Summary
The export of proteins from their site of synthesis in the cytoplasm across the inner membrane is an important aspect of bacterial physiology. Because the location of extracytoplasmic proteins is ideal for host-pathogen interactions, protein export is also important to bacterial virulence. In bacteria there are conserved protein export systems that are responsible for the majority of protein export: the general secretion (Sec) pathway and the twin-arginine translocation (Tat) pathway. In some bacteria, there are also specialized export systems dedicated to exporting specific subsets of proteins. In this review, we discuss a specialized export system that exists in some Gram-positive bacteria and mycobacteria – the accessory Sec system. The common element to the accessory Sec system is an accessory SecA protein called SecA2. Here we present our current understanding of accessory Sec systems in Streptococcus gordonii, Streptococcus parasanguinis, Mycobacterium smegmatis, Mycobacterium tuberculosis, and Listeria monocytogenes, making an effort to highlight apparent similarities and differences between the systems. We also review the data showing that accessory Sec systems can contribute to bacterial virulence.
The conserved general secretion (Sec) pathway
In bacteria, the bulk of protein export across the cytoplasmic membrane is carried out by the general secretion (Sec) pathway (Driessen and Nouwen, 2007; Murphy and Beckwith, 1996). The final destination of Sec exported proteins can be the cell envelope or the extracellular space. Since the Sec pathway exports numerous substrates, many with critical roles in cell physiology, it is not surprising that it is an essential process in all bacteria tested.
The Sec pathway is well-characterized through studies with Escherichia coli and Bacillus subtilis (for comprehensive recent reviews see Driessen and Nouwen, 2007; Papanikou et al., 2007). At the core of the Sec pathway is a membrane-spanning translocase channel composed of the integral membrane proteins SecY, SecE, and SecG (Brundage et al., 1990). Working closely with SecYEG is SecA, a multi-functional protein that participates in nearly every step of the pathway (Economou, 1998). SecA binds to cytoplasmic precursor proteins destined for export and delivers them to the translocase through its ability to bind both membrane phospholipids and the translocase itself. SecA is also an ATPase that provides energy for translocation. Through repeated cycles of ATP binding and hydrolysis, SecA undergoes conformational changes that drive stepwise export of a precursor protein through the core of the Sec translocon and across the membrane (Economou and Wickner, 1994; Hartl et al., 1990). This transport is post-translational and chaperones play an important role in maintaining precursors in a translocation-competent unfolded state.
Sec-exported proteins are synthesized as precursors with a signal sequence at the amino-terminus. Sec signal sequences have a tripartite domain structure: a positively charged N domain, a hydrophobic H domain, and a polar C domain (Driessen and Nouwen, 2007). A signal sequence cleavage site is located at the junction between the C domain and the mature protein. During or immediately after transport across the membrane, the signal sequence is cleaved from the precursor by a signal peptidase (Tokunaga et al., 1984; van Roosmalen et al., 2004; Zwizinski and Wickner, 1980). The resulting mature protein then folds into its final conformation.
Accessory SecAs in bacteria
It was long believed that all bacteria possess a single essential SecA (Economou, 1999). It is now recognized that some, though not all, Gram-positive bacteria and mycobacteria have two SecAs. Some bacteria with extra SecAs are pathogens (e.g. Streptococcus gordonii, S. parasanguinis, S. pneumoniae, Staphylococcus aureus, Mycobacterium tuberculosis, Listeria monocytogenes, and Bacillus anthracis) but accessory SecAs are also found in nonpathogenic bacteria (e.g. Mycobacterium smegmatis, Listeria innocua, and Corynebacterium glutamicum) (Figure 1).
Fig 1. Evolutionary relationships of SecA2 proteins.
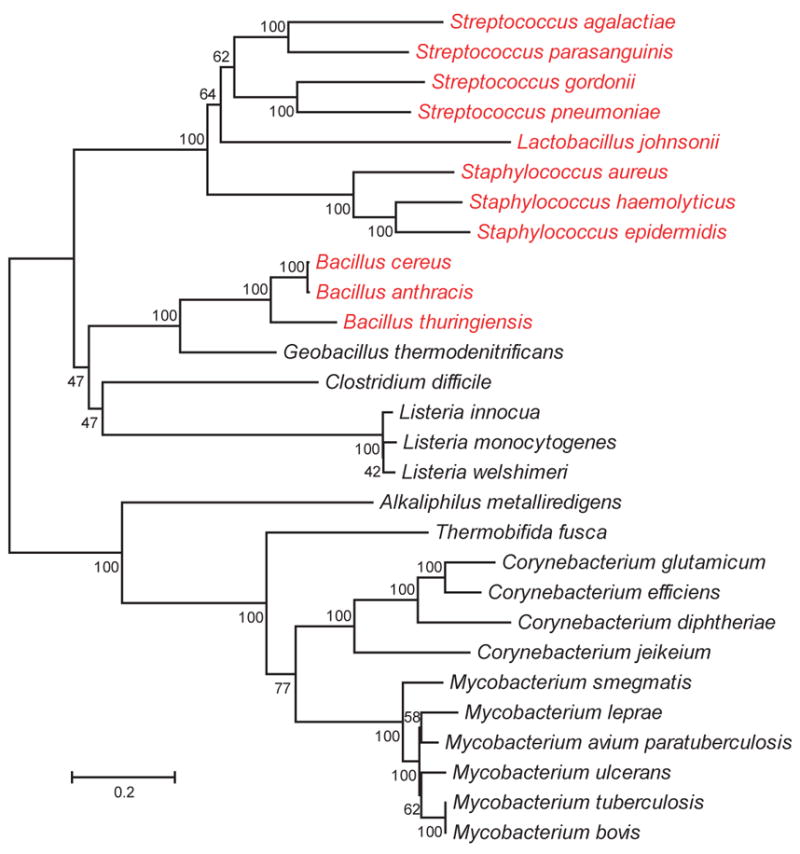
The phylogenetic tree was generated in MEGA4 using the Neighbor-Joining method. The length of the branches reflects the number of amino acid changes between different SecA2s, as indicated by the bar. Bootstrap values are shown at the junctions. SecA2 sequences were obtained by searching all bacterial genome and protein databases available from NCBI and TIGR (as of April 2008) for the term SecA. For organisms with two SecA sequences, the one least like B. subtilis SecA was defined as SecA2 and used to build the tree. For simplicity, only one species of Lactobacillus with a SecA2 is included on the tree. Organisms in red possess two SecY homologs.
