kinase that functions downstream of Ste4p/Ste18p. The gpa1Δste7Δ mutant strain (in the BY4741 background) was kindly provided by Dr. Henrik Dohlman (University of North Carolina, Chapel Hill).[32] The STY50 strain lacking the HIS4 gene [33] was kindly provided by Dr. Gunnar von Heijne (Stockholm University).
Plasmids and primers
Human PAQRs were cloned into various vectors by PCR from commercially available cDNAs, the source and names of which are listed in the Supplemental Data. PAQRs were cloned into the pYES260 [34], pRS316 [35] or pGREG536 [36] expression vectors using gap repair. The pJK90 plasmid containing a Dual Topology Reporter (DTR)-tagged OST4 gene driven by the constitutive TPI1 promoter was kindly provided by Dr. Gunnar von Heijne (Stockholm University). [37] The OST4 open reading frame in pJK90 was replaced with the PAQR6 and mPRα open reading frames by gap repair. Primers for all cloning reactions are listed in the Supplemental Data. Proteins expressed in pYES260 have an N-terminal 6x-histidine tag followed by a TEV protease cleavage site. Proteins expressed in pGREG536 have an N-terminal 7x-HA tag. Proteins expressed in the pRS316 vector are untagged. The pYES260, pRS316 and pGREG536 vectors allow for galactose-inducible expression via the GAL1 promoter. We see no vector-dependent difference in the functionality of any PAQR receptor, indicating that receptors in this family seem to be highly tolerant of N-terminal modifications. The FET3-lacZ plasmid was previously described.[30] Plasmids derived from pADM4 carrying hyperactive alleles of Gpa1p (Gpa1pQ323L) [38] and Gpa2p (Gpa2pQ300L) [39] driven by the ADH1 promoter were kindly provided by Dr. Henrik Dohlman (UNC Chapel Hill). A pRS316-based plasmid containing the STE2 gene under the control of the GAL1 promoter [40] was kindly provided by Dr. Nicholas G. Davis (Wayne State University).
Assays and growth conditions
Strains were maintained using standard protocol and grown in synthetic defined (SD) media with the appropriate amino acids to match the auxotrophies of the individual strains. Low Iron Medium (LIM) contains EDTA to limit iron-bioavailability and its composition has been previously described.[30] When supplemented with only 1 µM Fe3+, LIM is considered iron-deficient and the FET3 gene is fully induced under these conditions. β-galactosidase (lacZ) assays were performed as previously described.[30] In brief, overnights of cells grown in SD-glucose media were re-inoculated into iron-deficient LIM to induce the expression of FET3. 2% galactose was used as a carbon source to induce full expression of PAQR genes driven by the GAL1 promoter, while 0.05% galactose/1.95% raffinose was used for reduced PAQR expression. All ligands were added to the growth medium upon re-inoculation into LIM. For experiments in which steroids were added as ligands, steroids were added from ethanol stocks and "untreated" controls are actually treated with an equal volume of ethanol to control for vehicle effects. Cells were allowed to grow to mid-log phase at which time the activity of the FET3 gene was monitored using a FET3-lacZ promoter-reporter construct. lacZ activities are presented as a percentage of activity seen in untreated cells expressing the appropriate empty expression vector control. For individual experiments, each data point has been done in triplicate and the error bars represent +/− 1 standard deviation. All experiments were performed at least three times and a representative experiment is shown. EC50 values were obtained using the web-based BioDataFit software using sigmoidal curve-fitting. (http://www.changbioscience.com/stat/ec50.html)
Total membrane protein preparations were prepared as previously described.[30] Protein concentrations were determined and equal amounts were loaded onto SDS-PAGE gels for Western blotting. Western blots were performed using standard protocol using a rabbit polyclonal anti-HA primary antibody (Santa Cruz Biotechnology) and a goat anti-rabbit IgG-HRP conjugate as the secondary antibody (Santa Cruz Biotechnology). Proteins were detected by chemiluminescence. Determination of glycosylation with EndoH treatment was performed as published.[41]
Sequence analysis
Multiple sequence alignments and bootstrapped phylogenetic trees were produced using ClustalX with default parameters.[42] Trees were visualized using Tree View [43] and TreeView X (http://darwin.zoology.gla.ac.uk/~rpage/treeviewx/index.html). Pairwise sequence analysis was performed at the NCBI website by Blast 2 Sequences.[44] Hydropathy plots were first generated with TopPredictII 1.2 [45] using the Kyte-Doolittle algorithm and default parameters. Data was downloaded into a spreadsheet and the hydropathy values were aligned based on the multiple sequence alignment produced by ClustalX. Average hydropathy across the entire set of aligned proteins was then calculated. Sequences used for these comparisons are listed in the Supplemental Data. Most sequences were obtained from the NCBI database, however, sequences from Trichoplax adhaerens, Nematostella vectensis, Lottia gigantea, Capitella sp., Branchiostoma floridae and Ciona intestinalis were obtained from the Joint Genome Institute website (http://genome.jgi-psf.org/euk_home.html).
Results
Heterologous expression of human mPRs in yeast
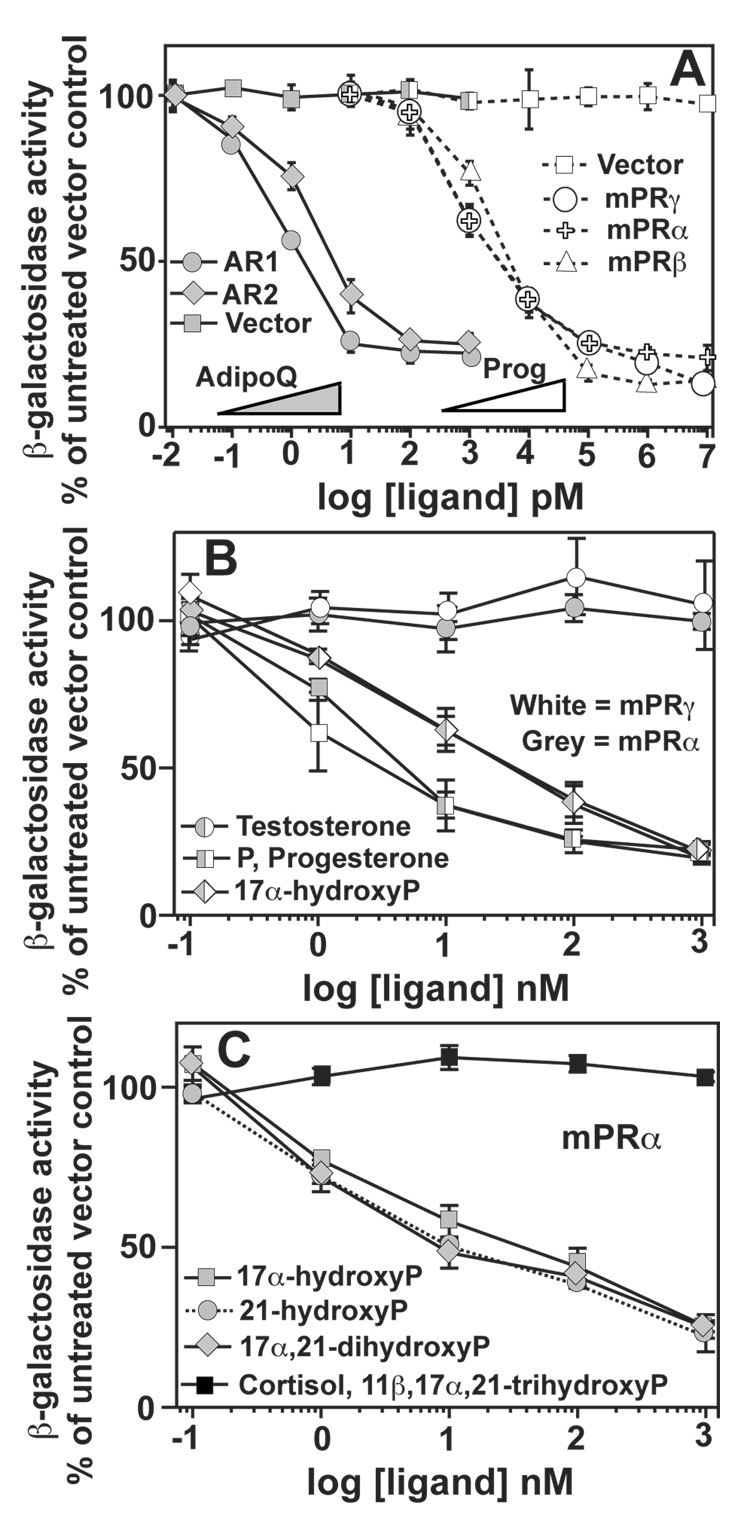
When grown in iron-deficient medium, the FET3 gene is induced to facilitate the uptake of exogenous iron. We previously demonstrated that the yeast osmotin receptor (Izh2p) activated a pathway that resulted in the constitutive repression of FET3 in iron-deficient conditions.[30] Control experiments demonstrated that this effect was not an artifact of the FET3-lacZ reporter and that the expression of FET3 was, indeed, regulated by Izh2p. We also demonstrated that the human adiponectin receptors (AdipoR1 and AdipoR2) could be functionally expressed in yeast. In these experiments, expression of the FET3 gene responded reciprocally to the amount of adiponectin in the medium in cells expressing AdipoR1 and AdipoR2.[30] The EC50 values for adiponectin were 0.7 and 2.4 pM for AdipoR1 and AdipoR2, respectively. (Table 1) This data is re-plotted in Figure 1A to demonstrate two important concepts. First, the expression of the FET3 gene can be used as a reporter for the activity of human PAQR receptors. Second, PAQR receptors from diverse sources activate the same pathway in yeast. This led us to predict that this system could be used to address whether mPRα, mPRβ and mPRγ could sense and respond to progesterone. We cloned all three human receptors into GAL1 driven expression plasmids and grew them in iron deficient LIM containing 0.05% galactose. In cells expressing the mPRs the expression of the FET3 gene responded reciprocally to progesterone in a dose dependent manner. (Figure 1A) The EC50 values for progesterone were 1.3, 2.3 and 1.6 nM for mPRγ, mPRα and mPRβ, respectively. (Table 1)
Table 1.
