Abstract
Oxidation of deoxyribose in DNA produces a variety of electrophilic residues that are capable of reacting with nucleobases to form adducts such as M1dG, the pyrimidopurinone adduct of dG. We now report that deoxyribose oxidation in DNA leads to the formation of oxadiazabicyclo(3.3.0)octaimine adducts of dC and dA. We previously demonstrated that these adducts arise in reactions of nucleosides and DNA with trans-1,4-dioxo-2-butene, the β-elimination product of the 2-phosphoryl-1,4-dioxobutane residue arising from 5′-oxidation of deoxyribose in DNA and with cis-1,4-dioxo-2-butene, a metabolite of furan. Treatment of DNA with enediyne antibiotics capable of oxidizing the 5′-position of deoxyribose (calicheamicin and neocarzinostatin) led to a concentration-dependent formation of oxadiazabicyclo(3.3.0)octaimine adducts of dC and dA, while the antibiotic bleomycin, which is capable of performing only 4-oxidation of deoxyribose, did not give rise to the adducts. The non-specific DNA oxidant, γ-radiation, also produced the adducts that represented ~0.1% of the 2-phosphoryl-1,4-dioxobutane residues formed during the irradiation. These results suggest that the oxadiazabicyclo(3.3.0)octaimine adducts of dC and dA could represent endogenous DNA lesions arising from oxidative stresses that also give rise to other DNA adducts.
INTRODUCTION
The base and deoxyribose moieties of DNA are subject to attack by reactive radical species from a variety of sources, including reactive nitrogen and oxygen species, xenobiotics and ionizing radiation. Relative to direct damage to DNA bases, such as the formation of 8-oxo-dG, the genetic toxicology of oxidation of deoxyribose in DNA has only recently become the focus of significant study. Deoxyribose oxidation results in the formation of oxidized abasic sites, strand breaks terminated with a variety of sugar residues, and freely diffusible degradation products. Many of these products are electrophilic and capable of reacting with local nucleophiles to form both DNA and protein adducts. Examples include the formation of DNA-protein cross-links when repair proteins interact with the deoxyribonolactone abasic site arising from 1′-oxidation of deoxyribose in DNA (1, 2), the formation of M1dG, the pyrimidopurinone adduct of dG, in reactions with base propenals arising from 4′-oxidation of deoxyribose (3–5), and the formation of glyoxal adducts of dG from the 3′-phosphoglycolaldehyde residue that arises from 3′-oxidation of deoxyribose (6). We now extend these observations to include DNA adducts derived from electrophilic products of 5′-oxidation of deoxyribose in DNA (7).
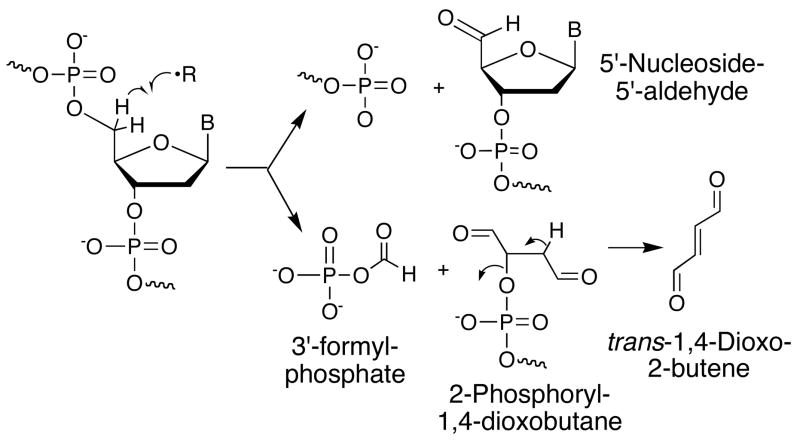
As shown in Scheme 1, the chemistry of 5′-oxidation of deoxyribose in DNA by a variety of DNA-cleaving agents, including enediynes (8) and manganese porphyrins (9), partitions to form strand breaks containing 3′-phosphate and 5′-nucleoside-5′-aldehyde residues, or a 3′-formylphosphate species accompanied by a four-carbon fragment that we and others have identified as a 5′-(2-phosphoryl-1,4-dioxobutane) residue (7, 10). We recently showed that this dialdehydic residue undergoes β-elimination to form trans-1,4-dioxo-2-butene (7), a highly reactiveα,β-unsaturated dicarbonyl species. Further, the cis-isomer of 1,4-dioxo-2-butene is a metabolite of furan (11), an industrial solvent and component of food, smog and cigarette smoke (12, 13) that has been shown to be carcinogenic in rodents (12, 13). Interestingly, analogous species were identified in the γ-radiolysis of thymidine by Dizdaroglu et al. (14). We have demonstrated that both of these electrophilic isomers react with nucleophiles in DNA in vitro to form novel, stable oxadiazabicyclo(3.3.0)octaimine adducts of dC, dG and dA under biologically relevant conditions (15–17) (Scheme 2). The dC and dA adducts were also detected in DNA from bacteria exposed to mutagenic concentrations of cis-1,4-dioxo-2-butene, indicating that these adducts are likely to be mutagenic (18, 19).
