Abstract
Published reports implicate a variety of mechanisms that may contribute to drug resistance in ovarian cancer. The chief aim of this study is to understand the relationship between overexpression of drug resistance associated genes and multidrug resistance in ovarian cancer. Using lentiviral short hairpin RNA (shRNA) collections targeting 132 genes identified from transcriptional profiling of drug resistant cancer cell lines, individual knockdown experiments were performed in the presence of sublethal doses of paclitaxel. Specific genes whose knockdown was found to be associated with cellular toxicity included MDR1 (ABCB1), survivin and PRP-4 (Pre-mRNA Processing factor-4). These genes, when repressed, can reverse paclitaxel resistance in the multidrug resistant cell line SKOV-3TR and OVCAR8TR. Both MDR1 and survivin have been previously reported to play a role in multidrug resistance and chemotherapy induced apoptosis; however, the effect of PRP-4 expression upon drug sensitivity is currently unrecognized. PRP-4 belongs to the Ser/Thr protein kinase family, plays a role in pre-mRNA splicing and cell mitosis, and interacts with CLK1. Northern analysis demonstrates that PRP-4 is overexpressed in several paclitaxel resistant cell lines and confirms that PRP-4 expression could be significantly repressed by PRP-4 lentiviral shRNA. Both clonogenic and MTT assays confirm that transcriptional repression of PRP-4 could reverse paclitaxel resistance 5–10 fold in SKOV-3TR. Finally, overexpression of PRP-4 in drug sensitive cells could induce a modest level of drug resistance to paclitaxel, doxorubicin and vincristine.
Keywords: shRNA, PRP-4, drug resistance, lentivirus, ovarian cancer
Introduction
Standard chemotherapy for newly diagnosed and recurrent ovarian cancer includes a combination of paclitaxel and carboplatin. Although objective responses and survival benefits are seen, the efficacy of both of these agents is limited by the eventual development of multidrug resistance (MDR) (1, 2). There is little understanding of how ovarian tumors, as well as other tumors, develop drug resistance. Published reports from several laboratories implicate several mechanisms that contribute to the drug resistance phenotype. Reversing drug resistance has been an important goal of clinical and investigational oncology (3–7).
Gene expression profile analysis is an efficient technology that allows screening for correlations between expression of many genes and acquisition of multidrug resistance. In an attempt to identify novel genes that are differentially expressed between paclitaxel-resistant and paclitaxel-sensitive cells, the paclitaxel-sensitive ovarian cancer cell lines, SKOV-3 and OVCAR8, and the paclitaxel-sensitive breast cancer cell line, MCF-7, were exposed to incrementally increasing concentrations of paclitaxel. This procedure resulted in the establishment of three paclitaxel resistant daughter cell lines, SKOV-3TR, OVCAR8TR and MCF-7TR, respectively (4, 8). Gene expression profiles in these three paclitaxel-resistant cell lines and their corresponding paclitaxel-sensitive parental lines (SKOV-3 vs SKOV-3TR, OVCAR8 vs OVCAR8TR, MCF-7 vs MCF-7TR) were characterized using Affymetrix microarray technology. A large number of transcripts was identified as differentially expressed between pairs of sensitive and resistant cell lines. 790 (SKOV-3TR), 689 (OVCAR8TR) and 964 (MCF-7TR) gene transcripts demonstrated more than a two-fold overexpression in the paclitaxel resistant lines relative to their expression in the parental lines (8). Although SKOV-3TR, OVCAR8TR and MCF-7TR all demonstrate a paclitaxel-resistant phenotype, the transcripts identified with altered expression in each cell line pair were largely non-overlapping and encoded proteins with a wide variety of biochemical functions. Establishing a strong correlation between drug sensitivity and expression of a particular gene has been challenging (9).
The ability to use RNA interference (RNAi) as a tool for functional gene silencing in tumor cells has enabled us to perform genetic loss-of-function studies in tissue culture systems (10, 11). But while siRNA has been shown to be effective for short-term gene inhibition in mammalian cell lines, there is a clear problem in its use for stable transcript knockdown (12). Recently, short hairpin RNA (shRNA) libraries in lentiviral vectors have been described and used in stable cell lines and in transgenic mice (13, 14). Our aim was to identify genes essential for drug resistance by screening for shRNAs that selectively reverse drug resistance in cancer cell lines. This type of screen holds promise for the discovery of novel targets in reversing drug resistance in cancer therapy and for genetically validating combination therapies.