EC50 values for various PAQR ligands.
| log EC50 pM (EC50) | ||
|---|---|---|
| Adiponectin | Progesterone | |
| AdipoR1 | −0.13 ± 0.08 (0.7 pM) |
N.R. |
| AdipoR2 | −0.38 ± 0.17 (2.4 pM) |
N.R. |
| mPRγ | N.R. | 3.11 ±0.44 (1.3 nM) |
| PAQR6 | N.R. | 3.42 ± 0.88 (2.6 nM) |
| mPRα | N.R. | 3.36 ± 0.08 (2.3 nM) |
| mPRβ | N.R. | 3.21 ± 0.61 (1.6 nM) |
| PAQR9 | N.R. | 4.14 ± 0.86 (13.8 nM) |
N.R. No response
Figure 1. Functional expression of human mPRα, mPRβ and mPRγ in yeast.
All cells are wild type and are grown in iron-deficient LIM. Medium in panels (A) and (B) contains 0.05% galactose/1.95% raffinose and medium in panel (C) contains 2% galactose. In all boxes, the activity of the FET3 gene is monitored by measuring β-galactosidase activity produced by the FET3-lacZ reporter. (A) All PAQRs are cloned into the pYES260 vector except AdipoR2, which is cloned into pGREG536. White symbols show the effect of progesterone on FET3 in cells carrying either empty expression vector or vectors that express mPRα, mPRβ or mPRγ. Grey symbols show the effect of adiponectin on FET3 in cells carrying either empty expression vector or vectors that express AdipoR1 or AdipoR2. (B) The effect of various steroids on the FET3 gene cells expressing either mPRγ or mPRα from the pYES260 plasmid. (C) The effect of various steroids on FET3 in cells expressing mPRα from the pRS316 plasmid. When data points overlap or are very close to overlapping, combined symbols are used.
Wild type cells carrying empty expression plasmid did not respond to either adiponectin or progesterone indicating that the effects described above cannot be attributed to an endogenous yeast protein, such as a yeast PAQR receptor. Furthermore, AdipoR1 and AdipoR2 do not respond to progesterone at concentrations as high as 10 µM (Figure 4A), demonstrating that progesterone specifically affects mPRα, mPRβ and mPRγ and does not generally affect yeast expressing non-native PAQR receptors.
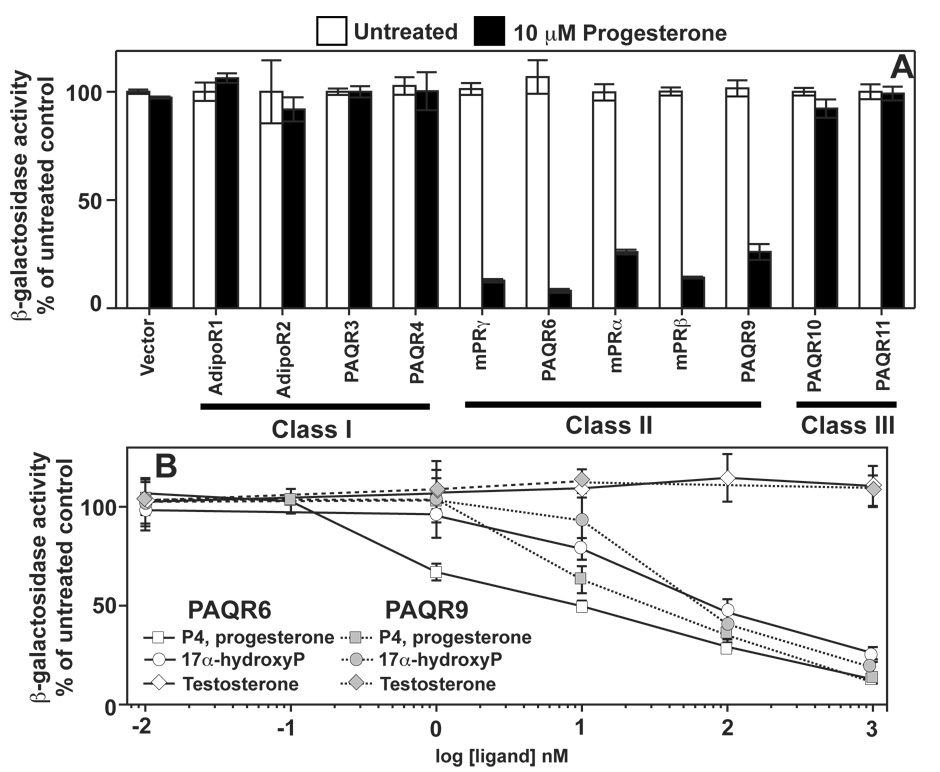
Figure 4. Identification of additional membrane progesterone receptors.
In all panels except (C) cells were grown in medium containing 0.05% galactose/1.95% raffinose and FET3 activity is measured using the FET3-lacZ construct as a reporter. All PAQRs are cloned into the pYES260 vector except AdipoR2, which is cloned into pGREG536. (A) Response FET3 in cells expressing all 11 human PAQR proteins to 10 µM progesterone. (B) Response of FET3 to various steroids in wild type cells expressing either PAQR6 (white symbols) or PAQR9 (grey symbols).
Specificity of mPRα, mPRβ and mPRγ
We also investigated the steroid specificity of mPRα and mPRγ and found that 17α-hydroxyprogesterone was an effective activator of both receptors, albeit with a lower EC50. (Figure 1B, Table 2) A further analysis of mPRα revealed that the receptor was largely permissive of alterations at the 17 and 21 positions of the pregnane ring. 17α-hydroxyprogesterone, 21-hydroxyprogesterone and 17α,21-dihydroxyprogesterone were all similarly effective activators of mPRα. However, 11β,17α,21-trihydroxyprogesterone (cortisol) was an ineffective agonist. (Figure 1C)
Table 2.
EC50 values for various PAQR ligands.
| log EC50 nM (EC50) | ||
|---|---|---|
| mPRγ | mPRα | |
| Progesterone | 0.04 ± 0.25 (1.1 nM) |
0.34 ± 0.08 (2.2 nM) |
| 17α-hydroxyprogesterone | 0.97 ± 0.63 (9.3 nM) |
1.01 ± 0.44 (10.2 nM) |
| RU-486 | 3.83 ± 0.23 (6.8 µM) |
4.04 ± 0.26 (11.0 µM) |
| Progesterone + 10 µM Testosterone |
1.03 ± 0.53 (10.7 nM) |
N.D. |
| Progesterone + 100 nM RU-486 |
1.19 ± 0.12 (15.5 nM) |
N.D. |
N.D. Not determined
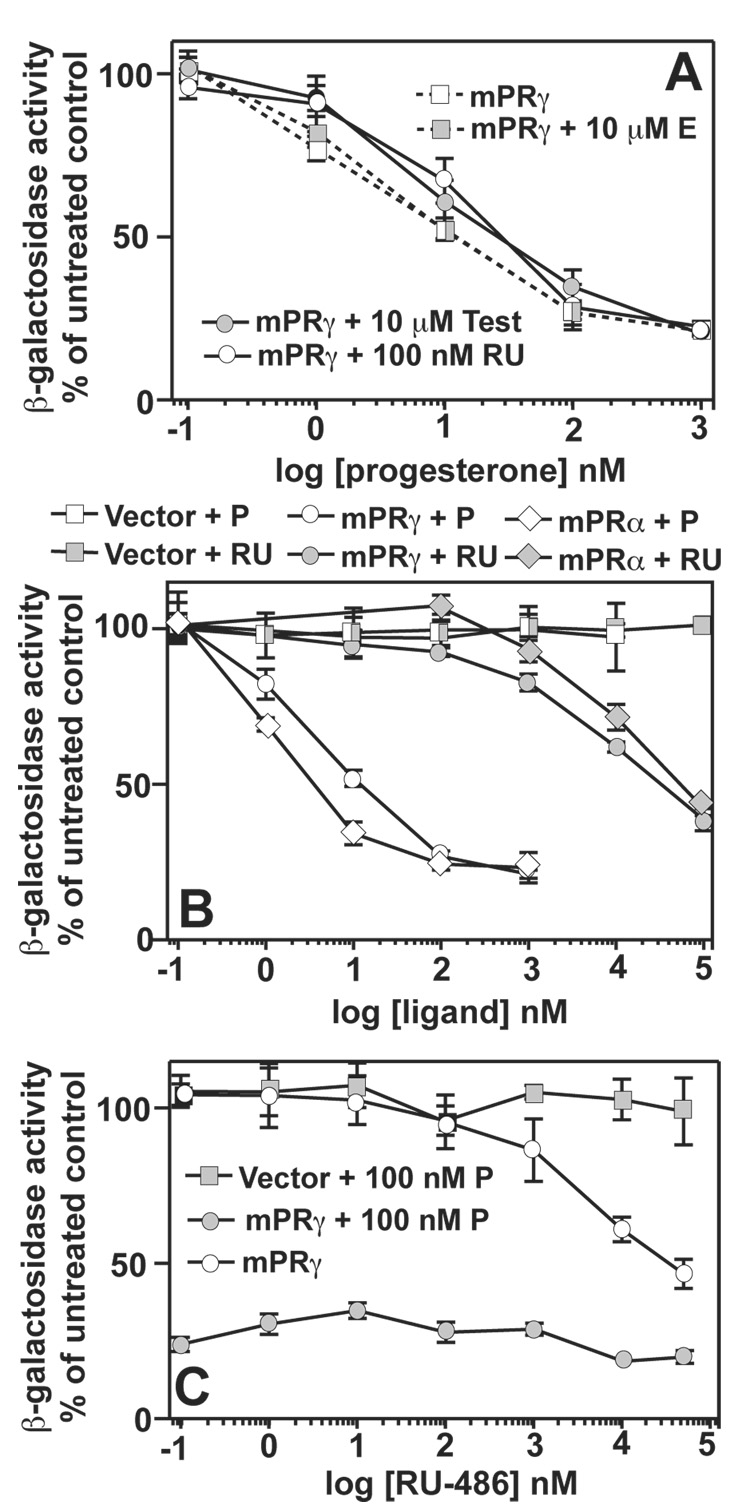
While testosterone is closely related in structure to progesterone, it is a poor agonist of the non-genomic pathways of progesterone signaling.[46] However, testosterone does seem to competitively inhibit progesterone binding to mPRγ and mPRα in mammalian cells.[19, 10, 11] and our data suggests that mPRγ and mPRα are tolerant to substitutions at the 17 position of the pregnane ring where testosterone and progesterone differ. This raised the possibility that testosterone might function as an antagonist of the mPRs. Figure 1B shows that testosterone had no agonist activity against either mPRγ or mPRα in concentrations up to 10 µM. However, when the response of mPRγ to progesterone was measured in the presence of 10 µM testosterone, the EC50 of progesterone increased 10-fold. (Figure 2A, Table 2) Exposure of cells to 10 µM estradiol had no effect on the EC50 of progesterone, indicating that this effect was specific for testosterone and not a non-specific effect of steroids on the assay system. (Figure 2A)
Figure 2. Steroid specificities for mPRγ and mPRα.