In bacteria with two SecA homologues (Table 1), both SecAs are similar across the entire length of the protein to the well-characterized E. coli and B. subtilis SecA. In all cases, the SecA with the slightly higher degree of sequence similarity to the canonical SecA is named SecA or SecA1. The SecA/SecA1 protein is the presumed essential ‘housekeeping’ export factor. The other SecA, with a slightly lower degree of similarity to E. coli SecA, is named SecA2. With one exception (Corynebacterium glutamicum) (Caspers and Freudl, 2008), SecA2 is not essential (Bensing and Sullam, 2002; Braunstein et al., 2001; Braunstein et al., 2003; Chen et al., 2004; Lenz and Portnoy, 2002).
Table 1.
Percent amino acid similarity to B. subtilis SecA
| SecA/SecA1 | SecA2 | |
|---|---|---|
| B. subtilis | 100 | N/A |
| S. gordonii | 74 | 59 |
| S. parasanguinis | 73 | 60 |
| M. tuberculosis | 63 | 52 |
| L. monocytogenes | 81 | 65 |
All SecA2 proteins possess conserved domains of SecA proteins: the DEAD-like ATPase motor domain, which includes ATP-binding Walker A and B boxes, the IRA1 (intramolecular regulator of ATP hydrolysis) domain, and the preprotein binding domain (Karamanou et al., 1999; Kimura et al., 1991; Mitchell and Oliver, 1993). A distinction between SecA2 and the canonical SecA is that SecA2 proteins are smaller in size, due in part to a truncation in the C-terminal linker (CTL) (Rigel and Braunstein unpublished). At least in E. coli, the CTL functions in binding phospholipids, the SecB chaperone, and Zn2+ (Breukink et al., 1995; Dempsey et al., 2004; Fekkes et al., 1997; Matousek and Alexandrescu, 2004).
SecA2/SecY2 systems
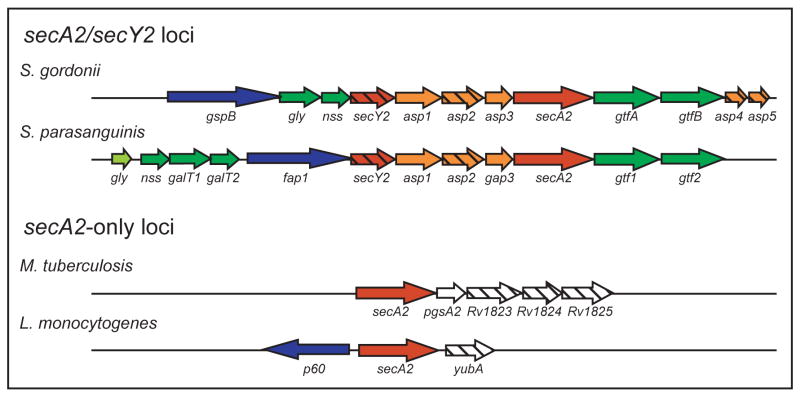
The accessory SecA systems can be divided into two groups: those that include an accessory SecY2 and those that do not (Figure 1). In some bacteria with an accessory SecA2 and SecY2, (e.g. S. gordonii, S. parasanguinis, S. pneumoniae, and S. aureus) the genomic locus contains conserved genes with a similar organization (Figure 2). As discussed later, genes in this locus have roles in export and glycosylation of SecA2-dependent substrates and encode an exported substrate. Some Bacillus sp., most notably B. anthracis, also contain genes predicted to encode an accessory SecA2 and SecY2. However, B. anthracis does not exhibit similar genomic organization to the SecA2/SecY2 systems studied in Streptococcus sp.
Fig 2. Organization of accessory sec loci.
Similar colouring indicates encoded proteins with homology or similar properties. Genes encoding SecA2-dependent substrates are in blue, glycosyltransferases in green, accessory Sec proteins in red, accessory SecA2-dependent secretion factors in orange. Any gene that does not fit in the above categories is white. Genes encoding proteins with predicted transmembrane domains are hatched.
SecA2-only systems
For bacteria with a SecA2 but lacking SecY2, the genomic region is not conserved (Figure 2). In mycobacteria, the gene immediately downstream of secA2 is pgsA2, which encodes a phosphatidylglycerolphosphate synthase homologue predicted to function in production of acidic phospholipids, which have multiple cellular functions including a role in Sec export (de Vrije et al., 1988; Jackson et al., 2000). Three genes further downstream encode small putative membrane proteins of unknown function. In the SecA2 system of L. monocytogenes, secA2 is adjacent to a gene encoding the p60 protein. These two genes appear to be divergently transcribed. The p60 protein is exported by the L. monocytogenes SecA2 system (Lenz and Portnoy, 2002). Downstream of secA2 is yubA, which encodes a predicted membrane protein with no informative homology.
Evolution of accessory SecA systems in bacteria
For the SecA2/SecY2 systems of Streptococcus sp. and Staphylococcus sp., the high level of genomic similarity in the locus suggests a common source. However, not all Streptococcus sp. and Staphylococcus sp. contain an accessory sec locus. This could be indicative of recent horizontal gene transfer. In M. tuberculosis and L. monocytogenes, which have the SecA2-only systems, the genomic region is not conserved. We propose that these SecA2-only systems are not directly related to each other or to the SecA2/SecY2 systems, and they evolved independently.
The basic questions of how the SecA2 systems operate are just beginning to be asked. What proteins interact and work with SecA2 in export? What proteins are exported by SecA2 and what features define them? What roles do accessory Sec systems play in virulence? Below, we summarize our current understanding of accessory Sec systems of bacteria.
SecA2/SecY2 systems
S. gordonii and S. parasanguinis
The SecA2/SecY2 systems of the oral pathogens S. gordonii and S. parasanguinis have many similarities. In S. gordonii strain M99, the SecA2 system is responsible for exporting GspB, a large serine-rich glycoprotein to the cell surface (Bensing and Sullam, 2002). Exported GspB promotes S. gordonii binding to platelets. Therefore, the SecA2 system is likely to contribute to pathogenesis, as platelet binding is believed to be important for S. gordonii attachment to damaged cardiac tissue and subsequent infective endocarditis. In S. parasanguinis, the SecA2 system exports a similarly large serine-rich glycoprotein Fap1 (Chen et al., 2004). Adherence of S. parsanguinis to saliva-coated hydroxylapatite, a model for the tooth surface, requires long fimbriae built from Fap1 subunits (Wu et al., 1998). Because export of Fap1 is required for optimal adherence of S. parasanguinis, the accessory Sec pathway in this bacterium is believed to be important to dental plaque formation, which is linked to caries and periodontal disease.