Scheme 1.
Generation of trans-1,4-dioxo-2-butene from 5′-oxidation of deoxyribose in DNA.
Scheme 2.
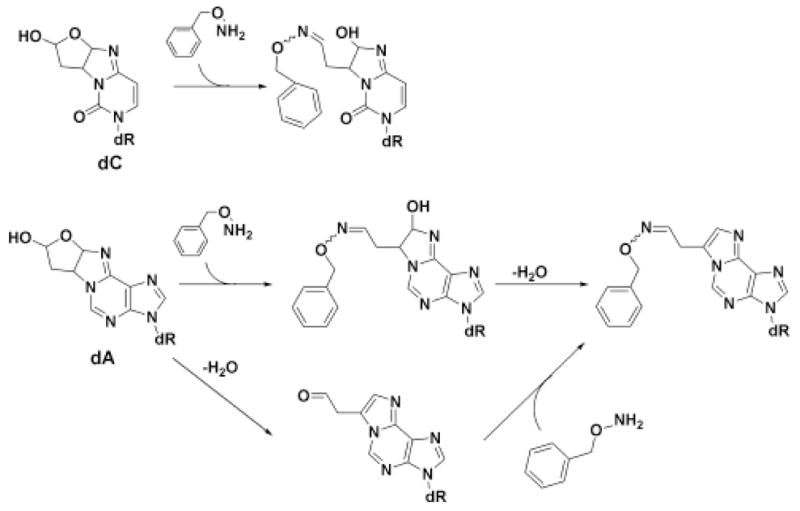
1,4-Dioxo-2-butene-derived adducts of dC and dA, and the formation of the benzylhydroxylamine derivatives.
Using a recently developed LC/MS method (18), we have now observed and quantified the formation of the 1,4-dioxo-2-butene-derived oxadiazabicyclo(3.3.0)octaimine adducts of dC and dA (Scheme 2) in DNA oxidized by agents known to produce 5′-oxidation of deoxyribose in DNA. These results broaden the repertoire of DNA-derived electrophiles that react with DNA to form adducts and further complicate the spectrum of potentially endogenous DNA lesions that contribute to the mutagenic burden of human cells.
EXPERIMENTAL PROCEDURES
Materials
All chemicals and reagents were of highest purity available and were used without further purification unless noted otherwise. Calf thymus DNA was purchased from Sigma Chemical Company (St. Louis, MO) and dialyzed four times against potassium phosphate buffer (50 mM, pH 7.4, Chelex-treated and autoclaved; ref. 20) at 4 °C. Bleomycin was purchased from BioWorld (Dublin, OH). Calicheamicin was obtained from Wyeth Research (Cambridge, MA) and neocarzinostatin was obtained from Kayaku Co., Ltd. (Tokyo, Japan; no longer available from Kayaku, but now available from Sigma Chemical Co., St. Louis, MO). Acid phosphatase, deoxyribonuclease II, and phosphodiesterase II were purchased from MP Biomedical (Irvine, CA). The isotopically labeled standards for the O-benzyloxime derivatives of the dA and dC adducts, [15N3,13C4]-11-(acetaldehyde-O-benzyloxime)-1,N6-etheno-2′-deoxyadenosine and [15N3,13C4]-7-hydroxy-8-(acetaldehyde-O-benzyloxime)-3,N4-ethano-2′-deoxycytidine, respectively, were synthesized as previously described (18).