In all cases, FET3 expression is measured using the FET3-lacZ reporter. All PAQR are cloned into the pGREG536 vector. All cells are wild type and are grown in iron-deficient LIM containing 0.05% galactose/1.95% raffinose. (A) The dose response of FET3 in cells expressing mPRγ plasmid to progesterone either alone or in the presence of 10 µM β-estradiol (E), 10 µM testosterone (test) or 100 nM RU-486 (RU). (B) Dose response of FET3 to either progesterone (P) or RU-486 (RU) in cells carrying either empty expression vector or vectors that express mPRγ or mPRα. (C) Dose response of FET3 to RU-486 in cells carrying either empty expression vector or a vector that expresses mPRγ. Cells are either treated with RU-486 alone or in the presence of 100 nM progesterone (P). When data points overlap or are very close to overlapping, combined symbols are used.
Mifepristone, also known as RU-486, is a clinically important antagonist of the nuclear progesterone receptor.[47] We examined the ability of this synthetic steroid to antagonize mPRγ and mPRα Surprisingly, RU-486 functioned as a weak agonist of both receptors with EC50 values in the low µM range. (Figure 2B, Table 2) RU-486 had no effect on yeast carrying empty expression vector. (Figure 2B) We also demonstrated that RU-486 functions as a weak antagonist of mPRγ at lower concentrations. Figure 2C shows that RU-486 weakly antagonizes mPRγ in the presence of 100 nM progesterone and that this effect disappears at higher concentrations of RU-486. Furthermore, exposure of cells to 100 nM RU-486 causes a 15-fold increase in the EC50 of progesterone for the activation of mPRγ. (Figure 2A, Table 2)
Sequence analysis of the mPR family
The human genome contains 11 genes encoding proteins in the PAQR family,[14] all of which are highly conserved in the vertebrate lineage. PAQR5, PAQR7 and PAQR8 encode the putative membrane progesterone receptors and their gene products have been named mPRγ, mPRα and mPRβ, respectively. A bootstrapped phylogenetic tree (Supplemental Figure 1A) shows the relatedness of the mPRs to other receptors in the PAQR family including the human adiponectin receptors and the yeast osmotin receptor homologues, Izh1p, Izh2p and Izh3p. We included sequences from a variety of species to demonstrate conservation throughout the vertebrate lineage. This tree clearly demonstrates that the PAQR family can be grouped into three distinct clades. Human PAQR10 and PAQR11 belong to a highly divergent clade of PAQRs that contains bacterial proteins been previously characterized as hemolysins.[48] We designated this clade as Class III and used it as an outgroup to root the tree. A second clade includes human PAQR1, PAQR2, PAQR3 and PAQR4 as well as the yeast PAQR homologues. We designated this clade as Class I. Homologs of PAQR1, PAQRQ2 and PAQR3 can be found in fungi. A homolog of PAQR4 can be found in Nematostella vectensis, but not Trichoplax adhaerens, suggesting that this gene may have originated in eumetazoans. The three mPRs belong to a third clade designated as Class II. This tree indicates that Class II receptors can be subdivided into 2 subgroups - one that contains mPRγ and one that contains mPRα and mPRβ. In addition to mPRγ, the human genome encodes 2 additional PAQRs (PAQR6 and PAQR9) that belong to the mPRγ subgroup.
We could not identify a PAQR6 homologue in chicken or an mPRα homologue in frog. The absence of such sequences in the databases may be due to gaps in G. gallus, X. tropicalis or X. laevis genomes, however, we could find neither supporting cDNAs nor ESTs, suggesting that these genes may have been lost in these species. The PAQR9 gene in the G. gallus genome is interrupted by an unsequenced gap, therefore the full sequence is not shown in the Supplemental Data. While the genomes of higher vertebrates contain only one copy of each of the mPRs, teleost fish genomes (herein represented by zebrafish) encode three distinct mPRγ isoforms, two of which are 100% identical and found in tandem on chromosome 7. The zebrafish genome also encodes multiple paralogs of mPRα, AdipoR1, PAQR4 and PAQR10. The multiple PAQR paralogs in zebrafish fish are consistent with the genome duplication event that is believed to have led to the evolution of teleost fish.[49]
The mPRs have been proposed to be a novel class of GPCR. Another bootstrapped phylogenetic tree (Supplemental Figure 1B) that contains the human and yeast PAQRs as well as several proteins belonging to the GPCR [50] and alkaline ceramidase [51] protein families. The tree is rooted with the GPCRs as an outgroup. The PAQRs, GPCRs and alkaline ceramidases are alike in that all three families have a core of seven transmembrane domains (TM). This tree demonstrates that the PAQR family is no more similar to GPCRs than they are to alkaline ceramidases, suggesting that any structural similarity between the PAQR and GPCR families is superficial. It should be noted, however, that the GPCR family is highly divergent and that there are no amino acid motifs that unify the entire family.[50] Thus, it is still possible that the PAQRs represent a unique class of highly divergent GPCR.
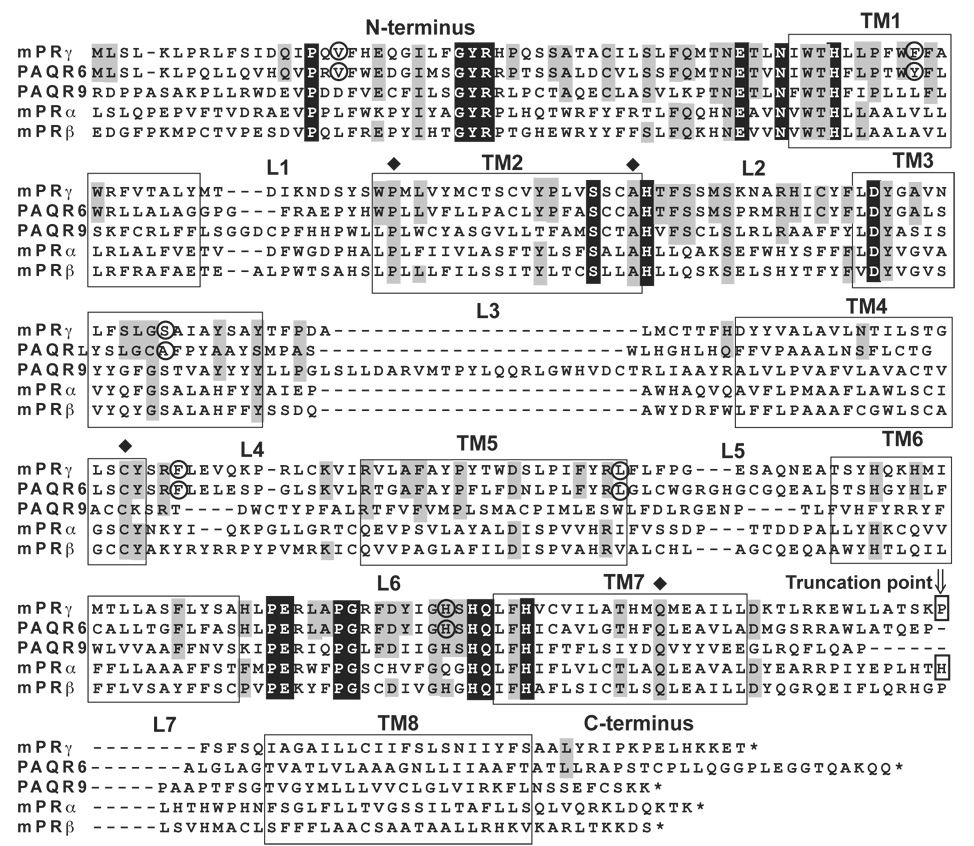
Figure 3 shows a multiple sequence alignment of the five human Class II PAQRs. The entire PAQR family is unified by the presence of seven predicted TMs and three conserved regions. (see alignment in ref [27]) First, there is a conserved motif that precedes TM1. This motif has the consensus PxnGYRxnEx2Nx3H, although this motif is truncated to Ex2–3Nx3H in Class III PAQRs. A second motif spans the end of TM2 and the beginning of TM3 and has the consensus sequence Sx3HxnD. A third motif spans the loop preceding TM7 and has the motif PEx3PGxnHQx2H, although this is also truncated to Hx3H in Class III proteins. The seven TM core and these three short motifs are all that unify the entire PAQR family. Class II PAQR receptors are unique in that they contain an eighth predicted hydrophobic motif that is C-terminal to the conserved PAQR core. (See Figure 5 for hydropathy plots)
Figure 3. Multiple sequence alignment of human Class II PAQRs.