1) Discovery of the accessory SecAs
In both S. gordonii and S. parasanguinis, the accessory sec locus was initially discovered in genetic screens. In S. gordonii, a deletion of the putative operon that includes secA2 and secY2 was identified in a mutant defective in platelet binding (Bensing and Sullam, 2002). It was hypothesized that the mutant phenotype reflects a failure to export a platelet binding protein. In an elegant experimental approach using immune serum enriched for antibodies to SecA2-dependent proteins, GspB was identified as a protein exported to the cell wall and released into culture media in a SecA2-dependent manner (Bensing and Sullam, 2002). Later, GspB was recognized as being encoded by a gene in the accessory sec locus. GspB is a large (286 kDa) heavily glycosylated protein (Bensing et al., 2004b).
In S. parasanguinis, transposon insertions at the 3′ end of secA2 were identified in a screen for mutants that fail to glycosylate Fap1 (Chen et al., 2004). It was later shown that SecA2 is required for export of Fap1 to the cell wall and for its release into culture media. In addition to these extracytoplasmic locations, some Fap1 is also detected in cytosolic and membrane fractions of wild-type cells (Chen et al., 2004). Like GspB, the large (263 kDa) glycosylated Fap1 protein is also encoded in the accessory sec locus.
2) Proteins that function in export and glycosylation of SecA2-dependent proteins
From mutation analysis of the genes in the accessory sec loci of S. gordonii and S. parasanguinis (Figure 2), genes can be placed in one of two groups: 1) genes required for export and 2) genes required for glycosylation. As detailed below, the current data reveal many similarities but also some differences between the S. gordonii and S. parasanguinis systems.
Export Factors
Disruption of secA2 or secY2 individually in S. gordonii prevents GspB export to the cell wall and release into culture media (Bensing and Sullam, 2002). In the absence of export, GspB protein accumulates on the cytosolic face of the membrane where it is protected from attack by externally added protease (Bensing and Sullam, 2002; Takamatsu et al., 2004a). Similarly, Fap1 is not exported in a secA2 deletion mutant of S. parasanguinis and non-exported Fap1 accumulates primarily in the membrane fraction (Chen et al., 2004). One of the differences between the SecA2/SecY2 systems is that in S. parasanguinis a secY2 mutation does not eliminate Fap1 export (Wu et al., 2007a).
In S. gordonii, the three open reading frames that separate secY2 and secA2 are named asp1–3 (for accessory secretory protein) (Figure 2). Mutations in these asp genes result in a GspB export defect. Asp2 contains a predicted transmembrane domain and could function as part of a channel in the cytoplasmic membrane. Asp1 and Asp3 do not have signal sequences or transmembrane domains, suggesting intracellular functions that are not readily predicted from the protein sequence. Two additional genes in the secA2/secY2 locus, asp4 and asp5, are required for GspB export (Takamatsu et al., 2005). Asp4 and Asp5 share sequence homology with B. subtilis SecE (52% similar) and SecG (55% similar). Thus, Asp4 and Asp5 might function as components of a membrane translocase as is the case for SecE and SecG, making the accessory Sec system completely independent of the canonical Sec system components. This possibility is worthy of further investigation.
A different result is obtained with an asp3 mutation in S. parasanguinis (Peng et al., 2008; Wu et al., 2007b). Like a secY2 S. parasanguinis mutant, an asp3 (renamed gap3 for glycosylation associated protein 3) mutant still exports Fap1, but in both of these mutants the protein runs at a higher apparent molecular weight in an SDS-PAGE gel and is not correctly glycosylated (Peng et al., 2008; Wu et al., 2007b). It is highly unlikely that SecY2 and Gap3 are directly involved in Fap1 glycosylation. Rather, SecY2 and Gap3 may be indirectly involved. For example, full glycosylation of Fap1 might be coupled to its normal route of export through a SecY2-containing translocase.
Glycosylation factors
Two genes downstream of secA2, gtfA, and gtfB (for glycosyltransferase) in S. gordonii or gtf1 and gtf2 in S. parasanguinis function in GspB and Fap1 glycosylation, respectively. In gtf mutants, glycosylation of a GST-GspB fusion protein or Fap1 protein does not occur (Bu et al., 2008; Takamatsu et al., 2004b; Wu et al., 2007a). Further, glycosylation of these proteins can be achieved when they are co-expressed in E. coli with GtfA and GtfB or Gtf1 and Gtf2 (Bu et al., 2008; Takamatsu et al., 2004b). For GspB, glycosylation is shown to influence protein stability and/or solubilization (Bensing et al., 2005; Takamatsu et al., 2004b).
Also found in the accessory sec loci of both S. gordonii and S. parasanguinis are additional genes encoding proteins that share amino acid homology with glycosyltransferases and nucleotide sugar synthetases. Examples in S. gordonii are the gly and nss genes. Disruption of either of these genes does not prevent glycosylation or export of GspB, but it does alter the lectin binding properties and carbohydrate composition of GspB (Takamatsu et al., 2004a). In S. parasanguinis, there are gly, galT1, galT2, and nss genes. Of these genes, galT2, which does not have a direct homologue in S. gordonii, is directly tested. A S. parasanguinis galT2 mutant exhibits a partial defect in Fap1 glycosylation (Wu et al., 2007b). Thus, it seems these additional glycosylation genes function in defining the carbohydrate composition of the glycan on GspB and Fap1.
Interestingly, glycosylation of GspB and Fap1 seems to occur prior to export. When GspB is not exported in a secA2 or secY2 mutant or when the signal sequence of GspB is removed, the protein retained in the cytosol is still glycosylated (Bensing et al., 2004a). Fap1 protein detected in the cytosol also appears to be glycosylated, as shown with antibodies specific for a Fap1 glycan (Chen et al., 2004). In eukaryotes, protein glycosylation occurs only when proteins are exported into the endoplasmic reticulum (Chavan and Lennarz, 2006). In M. tuberculosis and Campylobacter jejuni, where glycosylation has been studied, glycosylation is similarly linked to export across the cytoplasmic membrane (Nita-Lazar et al., 2005; VanderVen et al., 2005). Glycosylation of GspB and Fap1 in the cytosol and in the absence of export is unusual and suggests that the SecA2 system might be adapted to exporting proteins that are post-translationally modified.