Treatment of DNA with oxidants
Solutions of calf thymus DNA (0.5 ml, 300 μg) in Chelex-treated potassium phosphate buffer (50 mM, pH 7.4) were aliquoted into silylated glass vials and treated with oxidants under the following conditions. For γ-radiation, solutions of calf thymus DNA were irradiated in a 60Co source for various times (0–256 min) to produce doses up to 300 Gy. For calicheamicin and neocarzinostatin, aliquots (10 μL) of methanolic stock solutions of the drugs were added to calf thymus DNA solutions containing 5 mM glutathione (drug activator) and the reaction allowed to proceed at ambient temperature for 1 h in the dark. For bleomycin, aliquots of a stock of bleomycin freshly mixed with 0.9 molar equivalents of Fe(NH4)2(SO4)2 were added to the calf thymus DNA solutions and the reactions allowed to proceed at ambient temperature for 1 h, followed by Sephadex G25 chromatography to remove the Fe-bleomycin species. In all cases, DNA was precipitated with ethanol (0.3 M sodium acetate, pH 7), resuspended in water and frozen at −80 °C.
Quantification of the oxadiazabicyclo(3.3.0)octaimine adducts of dA and dC
These adducts were quantified using a recently developed LC-MS method (18) in which DNA (0.5 mg) was reacted with O-benzylhydroxylamine (2.9 μmol) in potassium phosphate buffer (44 mM, pH 8) in a total volume of 572 μL at 37 °C for 18 h. DNA was recovered by precipitation with ethanol and washed twice with 70% ethanol. Enzymatic digestion of DNA (0.5 mg/mL) was performed in ammonium acetate buffer (83 mM, pH 5) containing 174 mM magnesium sulfate, 392 mM ammonium chloride, 0.26 mg/mL deoxyribonuclease II, and 0.15 U/mL phosphodiesterase II in a total volume of 1 mL. After 20 min at 37 °C, wheat germ acid phosphatase type I (0.55 μg) and the internal standards (150 fmol each) were added to each sample and they were incubated for an additional 2 h. The samples were then filtered through an acrodisc LC 13 mm syringe filter with a 0.45 μm PVDF membrane (PALL, East Hills, NY). A portion (50 μL) of this solution was reserved for dG analysis. The DNA adducts were isolated by solid phase extraction (Strata-X 30 mg cartridges, Phenomenex, Torrence, CA) with sequential water, 20% methanol, and 100% methanol washes (1 mL each). The methanol wash, which contained the adducts, was concentrated to approximately 30 μL under reduced pressure for analysis by capillary LC-MS (18). Capillary LC-MS analyses were performed with an Agilent capillary LC system linked to a ThermoFinnigan Quantum triple quadrupole mass spectrometer with an electrospray ionization source in positive mode as previously described (18).
The amount of adduct in each sample was determined from the product of the area ratio (unlabeled/labeled) and the amount of internal standard added, as described in detail elsewhere (18). The concentration of DNA in each sample was determined by measuring the concentration of dG in each DNA hydrolysate by HPLC with UV detection (21). Adduct levels were then normalized to the total amount of unmodified dG present in the sample.
RESULTS
To explore the formation of oxadiazabicyclo(3.3.0)octaimine adducts of dA and dC in DNA following 5′-oxidation of deoxyribose in DNA, levels of these adducts were measured in DNA that had been treated with several agents known to oxidize deoxyribose in DNA with different defined chemistries. For example, the diradical forms of calicheamicin and neocarzinostatin cause oxidation of the 4′- and 5′-positions of deoxyribose in double-stranded DNA (8, 22, 23), while bleomycin causes only 4′-oxidation (8). γ-Radiation is known to oxidize all positions in deoxyribose (24), but the spectrum of products differs from the enediynes and bleomycin (3, 8, 25).
Adduct levels were determined by LC-MS/MS using the method recently established in our laboratory (18). The treated DNA was reacted with O-benzylhydroxylamine, which forms a stable oxime with the aldehydic functionality of the adducts (Scheme 2). Under the conditions of the derivatization reaction, the dA adducts undergo dehydration to form a substituted etheno adduct and are detected as 11-(acetaldehyde-O-benzyloxime)-1,N6-etheno-2′-deoxyadenosine.(Scheme 2). The dC adduct does not undergo the final dehydration (18). Enzyme hydrolysates of these samples were then analyzed by capillary electrospray tandem mass spectrometry (LC-ESI/MS/MS) with selected reaction monitoring for the O-benzyloxime derivatives of the adducts. Under the conditions of the derivatization and enzymatic hydrolysis, we only detect the modified dA and dC adducts. The comparable dG adducts are unstable to the derivatization and hydrolysis conditions (18).