This alignment was originally performed using ClustalX but was modified manually afterwards. TM and loop regions (L) are numbered. Predicted TMs are also boxed. Black shading shows amino acids that are highly, but not universally, conserved in the entire PAQR family. Grey shading indicates amino acids that are conserved in all vertebrate members of the mPRγ/PAQR6 clade, although only human sequences are shown. Circled amino acids indicate the positions of intron/exon boundaries in the pre-mRNA. Arrow indicates the location of truncations in the mPRγ and mPRα proteins discussed in Figure 7.
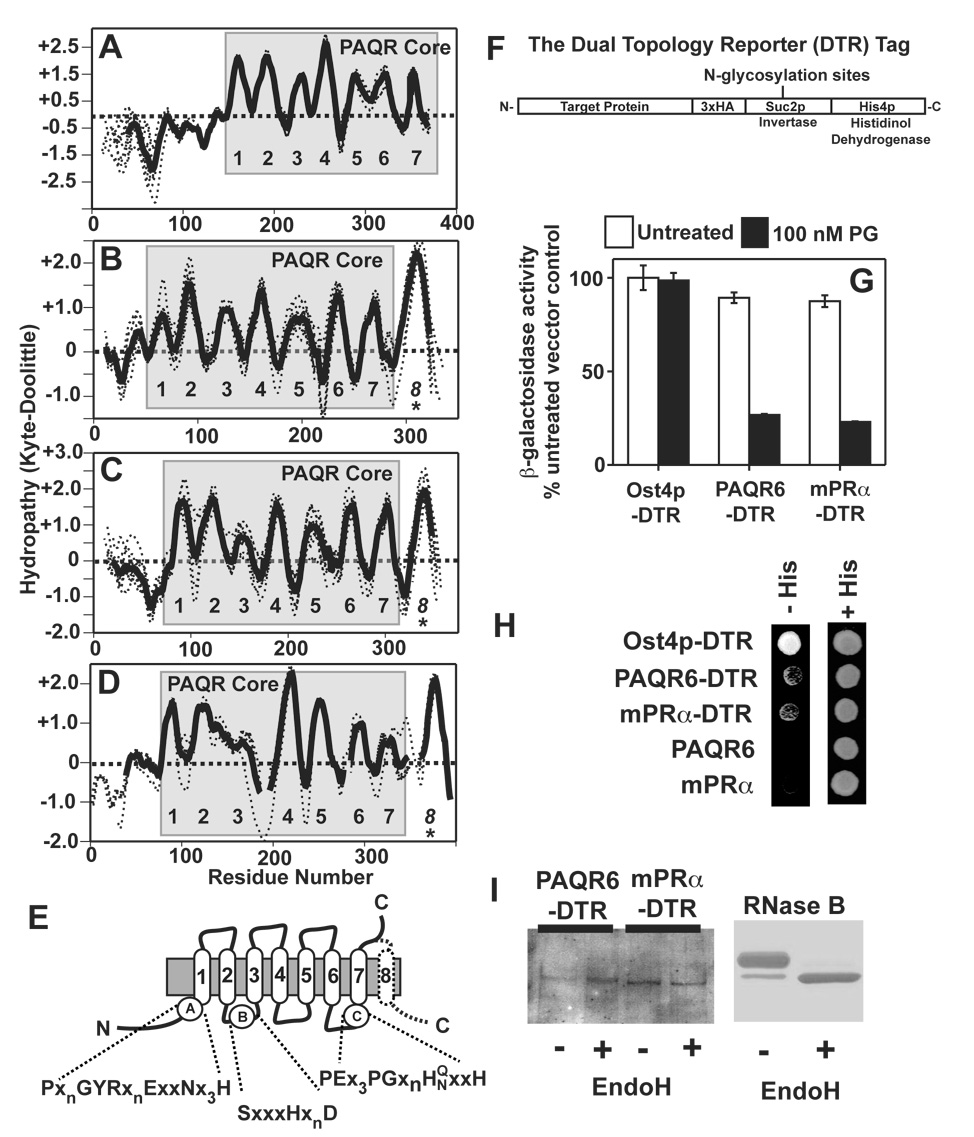
Figure 5. Topological analysis of Class I and Class II PAQRs. (A–D).
Hydropathy plots for the individual proteins analyzed in Figure 3 were generated and aligned (dotted lines). An average hydropathy plot for all members of each clade were generated (solid lines). The core PAQR motif is shaded in grey and the predicted TMs are numbered. All vertebrate members of the AdipoR1 and AdipoR2 clade (A), the mPRγ/PAQR6 clade (B), the mPRα/mPRβ clade (C) and the PAQR9 homologs (D). (E) Predicted topology of the core PAQR motif with the locations of the three highly conserved motifs shaded in black in Figure 4. The predicted eighth TM in the mPRs is shown with a dotted line. (F) The structure of the dual topology reporter tag. (G) DTR-tagged PAQR6 and mPRα expressed in the pJK90 plasmid repress FET3-lacZ in response to 100 nM progesterone in cells grown in medium containing 0.05% galactose/1.95% raffinose. (H) Rescue of the histidine auxotrophy of the STY50 strain by DTR-tagged PAQR6 and mPRα on plates containing histidinol. Plasmids expressing untagged versions of PAQR6 and mPRa are shown as negative controls. A plasmid expressing DTR-tagged Ost4p is shown as a positive control. (I) Membrane extracts from cells expressing DTR-tagged PAQR6 or mPRα are either left untreated (−) or treated with the endoglycosidase EndoH (+) and subsequently run on protein gels and transferred to nitrocellulose membranes for Western blot. anti-HA antibodies were used to detect the DTR tag. (left) Coomassie stained gel of EndoH treated RNase B under the same conditions as those used to treat the cell membrane extracts is shown on the right as a positive control.
Effect of progesterone on Class II receptors
mPRγ is more similar to PAQR6 and PAQR9 than it is to mPRα and mPRβ. Since mPRγ, mPRα and mPRβ all function as progesterone receptors, it is likely that PAQR6 and PAQR9 do as well. To test this, we cloned all 11 human PAQRs into GAL1-driven yeast expression vectors to determine if they could repress FET3 in response to progesterone treatment. (We have evidence for expression of all human PAQRs except PAQR10, data not shown) Figure 4A shows that 10 µM progesterone was an effective agonist of all five Class II PAQRs, but was ineffective against receptors in Classes I and III. The dose response of PAQR6 and PAQR9 to progesterone reveals EC50 values of 2.6 and 13.8 nM, respectively. (Figure 4B, Table 1) Figures 4B shows that, like mPRγ and mPRα, PAQR6 and PAQR9 can be activated by 17α-hydroxyprogesterone, but not testosterone.
Structural analysis of Class II PAQRs
In Figure 5A overlaid hydropathy plots of various vertebrate homologs of the human adiponectin receptors demonstrate the seven predicted TMs of the conserved PAQR core. The same work up for vertebrate proteins in the mPRα/β clade (Figure 5B) and the mPRγ clade (Figures 5C and 5D) clearly shows that the seven hydrophobic domains of the PAQR core are conserved with varying degrees of hydrophobicity. Topological studies of the yeast and human Class I receptors as well as bacterial Class III receptors indicated that the C-terminus of these receptors is extracellular.[52, 53, 41] This led to the topological model shown in Figure 5E showing the conserved PAQR core and the location of the three highly conserved motifs shown in Figure 3. If the additional hydrophobic domain at the C-termini of the Class II receptors is, indeed, a TM, then the C-termini of receptors in this class should be intracellular.