3) Features of SecA2-dependent proteins GspB and Fap1 that target them for export
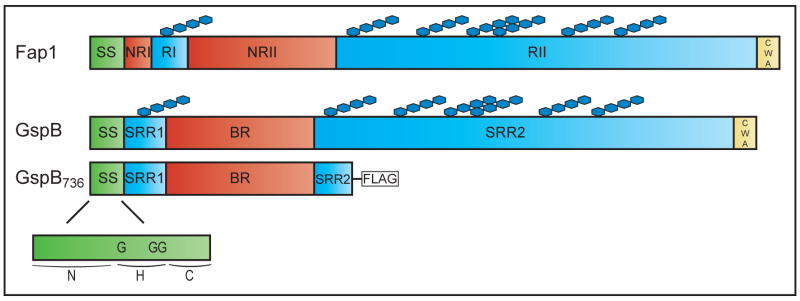
GspB and Fap1 are comprised of similar multi-domain structures (Figure 3) (Chen et al., 2007; Takamatsu et al., 2004b). Both GspB and Fap1 have an abnormally long amino-terminal signal sequence (90 and 68 amino acids, respectively). The signal sequence is followed by short and long serine rich regions that are the sites of glycosylation. In GspB, the serine rich domains are named SRR1 and SRR2 and in Fap1, they are named RI and RII. In GspB, the SRR domains are separated by a region rich in basic residues (BR). In Fap1, there are two interspersed non-repetitive regions named NRI and NRII. At the C-terminus of both GspB and Fap1 is a cell wall anchoring (CWA) domain containing a LPXTG motif that is required for attachment to the cell wall. The CWA domain is consistent with the cell wall localization of these proteins. Consequently, the GspB and Fap1 proteins additionally seen released into culture media may be a result of cell wall remodelling.
Fig. 3. Domain organization of SecA2-dependent serine rich glycoproteins.
Domains in full-length Fap1 and GspB are shown, with similar domains colour coded. Truncated GspB736-FLAG is also depicted, and the GspB signal sequence is shown, including the location of critical glycine (G3) residues.
Native GspB and Fap1 are not in the cell wall or released into culture media by a secA2 mutant; rather, they accumulate in the cytosol (Bensing and Sullam, 2002). This leads to two questions. What features of SecA2-exported proteins target them for export by the SecA2 pathway and what features prevent them from being exported by the canonical Sec pathway? In short, it appears that glycosylation in the mature domain and elements of the signal sequence are both important for determining export by SecA2 and preventing export by the canonical Sec pathway.
The effect of glycosylation
Glycosylation is not required for GspB export by secA2/secY2, but it does have a role in blocking GspB export by the canonical Sec pathway. Since full-length non-glycosylated GspB forms aggregates or is unstable, a truncated version of GspB known as GspB736-FLAG is used in many of the experiments that address this phenomenon (Figure 3). GspB736-FLAG is exported even in a gtfA mutant, which fails to glycosylate GspB (Bensing et al., 2005). Further, unglycosylated GspB can be exported by the SecA2 pathway as shown by reduced GspB736-FLAG export in a gtfA secA2 double mutant. However, the level of GspB736-FLAG export by gtfA secA2 double mutant is actually higher than that in the single secA2 mutant, which is totally deficient. Treatment with azide, a well-established inhibitor of E. coli SecA, diminishes the residual export of GspB736-FLAG in the double mutant. This argues that the canonical SecA is also able to export GspB in the absence of glycosylation. Because the canonical Sec pathway does not have an obvious role in exporting glycosylated GspB, this implies that glycosylation of GspB blocks recognition by the canonical pathway. One possibility is that the size or shape of glycosylated GspB is incompatible with the canonical SecYEG translocon. It also suggests that the accessory Sec pathway is adapted to export the modified protein.
With Fap1, a similar finding is that the glycosylated R1 and RII regions of the protein prevent its export by the canonical SecA pathway. Simultaneous deletion of RI and RII, or RII alone, enables Fap1 export in the absence of SecA2 (Chen et al., 2007). This export is sensitive to azide, implying these Fap1 deletion variants are exported by the canonical SecA. Thus, it seems that glycosylated regions of GspB and Fap1 have a similar effect of inhibiting export via the canonical SecA.
The signal sequence
The signal sequences of GspB and Fap1 are atypical. All elements of a standard Sec signal sequence are present, but the positively charged N domain is longer than normal. For GspB, deletions in the signal sequence eliminate export (Bensing et al., 2005; Bensing et al., 2007). In addition, N-terminal sequencing of exported GspB shows the signal sequence is cleaved at a site between residues 90 and 91. Thus, the signal sequence appears functional.
The signal sequence is a good candidate for specifying SecA2-dependent export. GspB variants with alterations in the signal sequence reveal an important role for three glycines in the hydrophobic (H) domain in directing GspB export (Bensing et al., 2007). Substitution of these three glycines (G3) with α-helix promoting amino acids in full-length glycosylated GspB completely blocks export in wild-type S. gordonii. This indicates that these glycines are essential to SecA2-dependent export.
When the same (G3) substitution is introduced into a GspB736-FLAG protein, a slightly different result is observed. Export of GspB736 (G3)-FLAG, which is mostly unglycosylated, is only slightly reduced in a wild-type strain. When expressed in a secA2 mutant, the level of GspB736 (G3)-FLAG is further reduced but some protein is still exported. Results of azide treatment of the secA2 mutant expressing GspB736 (G3)-FLAG suggest the residual export seen is by the canonical Sec pathway (Bensing et al., 2007). The different results obtained with (G3) mutations in native GspB and the GspB736-FLAG protein are likely due to the added influence of glycosylation preventing canonical Sec export. Full-length GspB is larger and more glycosylated than the truncated GspB736-FLAG (Figure 3). Interestingly, when tested in the absence of glycosylation and absence of SecA2 (in a gtfA secA2 double mutant) the GspB736 (G3)-FLAG is more efficiently exported than the non-mutated GspB736-FLAG. These results indicate that, in addition to being required for SecA2-dependent export, the (G3) residues in the GspB signal sequence function with glycosylation of the mature domain of GspB to inhibit export by the canonical SecA.
In light of the above results, it seemed possible that the GspB signal sequence alone might be sufficient to direct a protein to the accessory Sec pathway. On the contrary, the GspB signal sequence is unable to drive export of GspA, a heterologous surface protein with a consensus signal sequence (Bensing et al., 2005). This indicates that, in addition to the signal sequence, specific features of the mature protein are also required for compatibility with the accessory Sec pathway.
The Fap1 signal sequence was also tested for its ability to drive export of a heterologous protein. In this case, the Fap1 signal sequence is able to promote some export of the green fluorescent protein (GFP), although it was not determined if the exported GFP is functional (Chen et al., 2007). However, the export is independent of SecA2. This provides further indication that elements of the mature protein are important for defining SecA2-specificity. For Fap1 it is suggested that the signal sequence, NRI, and NRII are the minimal elements required for export by the SecA2-pathway (Chen et al., 2007).
4) Other SecA2-dependent proteins
In S. parasanguinis, the FimA adhesin is another protein reported to be exported in a SecA2-dependent manner (Chen et al., 2004). There are many differences between Fap1 and FimA. FimA has a classical signal sequence of normal length, it does not share homology with Fap1, and is not likely to be glycosylated. FimA is a predicted lipoprotein with a lipobox motif (Fenno et al., 1995). Unlike Fap1, FimA is still exported, though to a reduced extent, in a secA2 mutant (Chen et al., 2004). The residual export is presumably through the action of the canonical SecA. Thus, it seems that FimA differs from GspB and Fap1 in not being exclusively dependent on SecA2. Rather, it would appear that FimA is compatible with both the accessory and canonical Sec pathways. However, there has only been minimal study of FimA and additional work is needed to better define the relationship between FimA and SecA2.