Treatment of DNA with neocarzinostatin and calicheamicin led to concentration dependent increases in the 1,4-dioxo-2-butene-derived adducts of both dA and dC in the DNA (Figure 1A and B). Only minor amounts of these adducts were detected in DNA treated with bleomycin, a compound that does not oxidize the 5′-position of the sugar (Figure 1C). Relative to the enediynes, γ-radiation produced relatively modest amounts of the oxadiazabicyclo(3.3.0)octaimine adducts of dC (Figure 1D). In all cases, the levels of the dC adducts were substantially higher than the levels of the dA adduct, a phenomenon similar to that observed in DNA treated with cis-1,4-dioxo-2-butene (18). In addition, the dA adducts may be underestimated if there was not complete conversion of the initial oxime derivative of the oxadiazabicyclo(3.3.0)octaimine adduct of dA to the substituted etheno derivative of this adduct (18) (Scheme 2).
Figure 1.
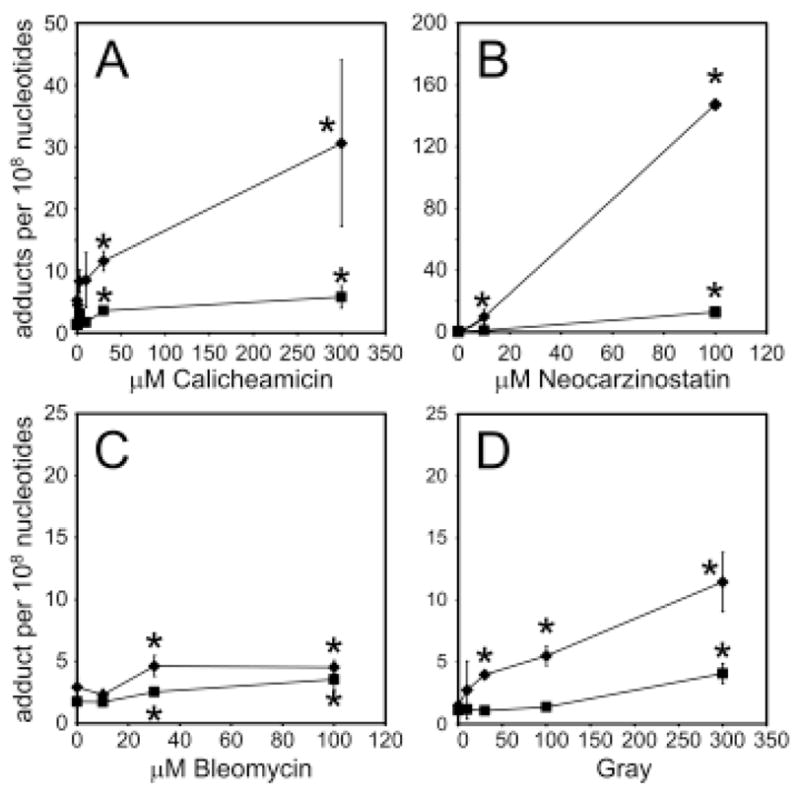
Levels of oxadiazabicyclo(3.3.0)octaimine adducts of dA (square) and dC (diamond) in DNA treated with (A) calicheamicin, (B) neocarzinostatin, (C) bleomycin or (D) γ-radiation. Each data point represents mean ± S.D. for three independent experiments, except for the 100 μM neocarzinostatin data point that represents an average of two samples. *Statistically significant difference from control levels (p < 0.05; Student’s t-test).
DISCUSSION
As we have demonstrated in previous studies, oxidation of deoxyribose in DNA leads to the formation of a variety of reactive electrophiles capable of forming DNA adducts (3–6, 15–17). The results of the present studies extend this phenomenon to include adducts arising from products of 5′-oxidation of deoxyribose in DNA. We demonstrated previously that both cis- and trans-1,4-dioxo-2-butene, the latter a β-elimination product of the 2-phosphoryl-1,4-dioxobutane product of 5′-oxidation of deoxyribose in DNA (Scheme 1) (7), react with dG, dA and dC to form oxadiazabicyclo(3.3.0)octaimine adducts (Scheme 2) (15–17). The present results are consistent with facile formation of these adducts in DNA exposed to a variety of oxidants capable of performing 5′-hydrogen atom abstraction in DNA.