To test this, we used the dual topology reporter method to probe the topology of mPRα and PAQR6 in yeast. This is the same method that was previously used to determine the topology of the yeast Izh2p receptor.[41] In this method, an HA-Suc2p-His4p tag is placed at the C-terminus of a protein of interest. (Figure 5F) The HA (hemagglutinin) part of the tag allows for detection of the chimera by Western blot. The His4p part of the tag encodes a functional histidinol dehydrogenase that is required for histidine biosynthesis. This enzyme requires cytosolic NADH as a cofactor and will only rescue the histidine auxotrophy of a his4Δ mutant strain if the tag is cytosolic. In addition, if the C-terminus is extracellular, it must pass through the lumen of the Endoplasmic Reticulum. The Suc2p fragment of the tag contains multiple N-glycosylation sites, which will be glycosylated if it passes through the ER lumen. This modification can be detected by a shift in molecular weight in Western blots after treatment with an endoglycosidase.
Consistent with the model shown in Figure 5E, C-terminally tagged Izh2p was previously shown to be unable to confer histidine prototrophy indicating that its C-terminus was extracellular. (see [41]) When this tag was placed at the C-terminus of PAQR6 and mPRα, the resulting chimeras could still respond to progesterone demonstrating that the tag did not affect functionality. (Figure 5G) Moreover, both chimeras could weakly rescue the histidine auxotrophy of a his4Δ strain, indicating that their C-termini are intracellular. (Figure 5H) Treatment of membrane extracts from cells expressing PAQR6 and mPRα with the endoglycosidase, EndoH, does not alter the mobility of either PAQR6 or mPRα in Western blots. (Figure 5I) A change in mobility for RNase B, a protein that is known to be N-glycosylated, is shown as a positive control. These findings indicate that the C-terminus of PAQR6 and mPRα does not pass through the ER lumen and that the C-terminus of Class II PAQRs does, indeed, contain an additional TM.
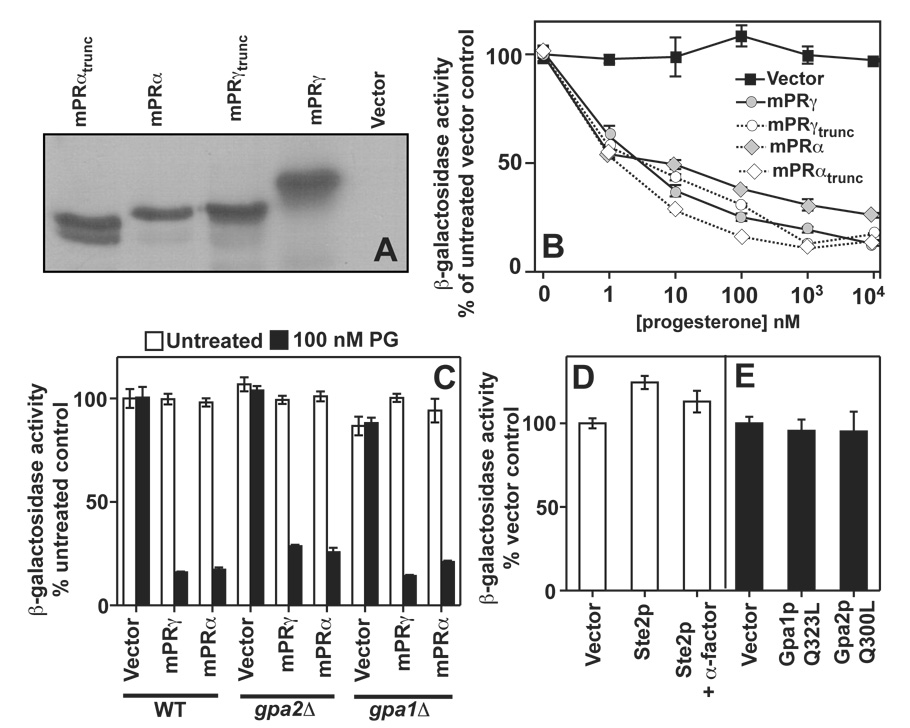
Thus, human Class II receptors are unified by the presence of an additional TM that is C-terminal to the conserved PAQR core and by the fact that all five respond to progesterone when expressed in yeast. This led us to postulate that this additional hydrophobic domain is responsible for progesterone sensing. Unfortunately, there are no amino acids in this TM that are conserved in all five receptors, (See Figure 3) hence, we did not undertake site directed mutagenesis to determine if any particular amino acid is involved in sensing. Instead we truncated the entire TM for mPRγ and mPRα. Western blots show that these truncation mutants are stably expressed in yeast. (Figure 6A) Unexpectedly, these truncations had no effect on the ability of these receptors to sense and respond to progesterone. (Figure 6B)
Figure 6. Truncation mutations and G-protein signaling.
In all cases, FET3 expression is measured using the FET3-lacZ reporter. (A) Full length and truncated mPRγ and mPRα were expressed in wild type cells grown in 2% galactose from the pGREG536 vector. The location of the C-terminal truncations is shown in Figure 4. All proteins possess an N-terminal 7x-HA tag. Proteins were detected by Western blot using an anti-HA antibody. (B) The dose response of FET3 to progesterone in cells expressing the full length or truncated mPRγ and mPRα and grown in 0.05% galactose/1.95% raffinose. (C) The ability of mPRα and mPRγ to respond to progesterone and repress FET3 is not impaired in either gpa2Δ or gpa1Δ cells (gpa1Δ cells also lack the STE7 gene, see text) (D) Overexpression of the Ste2p GPCR using the GAL1 promoter does not repress FET3 in wild type cells (mat a) grown in 2% galactose. Activation of overexpressed Ste2p via the addition of 1 µM of its agonist, α-factor, also has no effect on FET3 under these conditions. (C) Expression of constitutively active alleles of Gpa1p (Gpa1pQ323L) and Gpa2p (Gpa2pQ300L) from the GAL1 promoter had no effect on FET3 in cells grown in 2% galactose.
Lack of involvement of Gα-proteins in mPR-dependent signaling
It has been proposed that mPRα, mPRβ and mPRγ function as a novel class of GPCR.[18, 19, 10, 11] If this were true, then heterotrimeric G-proteins would be required as intracellular second messengers downstream of the mPRs. Yeast possess only two heterotrimeric Gαβγ-protein complexes. The first includes Gpa1p (α), Ste4p (β) and Ste18p (γ) and is coupled to the Ste2p and mating pheromone-sensing GPCR.[32] The second is less well characterized and contains Gpa2p (α) and a non-canonical β-subunit, Acs1p. To date, no γ-subunit has been identified for Gpa2p. Gpa2p is coupled to the Gpr1p glucose-sensing GPCR.[54] Both mPRγ and mPRα were capable of sensing and responding to progesterone in strains lacking either Gpa1p or Gpa2p. (Figure 6C) In addition, we demonstrated that neither overexpression of the Ste2p GPCR from the GAL1 promoter nor its concomitant activation with α-factor pheromone could recapitulate the effect of the PAQRs on the FET3 gene. (Figure 6D) Moreover, expression of constitutively active alleles of Gpa1p (Gpa1pQ323L)and Gpa2p (Gpa2pQ300L) were incapable of causing repression of FET3. (Figure 6E)
Discussion
The definitive classification of mPRα, mPRβ and mPRγ as membrane progesterone receptors is still a matter of debate and the recent publication of conflicting data that directly challenged the role of mPRα, mPRβ and mPRγ in progesterone signaling [17] has reinforced the controversy. To address whether or not the mPRs are, indeed, membrane progesterone receptors, we expressed the mPRs in the tractable eukaryotic model organism, Saccharomyces cerevisiae. The obvious reason for this choice is that yeast do not make or use progesterone. Indeed, that progesterone has very low biological activity towards this organism [28] and that its genome does not encode proteins in the nuclear receptor family, [55] make S. cerevisiae an ideal model system to study progesterone receptors. In fact, recombinant yeast are routinely used to study the structure and function of vertebrate nuclear progesterone receptors.[29]
Data from this and previous publications demonstrate that Izh2p, an endogenous yeast PAQR receptor, activates a pathway that leads to the repression of a gene called FET3.[30] The physiological significance of this regulation, while an important aspect of fungal biology, is irrelevant for this discussion, which focuses solely on the development of this phenomenon into an assay for human PAQR receptor activation. To demonstrate proof of principle, we heterologously expressed the human adiponectin receptors, AdipoR1 and AdipoR2, in yeast and showed that they repress FET3 in an adiponectin-dependent manner.[30] The fact both yeast and human PAQRs repress FET3 in yeast suggests that PAQR receptors from various sources generate the same second messenger, which regulates the pathway leading to FET3 repression. It is important to state that we are only suggesting that receptors in the PAQR family generate similar second messengers, not that the signaling components and pathways downstream of PAQR receptors are conserved from yeast to humans. We are in the process of mapping the signaling components downstream of Izh2p in yeast and future research may yet reveal such conservation. In the meantime, we used the FET3 gene as a reporter to address two highly contentious issues surrounding mPRα, mPRβ and mPRγ. First and foremost, we addressed whether or not mPRα, mPRβ and mPRγ actually function as progesterone receptors. Second, we began to address models for how the mPRs convert extracellular progesterone into intracellular second messengers.