SecA2-only systems
Mycobacterium smegmatis and Mycobacterium tuberculosis
M. smegmatis is a fast-growing non-pathogenic mycobacterium that is commonly used as a model to study the general physiology of mycobacteria. M. tuberculosis is a slow-growing intracellular pathogen responsible for tuberculosis disease. Mycobacteria are sometimes classified as Gram-positive bacteria; however, mycobacteria are distinguished by acid-fast staining, a property imparted by the highly impermeable mycobacterial cell wall (Goren et al., 1978). The SecA2-exported proteins identified in M. smegmatis and M. tuberculosis include examples with and examples without signal sequences. While nothing is yet known about factors that work with mycobacterial SecA2 to recognize or translocate substrates, it is clear that the SecA2 system of M. tuberculosis is important to virulence and protective immunity in the host.
1) Discovery of the accessory SecA
The accessory sec locus of mycobacteria was discovered during evaluation of the M. tuberculosis genome sequence (Braunstein et al., 2001). The presence of two secAs appears to be a property shared by all Mycobacterium sp. including non-pathogenic M. smegmatis.
In mycobacteria, there is substantial evidence that SecA1 is the ‘housekeeping’ SecA that exports the majority of proteins. Like E. coli SecA, mycobacterial SecA1 is essential, as shown by the inability to delete the M. smegmatis secA1 gene unless secA1 is expressed from a plasmid (Braunstein et al., 2001). Furthermore, SecA1 depletion in M. smegmatis eliminates export of the Sec signal sequence-containing M. smegmatis porin (MspA) and is associated with loss of viability (Guo et al., 2007). secA1 also appears to be essential in M. tuberculosis (Sassetti et al., 2003). In contrast, the secA2 gene is not essential and secA2 mutants have been constructed in M. smegmatis, M. tuberculosis, and M. bovis BCG (Braunstein et al., 2001; Braunstein et al., 2003; Braunstein, unpublished). As described below, mycobacterial SecA2 functions in the export of a subset of proteins.
Although both mycobacterial SecAs function in protein export, the two SecAs are not functionally redundant or interchangeable. SecA2 cannot fulfil the essential role of SecA1 since, even when secA2 is overexpressed, the chromosomal copy of secA1 cannot be deleted (Braunstein et al., 2001). The converse is also true; SecA1 cannot fulfil the role of SecA2, since overexpression of secA1 does not rescue in vitro phenotypes of the M. smegmatis secA2 mutant. (Braunstein et al., 2001; Rigel and Braunstein, unpublished).
2) SecA2-exported proteins of M. smegmatis: Msmeg1704 and Msmeg1712 lipoproteins
By comparing cell envelope fractions of M. smegmatis using 2D-PAGE, two M. smegmatis SecA2 dependent proteins were identified as Msmeg1704 and Msmeg1712 (YtfQ) (Gibbons et al., 2007). In the absence of SecA2, these proteins are not exported to the cell wall and they accumulate in cytosolic and membrane fractions as slightly larger species that are presumed to be unprocessed precursors. Of note is that the export defect in the secA2 mutant is not complete (Gibbons et al., 2007).
Msmeg1704 and Msmeg1712 are encoded by genes in an apparent operon. Both proteins are homologous to periplasmic sugar binding proteins and possess amino-terminal signal sequences with a lipobox motif. Both proteins appear to be genuine lipoproteins, as shown by Triton X-114 extraction, a fractionation method that enriches for lipoproteins, and sensitivity to globomycin, an inhibitor of the lipoprotein signal peptidase LspA (Gibbons et al., 2007). The role of SecA2 in the export of these two lipoproteins is specific; other mycobacterial lipoproteins are not affected. There are no direct homologues of Msmeg1704 and Msmeg1712 in M. tuberculosis. However, when expressed in M. tuberculosis, Msmeg1704 is also influenced by SecA2. This indicates some degree of conservation of the accessory Sec systems of these mycobacteria.
It is interesting that both of these SecA2-dependent proteins are lipoproteins. Because glycosylation plays a role in defining SecA2 substrates of other systems, specific post-translational lipid modification of Msmeg1704 and Msmeg1712 might make them dependent on SecA2 for export and/or block effective export by the canonical SecA1.
3) SecA2-exported proteins of M. tuberculosis: SodA
A comparative proteomic analysis of M. tuberculosis secreted proteins also identified SecA2-dependent proteins (Braunstein et al., 2003). Only a small number of secreted proteins exhibit differences between wild-type and secA2 mutant M. tuberculosis on 2D-PAGE. Three protein spots were less abundant in culture filtrates of a M. tuberculosis secA2 mutant: SodA (Iron superoxide dismutase), Acr (Alpha-crystallin, HspX), and Rv0390 (protein of unknown function). All of these proteins lack signal sequences.
Of these proteins, only SodA was studied further. In the absence of SecA2, the amount of SodA protein and superoxide dismutase activity secreted into culture media by M. tuberculosis is reduced, as measured by immunoblot analysis and enzymatic assays (Braunstein et al., 2003; Hinchey et al., 2007). Furthermore, in the secA2 mutant SodA is observed to accumulate in the cell pellet. Secreted antioxidants like SodA could protect intracellular M. tuberculosis from the oxidative burst of macrophages and be important to virulence. Catalase-peroxidase (KatG) is another antioxidant detected in culture filtrates of M. tuberculosis. Immunoblot analysis with anti-KatG antibodies show its secretion is also dependent on SecA2 (Braunstein et al., 2003). Like SodA, KatG lacks a signal sequence.
These studies in M. smegmatis and M. tuberculosis show that SecA2 functions in the export of a subset of proteins. It remains to be clarified how proteins with and proteins without signal sequences can both be exported in a SecA2-dependent manner by mycobacteria. There might be dual roles for SecA2: one in the export of signal sequence-containing proteins and another in the export of substrates lacking signal sequences. Alternatively, the role of SecA2 in exporting proteins like SodA might be indirect. There may be unidentified signal sequence-containing proteins exported by SecA2, that are themselves components of a specialized export machinery, responsible for secreting unconventional proteins such as SodA. This latter possibility is worth serious consideration. There are examples of exported lipoproteins synthesized with signal sequences (i.e. YscJ) that function as components of specialized Sec-independent Type III secretion systems of didermic bacterial pathogens (Silva-Herzog et al., 2008).