Our conclusion that the formation of the oxadiazabicyclo(3.3.0)octaimine adducts of dA and dC is the result of 5′-oxidation of deoxyribose is supported by the negative results obtained with bleomycin. This DNA-cleaving antibiotic specifically oxidizes the 4′-position of deoxyribose in DNA (8, 26) and thus does not give rise to the 2-phosphoryl-1,4-dioxobutane product of 5′-oxidation. The minor amount of adducts observed with bleomycin may have resulted from DNA oxidation by Fe+2 released from or not chelated by bleomycin. Indeed, the enediynes calicheamicin and neocarzinostatin both cause 5′-oxidation of deoxyribose (7, 8) and both produce the 2-phosphoryl-1,4-dioxobutane precursor of trans-1,4-dioxo-2-butene, so it is not surprising that these agents caused the formation of the oxadiazabicyclo(3.3.0)octaimine adducts of dA and dC (Figure 1A and B). The levels of these adducts are likely to be reduced by the presence of glutathione in the reaction mixtures, since glutathione is an effective trap for 1,4-dioxo-2-butene (27, 28).
The efficiency of formation of the adducts of dA and dC can be estimated from previous studies of the efficiency of formation of the 2-phosphoryl-1,4-dioxobutane residues of 5′-oxidation of deoxyribose. Previous studies revealed that the 2-phosphoryl-1,4-dioxobutane formed to the extent of 6 lesions per 106 nt per Gy of γ-radiation (7). Considering that we observed dC and dA adduct formation in γ-irradiated DNA to the extent of ~8–20 per 109 nt per Gy (Figure 1D), we estimate that about one in every ~1000 2-phosphoryl-1,4-dioxobutane residues formed in the purified DNA leads to 1,4-dioxo-2-butene-derived DNA adducts. This value of 0.1% does not take into account any dG adducts that are likely to form, but we do not expect a dramatically higher yield of these adducts that those of dC (18). The mechanistic basis for this level of conversion of the 2-phosphoryl-1,4-dioxobutane residues to DNA adducts is unclear, but there are several possibilities. For example, sequence context could affect the local nucleobase and sugar residue orientations for the formation of these adducts. It is also possible that there are competing reactions of the 2-phosphoryl-1,4-dioxobutane residue or the eliminated trans-1,4-dioxo-2-butene, such reactions with glutathione or simply diffusion out of the helix. Whether this level of conversion of the 2-phosphoryl-1,4-dioxobutane residue to DNA adducts occurs in cells and tissues remains to be established.
The relatively high proportion of C adducts compared to A adducts produced by all of the oxidants used in the present studies (Figure 1) is similar to results obtained in DNA treated with cis-1,4-dioxo-2-butene (18). However, the level of C adducts obtained with neocarzinostatin is 2- to 4-fold higher than that observed with either calicheamicin or γ-radiation (Figure 1A, D). One possible explanation for the differences between the oxidants is sequence selectivity of the deoxyribose oxidation. Neocarzinostatin produces 5′-oxidation at the T of ACT and GCT sequences of double-stranded DNA lesions (8), unlike calicheamicin and γ-radiation for which the site of deoxyribose oxidation is relatively sequence non-specific (23, 29). Thus, it is possible that formation of the either the 2-phosphoryl-1,4-dioxobutane or trans-1,4-dioxo-2-butene residues adjacent to C leads to higher levels of the oxadiazabicyclo(3.3.0)octaimine adduct of C with neocarzinostatin compared to the other agents. This is similar to the higher levels of M1dG formed at the G of GC and GT sequence motifs in DNA following 4′-oxidation of the C and T by bleomycin (3–5).
In conclusion, we have demonstrated formation of the oxadiazabicyclo(3.3.0)octaimine adducts of dA and dC in DNA oxidized by a variety of agents. The results suggest that these lesions could also represent endogenous DNA lesions arising from oxidative stresses that also give rise to other DNA adducts. Since they can potentially cause DNA-DNA and DNA-protein cross-links (17) and are likely to miscode (18), they may contribute to the overall toxicological effects of oxidative stress.