Our data clearly demonstrate that mPRα, mPRβ and mPRγ, when heterologously expressed in yeast, mediate progesterone-dependent repression of the FET3 gene. Control experiments showed that progesterone did not affect FET3 in yeast carrying empty expression vector or the human adiponectin receptors, indicating that this effect was not (a) a non-specific effect of progesterone, (b) the result of expressing a foreign membrane protein or (c) a general effect mediated by any PAQR receptor. These results unambiguously demonstrate that mPRα, mPRβ and mPRγ are capable of conferring progesterone-responsiveness onto yeast cells. When coupled with previous studies showing progesterone binding by membranes from E. coli expressing mPRα, mPRβ and mPRγ, our data strongly argues that these proteins directly mediate the response to progesterone. That said, we cannot yet rule out the remote possibility that yeast express an unidentified progesterone-binding protein that interacts with the mPRs and functions as the ligand-binding component.
The EC50 for progesterone activation of mPRα, mPRβ and mPRγ in yeast is between 1–3 nM, values that are consistent with the Kd's for progesterone binding to mPRs (~ 5 nM) determined by Thomas et al.[56] These EC50 values are also close to the physiological concentration of progesterone in human serum, which has been estimated to be between 1–10 nM in men and non-pregnant women in the follicular phase of the menstrual cycle.[57, 58] Thus, our data suggests that human mPRα, mPRβ and mPRγ are most responsive to progesterone at physiologically relevant hormone concentrations and would likely function as legitimate progesterone receptors. This interpretation must be tempered by the possibility that other human proteins, not present in the yeast system, may modulate the response of these receptors to progesterone.
Analysis of the steroid specificity of mPRα and mPRγ activation indicates that they have distinct agonist profiles from nuclear progesterone receptor and provides evidence that their profiles are similar to those of the receptors that mediate non-genomic responses of progesterone.[46, 3, 31] To begin with, our data indicate that the steroid activation profile of the mPRs correlates well with a study that showed 17α-hydroxyprogesterone (17α-HP), 21-hydroxyprogesterone (21-HP) and 17α,21-dihydroxyprogesterone (17α,21-DHP) were effective agonists of the non-genomic progesterone signaling pathway but 11β,17α,21-trihydroxyprogesterone (cortisol) and testosterone were not.[46, 3] Another distinction between the mPRs and nPR is the fact that mifepristone (RU-486), a potent nPR antagonist, functions as a weak agonist for both mPRγ and mPRα at high concentrations RU-486 (EC50 > 5 µM) and, enigmatically, as only a very weak antagonist of mPRγ at lower concentrations (100 nM), shifting the EC50 for progesterone from ~ 1 nM to ~ 15 nM.
The ability of 17α-HP and mifepristone to activate mPRs is particularly intriguing because the former steroid has very low agonist activity towards nPR [46] and the latter is actually an antagonist of nPR.[47] Both, however, seem to be able to activate non-genomic signaling.[3, 31] In fact, the differential reactivity of these steroids towards the genomic and non-genomic pathways has been used as evidence for the existence of distinct membrane progesterone receptors.[3] Hence, the specificity profiles of the mPRs in yeast indicate that these receptors closely resemble the hypothetical non-genomic progesterone receptors.
The unique agonist profiles for the mPRs are also intriguing from a physiological standpoint. While the physiological levels of 17α-HP, 21-HP and 17α,21-DHP are low in human serum (~ 1 nM) they can be as high as 10–20 nM in pregnant women.[59] Considering that the EC50 values for the activation of mPRα by 17α-HP, 21-HP and 17α,21-DHP are similar (~ 10 nM), it is possible that these steroids may indeed be physiologically relevant ligands for this receptor. On the other hand, while testosterone seems to function as an antagonist of mPRγ, the levels of testosterone required for this effect (10 µM) seem to be far too high to be physiologically important.[60] The pharmacokinetics of RU-486 [47] indicate that low doses of RU-486 result in sustained circulating levels in the 100 nM range where it antagonizes mPRγ, while high doses result in sustained levels in the 5 µM range, where it agonizes mPRγ. Thus, RU-486 may have opposing effects on the mPRs depending on the dose given.
The second contentious issue that we attempted to address is the nature of the second messenger produced by the mPRs. When first discovered, structural analysis suggested that the mPRs had seven TMs.[10, 11] This naturally led people to postulate that the mPRs were similar to GPCRs despite the fact that the mPRs bear no more similarity to GPCRs than they do to other groups of heptahelical integral membrane proteins. (See Supplemental Figure 1B) Nevertheless, some preliminary evidence supports this conclusion [18, 19, 10, 11] and now this model seems to be either universally accepted or at least not seriously questioned. A major problem with the GPCR hypothesis is that there is no evidence for the involvement of G-proteins in signaling by any other class of PAQR receptor, suggesting that either there are significant differences between PAQR classes or the GPCR model needs revision.
Before discussing our data, it is necessary to discuss the evidence for G-protein involvement. First, there is a large body of evidence suggesting that Giα proteins are involved in some of the non-genomic effects of progesterone.[10, 11] The involvement of Giα in progesterone signaling naturally led to the hypothesis that the hypothetical mPR was a GPCR. The discovery that mPRα, mPRβ and mPRγ belonged to a family of proteins with seven TMs and that these receptors had intracellular C-termini bolstered this idea. More compelling evidence for the classification of mPRs as GPCRs came from experiments showing co-immunoprecipitation of a Giα-protein with mPRα.[18, 19] In addition, Thomas et al. showed that deletion of the soluble part of the C-terminus of mPRα abrogated the stimulation of 35S-γ-GTP to Giα in response to progestin treatment,[19] suggesting that activation of Giα requires the C-terminus of mPRα in a manner that is reminiscent of GPCRs.
Closer scrutiny of these findings reveals that none offer unequivocal evidence that the mPRs are GPCRs. First, co-immunoprecipitation experiments with membrane-associated proteins can be misleading since one is not actually precipitating a protein, but the entire membrane complex in which the protein is embedded. Since both mPRα and Giα associate with membranes,[61] it is not surprising that they co-precipitate. Consquently, a direct interaction between Giα and mPR is yet to be unambiguously demonstrated and the possibility that the involvement of Giα in progesterone signaling is indirect has not yet been ruled out. Second, we feel that the issue of receptor topology has introduced bias towards the characterization of these receptors as being GPCRs in such a way that the GPCR model has been accepted without concrete proof. Simply having seven TMs and an intracellular C-terminus does not make a protein a GPCR.
We have presented data that contradicts the characterization of these receptors as GPCRs. To begin with, data in this study and from other publications [52, 53, 41, 17, 19] support a model in which PAQRs from all Classes share an identical core structure of seven TMs with identical topologies and that Class II PAQRs possess an eighth TM that places the C-terminus in the cytoplasm. (See Figure 5E) Our data indicating that all PAQRs activate the same signaling pathway in yeast is consistent with a model in which all PAQRs have a similar core structure and mechanism of action. To be clear, however, no group has undertaken a rigorous mapping of the number and topology of TMs in any PAQR. A satisfactory resolution to the debate about whether or not the mPRs resemble GPCRs or other PAQRs will have to wait for such a study.
Regardless of whether or not the mPRs structurally resemble GPCRs, our data demonstrate that no human heterotrimeric G-proteins are essential for mPR-dependent signal transduction in response to progesterone. Since many human GPCRs have been shown to functionally couple to yeast Gα-proteins [62] it is possible that the endogenous Gα-proteins functionally substitute for their human homologs. However, we demonstrated that mPRγ and mPRα were also fully capable of sensing and responding to progesterone in cells lacking either endogenous Gα-protein. Moreover, we demonstrated that constitutive activation of the endogenous Gα-proteins did not result in activation of the same pathway as PAQR activation, as would be expected if the mPRs repressed FET3 by activating these Gα-proteins. Finally, we demonstrated that the C-termini of the mPRs, which seem to be essential for mPR-dependent activation with Gα-proteins in mammalian cells, is not absolutely essential for signal transduction by mPRα and mPRγ in response to progesterone. These findings indicate either that the mPRs are not directly coupled to G-proteins or that these receptors respond to progesterone by G-protein-dependent and -independent mechanisms. It is certainly possible that the C-terminal TM in Class II PAQRs allows these receptors to couple directly to G-proteins, but that this coupling is not the sole mechanism of signal transduction.
Our assay system has produced additional data that bears brief discussion. The phylogenetic analysis in Supplemental Figure 1A suggests that the mPRs are distinct from other PAQR receptors and form a group of PAQRs that we have called Class II. The human genome encodes two other Class II PAQR proteins called PAQR6 and PAQR9. Figure 3 shows that proteins in this class of PAQRs are unified by the presence of an eighth TM. We hypothesized that the presence of this eighth TM at the C-terminus of a PAQR is diagnostic of progesterone receptors, even though this additional TM does not seem to be required for progesterone sensing. Not surprisingly, both PAQR6 and PAQR9 sense and respond to progesterone in yeast and have similar steroid specificities to mPRα, mPRβ and mPRγ. PAQR9 is unique in that the EC50 for progesterone activation of this receptor is approximately 10-fold higher than the other four Class II receptors, although it is not yet clear if this is an inherent property of PAQR9 or an artifact of its expression in yeast. Because of their ability to sense and respond to progesterone, we propose the renaming of these receptors to mPRδ (PAQR6) and mPRε (PAQR9).