4) Role of SecA2 in virulence and immunity
Evaluation of the M. tuberculosis secA2 mutant in models of tuberculosis clearly demonstrates a role for SecA2 in virulence. In mice, the secA2 mutant exhibits a growth defect early in infection and mice infected with the mutant survive longer than wild-type infected animals (Braunstein et al., 2003; Kurtz et al., 2006). The secA2 mutant is also defective in its ability to grow in cultured macrophages (Kurtz et al., 2006). Because the secA2 mutant is defective in the export of the antioxidants SodA and KatG, the role of SecA2 in macrophages could be to protect against the oxidative burst. However, there must be additional roles for SecA2-dependent export in M. tuberculosis because the secA2 mutant is still defective for growth in macrophages that do not generate an oxidative burst. Macrophages infected with the secA2 mutant can exhibit more apoptosis than macrophages infected with wild-type M. tuberculosis, a phenotype that appears to reflect the SodA defect of the secA2 mutant (Hinchey et al., 2007). However, this effect on macrophage apoptosis does not account for the mutant phenotype in macrophages (Kurtz, McCann, and Braunstein unpublished). Macrophages infected with the secA2 mutant also release more proinflammatory cytokines, so another role for SecA2-dependent export might be to limit the host cell response (Kurtz et al., 2006).
Finally, the secA2 mutant of M. tuberculosis elicits better protective immunity in mice and guinea pigs to M. tuberculosis challenge than the current live attenuated M. bovis BCG vaccine (Hinchey et al., 2007). The increased protection is attributed to generation of an enhanced CD8+ T cell response in animals infected with the secA2 mutant. This could reflect the increased apoptosis phenotype mentioned above. These results reveal a role for M. tuberculosis SecA2 in evading protective immunity.
Listeria monocytogenes
L. monocytogenes is a Gram-positive intracellular pathogen responsible for the food-borne illness listeriosis. A larger collection of proteins is reported as exported by SecA2 of L. monocytogenes than in the above cases. This list includes examples of proteins with and without signal sequences. Like M. tuberculosis, SecA2 of L. monocytogenes is important to virulence and protective immunity.
1) Discovery of the accessory SecA
The accessory sec locus of L. monocytogenes was discovered during the study of mutants with rough colony morphologies (Lenz and Portnoy, 2002). In one such mutant, a transposon is inserted in secA2 while other rough mutants have point mutations in secA2. Complementation experiments demonstrate that mutations in secA2 cause the rough colony phenotype (Lenz and Portnoy, 2002; Machata et al., 2005).
2) SecA2-dependent proteins of L. monocytogenes
Comparison of surface and secreted proteins of wild-type and secA2 mutant L. monocytogenes by 1D gel analysis reveals that 8 out of 25 secreted proteins and 19 of 49 surface extractable proteins are either completely absent or significantly reduced in the fractions prepared from the secA2 mutant (Lenz and Portnoy, 2002; Lenz et al., 2003). Of these proteins, 17 have been identified (Lenz et al., 2003). An additional SecA2-dependent secreted protein was identified in a separate study (Archambaud et al., 2006). As is the case with mycobacteria, some of the SecA2-dependent proteins have signal sequences and some do not.
Proteins with signal sequences
The list of SecA2-dependent proteins with signal sequences includes the p60 autolysin, two predicted lipoproteins, two predicted transporters, and a penicillin binding protein (Lenz et al., 2003). Only p60 was studied further. The p60 protein (also known as CwhA and Iap) is a major secreted protein of L. monocytogenes. It is a cell wall hydrolase with bacteriolytic activity that can cleave peptidoglycan (Lenz et al., 2003; Machata et al., 2005; Wuenscher et al., 1993). p60 is abundant in cell wall and secreted protein fractions of wild-type L. monocytogenes (Lenz and Portnoy, 2002; Machata et al., 2005). Mutants lacking secA2 produce p60 but exhibit a 50% reduction in its secretion (Lenz et al., 2003; Machata et al., 2005). Thus, p60 is a protein that can be exported in a SecA2-dependent or SecA2-independent manner. In the secA2 mutant, p60 accumulates in membrane and cytosolic fractions (Machata et al., 2005). The gene encoding p60 is located upstream of secA2 in a divergent transcriptional unit.
Proteins without signal sequences
Like p60, MurA/NamA (Muramidase) is a cell wall hydrolase able to cleave peptidoglycan (Carroll et al., 2003). Based on our sequence analysis, MurA does not possess a canonical signal sequence. In a secA2 mutant MurA is not detected in surface-extractable or cell wall fractions (Lenz et al., 2003; Machata et al., 2005). It is striking that similar peptidoglycan hydrolases are SecA2-dependent. However, in a secA2 mutant MurA is not only absent from the cell wall but is also absent in all subcellular fractions (Machata et al., 2005). While this could reflect intracellular degradation of MurA when it is not being exported, other possibilities such as altered expression in a secA2 mutant are not yet ruled out.
Like SodA of M. tuberculosis, the manganese superoxide dismutase (MnSOD) of L. monocytogenes lacks a signal sequence and depends on SecA2 for export (Archambaud et al., 2006). Proteins like superoxide dismutase that lack signal sequences and have well-described cytosolic functions are not commonly thought of as being exported, but there are examples of such unconventional exported proteins in bacteria (Pancholi and Fischetti, 1992, 1998). Ten additional SecA2-dependent surface proteins lacking signal sequences and with predicted cytosolic functions are identified by 1D gel analysis and peptide mass fingerprinting of L. monocytogenes (Lenz et al., 2003).
For these examples, it is important to recognize that additional investigation is needed to determine if SecA2 is involved directly or indirectly in the export of these proteins or if the effect of SecA2 on these proteins is unrelated to export. As a starting point, experiments to demonstrate intracellular accumulation of unexported protein in a secA2 mutant should be conducted.
3) Role of SecA2 in virulence and in eliciting an immune response
SecA2 of L. monocytogenes clearly plays a role in virulence. The LD50 of a secA2 mutant is higher than that of the wild-type strain (Lenz and Portnoy, 2002) and the secA2 mutant is cleared from mice faster than wild-type bacteria (Lenz et al., 2003; Muraille et al., 2007).
Secretion of p60 might be one way SecA2 contributes to virulence, since a p60 mutant is also attenuated in mice (Lenz et al., 2003). The secA2 mutant is defective in cell to cell spread (Lenz et al., 2003) and, in at least one study, p60 appears to promote cell-cell spread (Pilgrim et al., 2003). Another possible explanation is that the muramyl peptides released by the p60 hydrolase limit protective host immune responses and thereby promote virulence (Lenz 2003). Because the attenuated phenotype of a p60 mutant is not as dramatic as that of a secA2 mutant, additional proteins must be involved. MurA and MnSOD are other SecA2-dependent proteins that could act in virulence (Lenz et al., 2003; Muraille et al., 2007).