Acknowledgments
This work was supported by National Institutes of Health grants CA103146 (PD), GM59790 (PD) and ES10577 (LP), and by a Center grant from the NIEHS (ES002109). We thank Brock Matter and Dr. Peter Villalta for their assistance with the mass spectral analysis. The mass spectral analyses were performed in the Analytical Biochemical Core at the Cancer Center, University of Minnesota, which is funded by National Cancer Institute Center Grant CA-77598.
Footnotes
Abbreviations Bp, base pair; dA, 2′-deoxyadenosine; dC, 2′-deoxycytidine; dG, 2′-deoxyguanosine; nt, 2′-deoxynucleotides; LC/MS, liquid chromatography/mass spectrometry.
References
- 1.Hashimoto M, Greenberg MM, Kow YW, Hwang JT, Cunningham RP. The 2-deoxyribonolactone lesion produced in DNA by neocarzinostatin and other damaging agents forms cross-links with the base-excision repair enzyme endonuclease III. J Am Chem Soc. 2001;123:3161–3162. doi: 10.1021/ja003354z. [DOI] [PubMed] [Google Scholar]
- 2.DeMott MS, Beyret E, Wong D, Bales BC, Hwang JT, Greenberg MM, Demple B. Covalent trapping of human DNA polymerase beta by the oxidative DNA lesion 2-deoxyribonolactone. J Biol Chem. 2002;277:7637–7640. doi: 10.1074/jbc.C100577200. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Taghizadeh K, Dedon PC. Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 4.Dedon PC, Plastaras JP, Rouzer CA, Marnett LJ. Indirect mutagenesis by oxidative DNA damage: Formation of the pyrimidopurinone adduct of deoxyguanosine by base propenal. Proc Natl Acad Sci USA. 1998;95:11113–11116. doi: 10.1073/pnas.95.19.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plastaras JP, Dedon PC, Marnett LJ. Effects of DNA structure on oxopropenylation by the endogenous mutagens malondialdehyde and base propenal. Biochemistry. 2002;41:5033–5042. doi: 10.1021/bi0113059. [DOI] [PubMed] [Google Scholar]
- 6.Awada M, Dedon PC. Formation of the 1, N2-glyoxal adduct of deoxyguanosine by phosphoglycolaldehyde, a product of 3′-deoxyribose oxidation in DNA. Chem Res Toxicol. 2001;14:1247–1253. doi: 10.1021/tx0155092. [DOI] [PubMed] [Google Scholar]
- 7.Chen B, Bohnert T, Zhou X, Dedon PC. 5′-(2-Phosphoryl-1,4-dioxobutane) as a product of 5′-oxidation of deoxyribose in DNA: elimination as trans-1,4-dioxo-2-butene and approaches to analysis. Chem Res Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 8.Dedon PC, Goldberg IH. Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 9.Angeloff A, Dubey I, Pratviel G, Bernadou J, Meunier B. Characterization of a 5′-aldehyde terminus resulting from the oxidative attack at C5′ of a 2′-deoxyribose on DNA. Chem Res Toxicol. 2001;14:1413–1420. doi: 10.1021/tx0100800. [DOI] [PubMed] [Google Scholar]
- 10.Kawabata H, Takeshita H, Fujiwara T, Sugiyama H, Matsuura T, Saito I. Chemistry of neocarzinostatin-mediated degradation of d(GCATGC). Mechanism of spontaneous thymine release. Tetrahedron Lett. 1989;30:4263–4266. [Google Scholar]
- 11.Chen LJ, Hecht SS, Peterson LA. Identification of cis-2-butene-1,4-dial as a microsomal metabolite of furan. Chem Res Toxicol. 1995;8:903–906. doi: 10.1021/tx00049a001. [DOI] [PubMed] [Google Scholar]
- 12.Maga JA. Furans in foods. CRC Crit Rev Food Sci Nutr. 1979;11:355–400. doi: 10.1080/10408397909527268. [DOI] [PubMed] [Google Scholar]
- 13.Egle JL, Jr, Gochberg BJ. Respiratory retention and acute toxicity of furan. Am Ind Hyg Assoc J. 1979;40:310–314. doi: 10.1080/15298667991429633. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M, Neuwald K, von Sonntag C. Radiation chemistry of DNA model compounds, IX. Carbohydrate products of _-radioloysis of thymidine in aqueous solution. The radical-induced scission of the N-glycosidic bond. Z Naturforsch. 1976;31b:227–233. [Google Scholar]