The fact that all five Class II human PAQRs function as progesterone receptors is intriguing from an evolutionary perspective. mPRγ belongs to a different subgroup of Class II receptors than mPRα and mPRβ, yet all three proteins are progesterone receptors. This suggests that either their functions have converged during evolution or that their last common ancestor was also a progesterone receptor. The phylogenetic tree in Supplemental Figure 1A includes a variety of proteins with the diagnostic eighth TM necessary for their inclusion in Class II. Intriguingly, the mPRγ clade of PAQRs, which includes mPRδ and mPRε, actually includes proteins found in tunicates, lancelets, echinoderms, molluscs, annelids, flatworms, cnidarians and even Trichoplax adhaerens, an organism at the very base of the metazoan lineage. On the other hand, the mPRα/β clade includes proteins from tunicates, lancelets, echinoderms, molluscs and annelids. It also should be noted that Class II proteins seemed to have been lost in ecdysozoans (nematodes, arthropods). This phylogenetic analysis suggests that mPRγ-like proteins evolved before mPRα/mPRβ-like proteins and that both families predate the evolution of vertebrates. In fact, the presence of mPRγ-like proteins in placozoans indicates that this subgroup of mPRs originated early in the evolution animals. In contrast, the nuclear progesterone receptor seems to have evolved in vertebrates.[63] Thus, the mPRs may represent the original progesterone receptors. Future experiments to test this hypothesis will involve cloning pre-vertebrate Class II PAQRs and testing their ability to respond to progesterone using our assay system.
In summary, we have presented data showing that the human mPRs are capable of sensing and responding to progesterone when expressed in yeast cells. Use of this heterologous expression system allowed us to probe the mechanism by which these receptors mediate the effects of progesterone and present data indicating that these proteins do not absolutely require heterotrimeric G-proteins to respond to progesterone. We also established a simple system that can be used to begin structure/function studies on human mPRs and to explore the structural features in the agonist steroid that are essential for mPR activation.
Supplementary Material
Phylogenetic analysis of the PAQR family. For each tree, the length of the tree branches is proportional to the calculated distance between sequences with the scale bar indicating 0.1 substitutions per site. Numbers at the nodes are confidence values that refer to the number times per 1000 trees drawn a particular grouping is made. (A) A bootstrapped phylogenetic tree showing the relationship between PAQR receptors from a variety of eukaryotic and prokaryotic sources. Lines delineate the three Classes of PAQR and grey shading indicates the distinct clades within Class II. The tree is rooted using the sequences in Class III as an outgroup. (B) A bootstrapped phylogenetic tree containing human, yeast and bacterial PAQRs as well as several sequences belonging to two other groups of proteins with a characteristic seven TM structure. One group includes five G-protein coupled receptor (GPCR) sequences and the other group includes five proteins in the alkaline ceramidase (AlkCer) family of enzymes. The tree is rooted with the clade containing the GPCRs. (C) Taxonomic grouping of the organisms from which the sequences in these trees were derived.
Acknowledgements
Funding for this study was provided by the National Institutes of Health (5 R21 DK074812-02 to TJL) and by the University of Florida Department of Chemistry.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors have nothing to disclose.
References
- 1.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. 1999;64(5):310–319. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 2.Correia JN, Conner SJ, Kirkman-Brown JC. Non-genomic steroid actions in human spermatozoa. Persistent tickling from a laden environment. Semin Reprod Med. 2007;25(3):208–219. doi: 10.1055/s-2007-973433. [DOI] [PubMed] [Google Scholar]
- 3.Blackmore PF, Fisher JF, Spilman CH, Bleasdale JE. Unusual steroid specificity of the cell surface progesterone receptor on human sperm. Mol Pharmacol. 1996;49(4):727–739. [PubMed] [Google Scholar]
- 4.Maller JL. Signal transduction. Fishing at the cell surface. Science. 2003;300(5619):594–595. doi: 10.1126/science.1083725. [DOI] [PubMed] [Google Scholar]
- 5.Boonyaratanakornkit V, Edwards DP. Receptor mechanisms mediating non-genomic actions of sex steroids. Semin Reprod Med. 2007;25(3):139–153. doi: 10.1055/s-2007-973427. [DOI] [PubMed] [Google Scholar]
- 6.Bagowski CP, Myers JW, Ferrell JE., Jr The classical progesterone receptor associates with p42 MAPK and is involved in phosphatidylinositol 3-kinase signaling in Xenopus oocytes. J Biol Chem. 2001;276(40):37708–37714. doi: 10.1074/jbc.M104582200. [DOI] [PubMed] [Google Scholar]
- 7.Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, et al. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol Cell. 2001;8(2):269–280. doi: 10.1016/s1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- 8.Martinez S, Pasten P, Suarez K, Garcia A, Nualart F, Montecino M, et al. Classical Xenopus laevis progesterone receptor associates to the plasma membrane through its ligand-binding domain. J Cell Physiol. 2007;211(2):560–567. doi: 10.1002/jcp.20964. [DOI] [PubMed] [Google Scholar]
- 9.Vallejo G, Ballare C, Baranao JL, Beato M, Saragueta P. Progestin activation of nongenomic pathways via cross talk of progesterone receptor with estrogen receptor beta induces proliferation of endometrial stromal cells. Mol Endocrinol. 2005;19(12):3023–3037. doi: 10.1210/me.2005-0016. [DOI] [PubMed] [Google Scholar]
- 10.Zhu Y, Bond J, Thomas P. Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc Natl Acad Sci U S A. 2003;100(5):2237–2242. doi: 10.1073/pnas.0436133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y, Rice CD, Pang Y, Pace M, Thomas P. Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc Natl Acad Sci U S A. 2003;100(5):2231–2236. doi: 10.1073/pnas.0336132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narasimhan ML, Coca MA, Jin J, Yamauchi T, Ito Y, Kadowaki T, et al. Osmotin is a homolog of mammalian adiponectin and controls apoptosis in yeast through a homolog of mammalian adiponectin receptor. Mol Cell. 2005;17(2):171–180. doi: 10.1016/j.molcel.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi T, Kamon J, Ito Y, Tsuchida A, Yokomizo T, Kita S, et al. Cloning of adiponectin receptors that mediate antidiabetic metabolic effects. Nature. 2003;423(6941):762–769. doi: 10.1038/nature01705. [DOI] [PubMed] [Google Scholar]
- 14.Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Emtage PC, et al. PAQR proteins: a novel membrane receptor family defined by an ancient 7-transmembrane pass motif. J Mol Evol. 2005;61(3):372–380. doi: 10.1007/s00239-004-0375-2. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes MS, Brosens JJ, Gellersen B. Honey, we need to talk about the membrane progestin receptors. Steroids. 2007 doi: 10.1016/j.steroids.2007.12.004. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 16.Ashley RL, Clay CM, Farmerie TA, Niswender GD, Nett TM. Cloning and characterization of an ovine intracellular seven transmembrane receptor for progesterone that mediates calcium mobilization. Endocrinology. 2006;147(9):4151–4159. doi: 10.1210/en.2006-0002. [DOI] [PubMed] [Google Scholar]
- 17.Krietsch T, Fernandes MS, Kero J, Losel R, Heyens M, Lam EW, et al. Human homologs of the putative G protein-coupled membrane progestin receptors (mPRalpha, beta, and gamma) localize to the endoplasmic reticulum and are not activated by progesterone. Mol Endocrinol. 2006;20(12):3146–3164. doi: 10.1210/me.2006-0129. [DOI] [PubMed] [Google Scholar]
- 18.Thomas P, Dressing G, Pan Y, Berg H, Tubbs C, Benninghoff A, et al. Progestin, estrogen and androgen G-protein coupled receptors in fish gonads. Steroids. 2006;71:310–316. doi: 10.1016/j.steroids.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Thomas P, Pang Y, Dong J, Groenen P, Kelder J, de Vlieg J, et al. Steroid and G protein binding characteristics of the seatrout and human progestin membrane receptor alpha subtypes and their evolutionary origins. Endocrinology. 2007;148(2):705–718. doi: 10.1210/en.2006-0974. [DOI] [PubMed] [Google Scholar]
- 20.Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92(1):73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 21.Rupprecht R, Reul JM, van Steensel B, Spengler D, Soder M, Berning B, et al. Pharmacological and functional characterization of human mineralocorticoid and glucocorticoid receptor ligands. Eur J Pharmacol. 1993;247(2):145–154. doi: 10.1016/0922-4106(93)90072-h. [DOI] [PubMed] [Google Scholar]
- 22.Peluso JJ, Pappalardo A, Fernandez G, Wu CA. Involvement of an unnamed protein, RDA288, in the mechanism through which progesterone mediates its antiapoptotic action in spontaneously immortalized granulosa cells. Endocrinology. 2004;145(6):3014–3022. doi: 10.1210/en.2004-0067. [DOI] [PubMed] [Google Scholar]