On the other end of the host pathogen spectrum, SecA2 promotes production of protective immunity to L. monocytogenes challenge. Unlike wild-type L. monocytogenes, the secA2 mutant fails to produce protective CD8+ T cells (Muraille et al., 2007). It is striking that opposite effects of SecA2 on the establishment of protective immunity are seen with L. monocytogenes and M. tuberculosis. In M. tuberculosis, SecA2 limits priming of antigen specific CD8+ T cells and protection in vivo, while in L. monocytogenes SecA2 promotes these effects.
Conclusion and future investigations
We now know of bacteria that possess accessory Sec systems dedicated to exporting a subset of proteins. In pathogens, the accessory Sec systems are linked to virulence. However, our understanding of SecA2-mediated export and how it compares among different bacteria is incomplete and many basic questions need to be answered.
The biochemical activities of SecA2 await investigation. SecA2 might function in a similar manner to the canonical SecA by delivering precursor proteins to a translocase and energizing export across the membrane. At least in mycobacteria, the two SecA proteins are not interchangeable (Braunstein et al., 2001). Furthermore, subcellular fractionation and immunoblot analysis shows that SecA/SecA1 partitions equally between cell envelope and cytosolic fractions, while SecA2 is found predominantly in cell envelope fractions in S. parasanguinis (Chen et al., 2006). These data reveal differences between SecA proteins and it emphasizes the need of direct study of SecA2 functions.
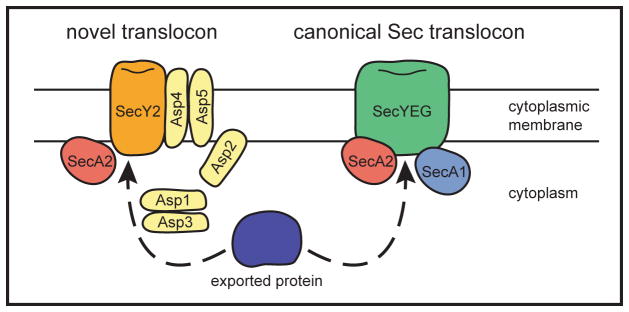
SecA2-interacting proteins need to be identified. This will greatly help understand the process of SecA2-mediated export. In two basic models, SecA2 either works with a novel translocase composed of dedicated accessory components or it works with the canonical SecA1/SecYEG translocase (Figure 4). While it is tempting to assume that the SecA2/SecY2 system works with a novel translocon and SecA2-only systems work with the canonical Sec translocase, these basic assumptions need to be experimentally addressed. Regarding the latter model, the role of SecA2 could be to recognize substrates that are normally overlooked for export and deliver them to the general SecYEG translocase. E. coli and B. subtilis SecA are known to form dimers (Ding et al., 2003; Jilaveanu et al., 2005), although whether or not the dimer is the translocation-active form of the protein remains controversial (de Keyzer et al., 2005; Jilaveanu et al., 2005; Or et al., 2005). One interesting possibility is that SecA2 forms heterodimers with SecA1 and/or interacts with other translocase components to promote export.
Fig. 4. Models for SecA2-dependent export.
In the various accessory Sec systems, a SecA2-exported protein (shown in blue) might be exported either through a novel translocon or the canonical SecA1/SecYEG translocon with the assistance of SecA2. The example of a novel translocon is modelled on the SecA2/SecY2 system of S. gordonii, which is a candidate for this type of pathway.
The studies of GspB and Fap1 provide a framework for understanding features that make a protein dependent on SecA2 for export. Two types of defining elements exist: signals that direct proteins to SecA2 and signals that block export by SecA/SecA1. In addition, there are some SecA2-dependent proteins that are still exported, albeit at a reduced level, in the absence of SecA2. We propose that exported proteins span a continuum from those with features that make them completely independent of SecA2 and exclusively reliant on SecA1 to those completely dependent on SecA2 and unaffected by SecA1. Future experiments are needed to understand how SecA1 and SecA2 substrates differ. Finally, for the SecA2-only systems it will be important to determine if the role of SecA2 in exporting proteins without signal sequences is direct or indirect.
Now that the existence and biological significance of accessory Sec pathways is well established, we anticipate that future studies will reveal the details of the export process itself. Given the sophisticated level of understanding of the general Sec pathway, we are optimistic rapid progress can be made in characterizing similarities and differences in the mechanism of accessory Sec export.
Acknowledgments
We would like to thank Hui Wu (University of Alabama) for sharing with us sequences of the accessory secA2/secY2 locus of S. parasanguinis. We also thank Tom Randall (UNC-Center for Bioinformatics) for help constructing the phylogenetic tree and members of the Braunstein lab for critical reading of the manuscript. This work was supported by NIH AI054540 (MB).