- 15.Gingipalli L, Dedon PC. Reaction of cis- and trans-2-butene-1,4-dial with 2′-deoxycytidine to form stable oxadiazabicyclooctaimine adducts. J Am Chem Soc. 2001;123:2664–2665. doi: 10.1021/ja0056421. [DOI] [PubMed] [Google Scholar]
- 16.Bohnert T, Gingipalli L, Dedon PC. Reaction of 2′-deoxyribonucleosides with cis-and trans-1,4-dioxo-2-butene. Biochem Biophys Res Commun. 2004;323:838–844. doi: 10.1016/j.bbrc.2004.08.164. [DOI] [PubMed] [Google Scholar]
- 17.Byrns MC, Predecki DP, Peterson LA. Characterization of nucleoside adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 2002;15:373–379. doi: 10.1021/tx0101402. [DOI] [PubMed] [Google Scholar]
- 18.Byrns MC, Vu CC, Neidigh JW, Abad JL, Jones RA, Peterson LA. Detection of DNA adducts derived from the reactive metabolite of furan, cis-2-butene-1,4-dial. Chem Res Toxicol. 2006;19:414–420. doi: 10.1021/tx050302k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson LA, Naruko KC, Predecki DP. A reactive metabolite of furan, cis-2-butene-1,4-dial, is mutagenic in the Ames assay. Chem Res Toxicol. 2000;13:531–534. doi: 10.1021/tx000065f. [DOI] [PubMed] [Google Scholar]
- 20.LaMarr WA, Sandman KM, Reeve JN, Dedon PC. Differential effects of DNA supercoiling on radical-mediated DNA strand breaks. Chem Res Toxicol. 1997;10:1118–1122. doi: 10.1021/tx970072c. [DOI] [PubMed] [Google Scholar]
- 21.Thomson NM, Mijal RS, Ziegel R, Fleischer NL, Pegg AE, Tretyakova NY, Peterson LA. Development of a quantitative liquid chromatography/electrospray mass spectrometric assay for a mutagenic tobacco specific nitrosamine-derived DNA adduct, O6-[4-Oxo-4-(3-pyridyl)butyl]-2′-deoxyguanosine. Chem Res Toxicol. 2004;17:1600–1606. doi: 10.1021/tx0498298. [DOI] [PubMed] [Google Scholar]
- 22.Dedon PC, Jiang ZW, Goldberg IH. Neocarzinostatin-mediated DNA damage in a model AGT·ACT site: Mechanistic studies of thiol-sensitive partitioning of C4′ DNA damage products. Biochemistry. 1992;31:1917–1927. doi: 10.1021/bi00122a004. [DOI] [PubMed] [Google Scholar]
- 23.Dedon PC, Salzberg AA, Xu J. Exclusive production of bistranded DNA damage by calicheamicin. Biochemistry. 1993;32:3617–3622. doi: 10.1021/bi00065a013. [DOI] [PubMed] [Google Scholar]
- 24.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor & Francis; New York: 1987. [Google Scholar]
- 25.Rashid R, Langfinger D, Wagner R, Schuchmann HP, Von Sonntag C. Bleomycin versus OH-radial-induced malondialdehyde-product formation in DNA. Int J Radiat Biol. 1999;75:101–109. doi: 10.1080/095530099140852. [DOI] [PubMed] [Google Scholar]
- 26.Stubbe J, Kozarich JW. Mechanisms of bleomycin-induced DNA degradation. Chem Rev. 1987;87:1107–1136. [Google Scholar]
- 27.Chen LJ, Hecht SS, Peterson LA. Characterization of amino acid and glutathione adducts of cis-2-butene- 1,4-dial, a reactive metabolite of furan. Chem Res Toxicol. 1997;10:866–874. doi: 10.1021/tx9700174. [DOI] [PubMed] [Google Scholar]
- 28.Peterson LA, Cummings ME, Vu CC, Matter BA. Glutathione trapping to measure microsomal oxidation of furan to cis-2-butene-1,4-dial. Drug Metab Dispos. 2005;33:1453–1458. doi: 10.1124/dmd.105.004432. [DOI] [PubMed] [Google Scholar]
- 29.Hayes JJ, Kam L, Tullius TD. Footprinting protein-DNA complexes with γ-rays. Methods Enz. 1990;186:545–549. doi: 10.1016/0076-6879(90)86148-o. [DOI] [PubMed] [Google Scholar]