- 23.Peluso JJ, Romak J, Liu X. Progesterone Receptor Membrane Component-1 (PGRMC1) Is the Mediator of Progesterone's Antiapoptotic Action in Spontaneously Immortalized Granulosa Cells As Revealed by PGRMC1 Small Interfering Ribonucleic Acid Treatment and Functional Analysis of PGRMC1 Mutations. Endocrinology. 2008;149(2):534–543. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falkenstein E, Tillmann HC, Christ M, Feuring M, Wehling M. Multiple actions of steroid hormones--a focus on rapid, nongenomic effects. Pharmacol Rev. 2000;52(4):513–556. [PubMed] [Google Scholar]
- 25.Johnson EF, Schwab GE, Dieter HH. Allosteric regulation of the 16 alpha-hydroxylation of progesterone as catalyzed by rabbit microsomal cytochrome P-450 3b. J Biol Chem. 1983;258(5):2785–2788. [PubMed] [Google Scholar]
- 26.Morrill GA, Kostellow AB, Askari A. Progesterone binding to the alpha1-subunit of the Na/K-ATPase on the cell surface: Insights from computational modeling. Steroids. 2008;73(1):27–40. doi: 10.1016/j.steroids.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons TJ, Villa NY, Regalla LM, Kupchak BR, Vagstad A, Eide DJ. Metalloregulation of yeast membrane steroid receptor homologs. Proc Natl Acad Sci U S A. 2004;101(15):5506–5511. doi: 10.1073/pnas.0306324101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee D, Pillai B, Karnani N, Mukhopadhyay G, Prasad R. Genome-wide expression profile of steroid response in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;317(2):406–413. doi: 10.1016/j.bbrc.2004.03.053. [DOI] [PubMed] [Google Scholar]
- 29.McEwan IJ. Bakers yeast rises to the challenge: reconstitution of mammalian steroid receptor signalling in S. cerevisiae. Trends Genet. 2001;17(5):239–243. doi: 10.1016/s0168-9525(01)02273-9. [DOI] [PubMed] [Google Scholar]
- 30.Kupchak BR, Garitaonandia I, Villa NY, Mullen MB, Weaver MG, Regalla LM, et al. Probing the mechanism of FET3 repression by Izh2p overexpression. Biochim Biophys Acta Mol Cell Res. 2007;1773(7):1124–1132. doi: 10.1016/j.bbamcr.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verikouki CH, Hatzoglou CH, Gourgoulianis KI, Molyvdas PA, Kallitsaris A, Messinis IE. Rapid effect of progesterone on transepithelial resistance of human fetal membranes: evidence for non-genomic action. Clin Exp Pharmacol Physiol. 2008;35(2):174–179. doi: 10.1111/j.1440-1681.2007.04803.x. [DOI] [PubMed] [Google Scholar]
- 32.Wu YL, Hooks SB, Harden TK, Dohlman HG. Dominant-negative inhibition of pheromone receptor signaling by a single point mutation in the G protein alpha subunit. J Biol Chem. 2004;279(34):35287–35297. doi: 10.1074/jbc.M404896200. [DOI] [PubMed] [Google Scholar]
- 33.Sengstag C. Using SUC2-HIS4C reporter domain to study topology of membrane proteins in Saccharomyces cerevisiae. Methods Enzymol. 2000;327:175–190. doi: 10.1016/s0076-6879(00)27275-3. [DOI] [PubMed] [Google Scholar]
- 34.Melcher K. A modular set of prokaryotic and eukaryotic expression vectors. Anal Biochem. 2000;277(1):109–120. doi: 10.1006/abio.1999.4383. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132(3):665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen G, Wu C, Schade B, Thomas DY, Whiteway M. Drag&Drop cloning in yeast. Gene. 2005;344:43–51. doi: 10.1016/j.gene.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Kim H, Yan Q, Von Heijne G, Caputo GA, Lennarz WJ. Determination of the membrane topology of Ost4p and its subunit interactions in the oligosaccharyltransferase complex in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100(13):7460–7464. doi: 10.1073/pnas.1332735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126(1):191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 39.Harashima T, Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol Cell. 2002;10(1):163–173. doi: 10.1016/s1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- 40.Feng Y, Davis NG. Feedback phosphorylation of the yeast a-factor receptor requires activation of the downstream signaling pathway from G protein through mitogen-activated protein kinase. Mol Cell Biol. 2000;20(2):563–574. doi: 10.1128/mcb.20.2.563-574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H, Melen K, von Heijne G. Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J Biol Chem. 2003;278(12):10208–10213. doi: 10.1074/jbc.M300163200. [DOI] [PubMed] [Google Scholar]
- 42.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 44.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174(2):247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 45.Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10(6):685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 46.Blackmore PF, Beebe SJ, Danforth DR, Alexander N. Progesterone and 17 alpha-hydroxyprogesterone. Novel stimulators of calcium influx in human sperm. J Biol Chem. 1990;265(3):1376–1380. [PubMed] [Google Scholar]
- 47.Sarkar NN. The potential of mifepristone (RU486) as a female contraceptive drug. Int J Clin Pract. 2002;56(2):140–144. [PubMed] [Google Scholar]
- 48.Baida GE, Kuzmin NP. Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta. 1996;1284(2):122–124. doi: 10.1016/s0005-2736(96)00168-x. [DOI] [PubMed] [Google Scholar]
- 49.Hurley I, Hale ME, Prince VE. Duplication events and the evolution of segmental identity. Evol Dev. 2005;7(6):556–567. doi: 10.1111/j.1525-142X.2005.05059.x. [DOI] [PubMed] [Google Scholar]
- 50.Surratt CK, Adams WR. G protein-coupled receptor structural motifs: relevance to the opioid receptors. Curr Top Med Chem. 2005;5(3):315–324. doi: 10.2174/1568026053544533. [DOI] [PubMed] [Google Scholar]
- 51.Mao C, Xu R, Szulc ZM, Bielawski J, Becker KP, Bielawska A, et al. Cloning and characterization of a mouse endoplasmic reticulum alkaline ceramidase: an enzyme that preferentially regulates metabolism of very long chain ceramides. J Biol Chem. 2003;278(33):31184–31191. doi: 10.1074/jbc.M303875200. [DOI] [PubMed] [Google Scholar]
- 52.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308(5726):1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 53.Deckert CM, Heiker JT, Beck-Sickinger AG. Localization of novel adiponectin receptor constructs. J Recept Signal Transduct Res. 2006;26(5–6):647–657. doi: 10.1080/10799890600920670. [DOI] [PubMed] [Google Scholar]
- 54.Zeller CE, Parnell SC, Dohlman HG. The RACK1 ortholog Asc1 functions as a G-protein beta subunit coupled to glucose responsiveness in yeast. J Biol Chem. 2007;282(34):25168–25176. doi: 10.1074/jbc.M702569200. [DOI] [PubMed] [Google Scholar]
- 55.Phelps C, Gburcik V, Suslova E, Dudek P, Forafonov F, Bot N, et al. Fungi and animals may share a common ancestor to nuclear receptors. Proc Natl Acad Sci U S A. 2006;103(18):7077–7081. doi: 10.1073/pnas.0510080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas P. Characteristics of membrane progestin receptor alpha (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front Neuroendocrinol. 2008 doi: 10.1016/j.yfrne.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nadjafi-Triebsch C, Huell M, Burki D, Rohr UD. Progesterone increase under DHEA-substitution in males. Maturitas. 2003;45(3):231–235. doi: 10.1016/s0378-5122(03)00147-6. [DOI] [PubMed] [Google Scholar]
- 58.Tang Y, Gong F, Lin G, Lu G. Early follicular progesterone concentrations and in vitro fertilization pregnancy outcomes. Fertil Steril. 2007;87(4):991–994. doi: 10.1016/j.fertnstert.2006.08.087. [DOI] [PubMed] [Google Scholar]
- 59.Sippell WG, Muller-Holve W, Dorr HG, Bidlingmaier F, Knorr D. Concentrations of aldosterone, corticosterone, 11-deoxycorticosterone, progesterone, 17-hydroxyprogesterone, 11-deoxycortisol, cortisol, and cortisone determined simultaneously in human amniotic fluid throughout gestation. J Clin Endocrinol Metab. 1981;52(3):385–392. doi: 10.1210/jcem-52-3-385. [DOI] [PubMed] [Google Scholar]
- 60.Kaufman JM, Vermeulen A. The decline of androgen levels in elderly men and its clinical and therapeutic implications. Endocr Rev. 2005;26(6):833–876. doi: 10.1210/er.2004-0013. [DOI] [PubMed] [Google Scholar]
- 61.Milligan G, Kostenis E. Heterotrimeric G-proteins: a short history. Br J Pharmacol. 2006;147 Suppl 1:S46–S55. doi: 10.1038/sj.bjp.0706405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Minic J, Persuy MA, Godel E, Aioun J, Connerton I, Salesse R, et al. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2005;272(2):524–537. doi: 10.1111/j.1742-4658.2004.04494.x. [DOI] [PubMed] [Google Scholar]
- 63.Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98(10):5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis of the PAQR family. For each tree, the length of the tree branches is proportional to the calculated distance between sequences with the scale bar indicating 0.1 substitutions per site. Numbers at the nodes are confidence values that refer to the number times per 1000 trees drawn a particular grouping is made. (A) A bootstrapped phylogenetic tree showing the relationship between PAQR receptors from a variety of eukaryotic and prokaryotic sources. Lines delineate the three Classes of PAQR and grey shading indicates the distinct clades within Class II. The tree is rooted using the sequences in Class III as an outgroup. (B) A bootstrapped phylogenetic tree containing human, yeast and bacterial PAQRs as well as several sequences belonging to two other groups of proteins with a characteristic seven TM structure. One group includes five G-protein coupled receptor (GPCR) sequences and the other group includes five proteins in the alkaline ceramidase (AlkCer) family of enzymes. The tree is rooted with the clade containing the GPCRs. (C) Taxonomic grouping of the organisms from which the sequences in these trees were derived.