References
- Archambaud C, Nahori MA, Pizarro-Cerda J, Cossart P, Dussurget O. Control of Listeria superoxide dismutase by phosphorylation. J Biol Chem. 2006 doi: 10.1074/jbc.M606249200. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Gibson BW, Sullam PM. The Streptococcus gordonii platelet binding protein GspB undergoes glycosylation independently of export. J Bacteriol. 2004a;186:638–645. doi: 10.1128/JB.186.3.638-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Lopez JA, Sullam PM. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect Immun. 2004b;72:6528–6537. doi: 10.1128/IAI.72.11.6528-6537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Takamatsu D, Sullam PM. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol. 2005;58:1468–1481. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- Bensing BA, Siboo IR, Sullam PM. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J Bacteriol. 2007;189:3846–3854. doi: 10.1128/JB.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Brown AM, Kurtz S, Jacobs WR., Jr Two nonredundant SecA homologues function in mycobacteria. J Bacteriol. 2001;183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Espinosa B, Chan J, Belisle JT, Jacobs WRJ. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Molecular Microbiology. 2003;48:453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- Breukink E, Nouwen N, van Raalte A, Mizushima S, Tommassen J, de Kruijff B. The C terminus of SecA is involved in both lipid binding and SecB binding. J Biol Chem. 1995;270:7902–7907. doi: 10.1074/jbc.270.14.7902. [DOI] [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–1266. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SA, Hain T, Technow U, Darji A, Pashalidis P, Joseph SW, Chakraborty T. Identification and characterization of a peptidoglycan hydrolase, MurA, of Listeria monocytogenes, a muramidase needed for cell separation. J Bacteriol. 2003;185:6801–6808. doi: 10.1128/JB.185.23.6801-6808.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspers M, Freudl R. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol. 2008 doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed] [Google Scholar]
- Chavan M, Lennarz W. The molecular basis of coupling of translocation and N-glycosylation. Trends Biochem Sci. 2006;31:17–20. doi: 10.1016/j.tibs.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wu H, Fives-Taylor PM. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol Microbiol. 2004;53:843–856. doi: 10.1111/j.1365-2958.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- Chen Q, Wu H, Kumar R, Peng Z, Fives-Taylor PM. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguis. FEMS Microbiol Lett. 2006;264:174–181. doi: 10.1111/j.1574-6968.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- Chen Q, Sun B, Wu H, Peng Z, Fives-Taylor PM. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J Bacteriol. 2007;189:7610–7617. doi: 10.1128/JB.00748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer J, van der Sluis EO, Spelbrink RE, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJ. Covalently dimerized SecA is functional in protein translocation. J Biol Chem. 2005;280:35255–35260. doi: 10.1074/jbc.M506157200. [DOI] [PubMed] [Google Scholar]
- de Vrije T, de Swart RL, Dowhan W, Tomassen J, de Kruijff B. Phosphatidylglycerol is involved in protein translocation across Escherichia coli inner membranes. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- Dempsey BR, Wrona M, Moulin JM, Gloor GB, Jalilehvand F, Lajoie G, Shaw GS, Shilton BH. Solution NMR structure and X-ray absorption analysis of the C-terminal zinc-binding domain of the SecA ATPase. Biochemistry. 2004;43:9361–9371. doi: 10.1021/bi0493057. [DOI] [PubMed] [Google Scholar]
- Ding H, Hunt JF, Mukerji I, Oliver D. Bacillus subtilis SecA ATPase exists as an antiparallel dimer in solution. Biochemistry. 2003;42:8729–8738. doi: 10.1021/bi0342057. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein Translocation Across the Bacterial Cytoplasmic Membrane. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- Economou A. Bacterial preprotein translocase: mechanism and conformational dynamics of a processive enzyme. Mol Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- Economou A. Following the leader: bacterial protein export through the Sec pathway. Trends Microbiol. 1999;7:315–320. doi: 10.1016/s0966-842x(99)01555-3. [DOI] [PubMed] [Google Scholar]
- Fekkes P, van der Does C, Driessen AJ. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. Embo J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol. 2007;189:5090–5100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MB, Cernich M, Brokl O. Some observations of mycobacterial acid-fastness. Am Rev Respir Dis. 1978;118:151–154. doi: 10.1164/arrd.1978.118.1.151. [DOI] [PubMed] [Google Scholar]
- Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. Silencing essential protein secretion in Mycobacterium smegmatis using tetracycline repressors. J Bacteriol. 2007 doi: 10.1128/JB.00216-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- Hinchey J, Lee S, Jeon BY, Basaraba RJ, Venkataswamy MM, Chen B, Chan J, Braunstein M, Orme IM, Derrick SC, Morris SL, Jacobs WR, Jr, Porcelli SA. Enhanced priming of adaptive immunity by a proapoptotic mutant of Mycobacterium tuberculosis. J Clin Invest. 2007;117:2279–2288. doi: 10.1172/JCI31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- Jilaveanu LB, Zito CR, Oliver D. Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci U S A. 2005;102:7511–7516. doi: 10.1073/pnas.0502774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli. J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- Kurtz S, McKinnon KP, Runge MS, Ting JP, Braunstein M. The SecA2 secretion factor of Mycobacterium tuberculosis promotes growth in macrophages and inhibits the host immune response. Infect Immun. 2006 doi: 10.1128/IAI.01022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz LL, Portnoy DA. Identification of a second Listeria secA gene associated with protein secretion and the rough phenotype. Mol Microbiol. 2002;45:1043–1056. doi: 10.1046/j.1365-2958.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci U S A. 2003;100:12432–12437. doi: 10.1073/pnas.2133653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machata S, Hain T, Rohde M, Chakraborty T. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol. 2005;187:8385–8394. doi: 10.1128/JB.187.24.8385-8394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matousek WM, Alexandrescu AT. NMR structure of the C-terminal domain of SecA in the free state. Biochim Biophys Acta. 2004;1702:163–171. doi: 10.1016/j.bbapap.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- Muraille E, Narni-Mancinelli E, Gounon P, Bassand D, Glaichenhaus N, Lenz LL, Lauvau G. Cytosolic expression of SecA2 is a prerequisite for long-term protective immunity. Cell Microbiol. 2007;9:1445–1454. doi: 10.1111/j.1462-5822.2007.00883.x. [DOI] [PubMed] [Google Scholar]
- Murphy CK, Beckwith J. Export of Proteins to the Cell Envelope in Escherichia coli. In: Neidhardt FC, editor. Escherichia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 967–978. [Google Scholar]
- Nita-Lazar M, Wacker M, Schegg B, Amber S, Aebi M. The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology. 2005;15:361–367. doi: 10.1093/glycob/cwi019. [DOI] [PubMed] [Google Scholar]
- Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3- phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Peng Z, Wu H, Ruiz T, Chen Q, Zhou M, Sun B, Fives-Taylor P. Role of gap3 in Fap1 glycosylation, stability, in vitro adhesion, and fimbrial and biofilm formation of Streptococcus parasanguinis. Oral Microbiol Immunol. 2008;23:70–78. doi: 10.1111/j.1399-302X.2007.00401.x. [DOI] [PubMed] [Google Scholar]
- Pilgrim S, Kolb-Maurer A, Gentschev I, Goebel W, Kuhn M. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect Immun. 2003;71:3473–3484. doi: 10.1128/IAI.71.6.3473-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Silva-Herzog E, Ferracci F, Jackson MW, Joseph SS, Plano GV. Membrane localization and topology of the Yersinia pestis YscJ lipoprotein. Microbiology. 2008;154:593–607. doi: 10.1099/mic.0.2007/013045-0. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol. 2004a;52:189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J Bacteriol. 2004b;186:7100–7111. doi: 10.1128/JB.186.21.7100-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. Two additional components of the accessory sec system mediating export of the Streptococcus gordonii platelet-binding protein GspB. J Bacteriol. 2005;187:3878–3883. doi: 10.1128/JB.187.11.3878-3883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Loranger JM, Wu HC. A distinct signal peptidase for prolipoprotein in Escherichia coli. J Cell Biochem. 1984;24:113–120. doi: 10.1002/jcb.240240203. [DOI] [PubMed] [Google Scholar]
- van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anne J. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- VanderVen BC, Harder JD, Crick DC, Belisle JT. Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science. 2005;309:941–943. doi: 10.1126/science.1114347. [DOI] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol. 2007a;189:1390–1398. doi: 10.1128/JB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 2007b;75:2181–2188. doi: 10.1128/IAI.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuenscher MD, Kohler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwizinski C, Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980;255:7973–7977. [PubMed] [Google Scholar]