In the present study, we utilized the above-mentioned collection of 132-lentiviral shRNA constructs targeting the expression of drug resistance associated genes to address whether loss of these transcripts impact paclitaxel sensitivity in vitro. This pre-selected shRNA library was drawn from our previous cDNA array studies; the screen has been carried out in the well-characterized ovarian cancer paclitaxel-resistant cell line SKOV-3TR (4, 7, 8). We show that inhibition of several target genes could sensitize SKOV-3TR to paclitaxel. Specifically, we observe that knockdown of human pre-messenger RNA processing (PRP-4) kinase in ovarian cancer multidrug resistant cells is associated with increased sensitivity to paclitaxel. Additionally, we observe that PRP-4 kinase is overexpressed in multi-drug resistant ovarian cancer cell lines. Furthermore, we have identified a set of drug resistance associated genes that, when silenced, cause cell sensitivity to paclitaxel, several of which (such as ABCB1/MDR1, MDM2, and survivin) have been previously shown to be involved in multidrug resistance (11, 15–17). This observation suggests that overexpression of PRP-4 is a new mechanism for the acquisition of drug resistance and therefore a potential target for the development of therapies treating drug resistance in ovarian cancer.
Materials and Methods
Cell Culture
The human ovarian cancer cell line SKOV-3 used in this study was obtained from the American Type Culture Collection (Rockville, MD). Dr. Patricia Donahoe (Massachusetts General Hospital, Boston, MA) provided the OVCAR8 human ovarian cancer cell line. The paclitaxel resistant SKOV-3TR and OVCAR8TR cell lines were established as previously reported (4, 7, 18) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin (all obtained from Invitrogen, Carlsbad, CA). Resistant sub-clones were continuously cultured in paclitaxel. Paclitaxel, doxorubicin, vincristine and cisplatin were obtained as unused residual clinical material at the Massachusetts General Hospital.
Drug Resistance Associated Genes Lentiviral shRNA Library
The genes were chosen based on their relevance to multidrug resistance in human ovarian cancer from our previous studies (4, 8). Based on 291 gene transcripts that demonstrated over-expression in at least two resistant cell lines, we constructed a lentiviral shRNA library composed of 132 genes at Sigma-Aldrich Research Biotechnology (St. Louis, MO). Details of the library-production methods can be found at the Sigma web site (http://www.sigmaaldrich.com/Area_of_Interest/Life_Science/Functional_Genomics_and_RNAi/shRNA). In particular, these genes were pre-selected because they also included full-length open reading frames (ORF). These ORF will be useful for confirmatory functional studies in which full-length transcripts would be transfected back into sensitive cell lines.
Lentiviral Infection
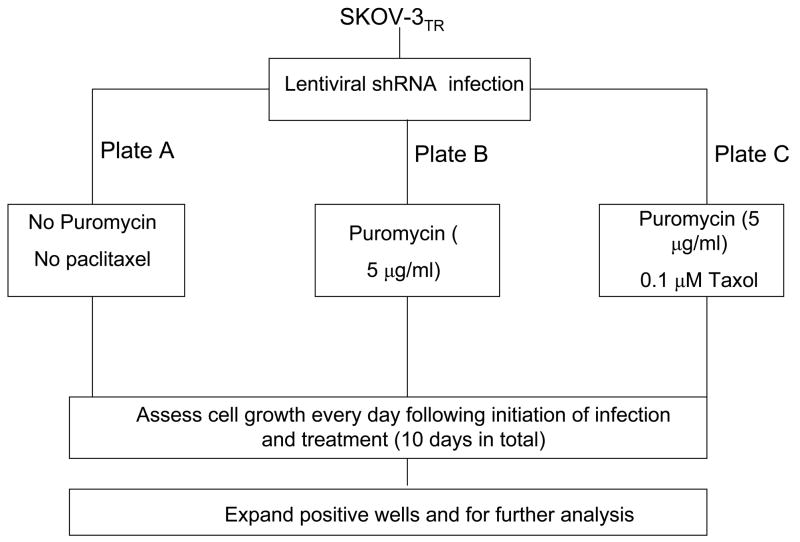
Infection conditions in SKOV-3TR were optimized with MISSON® TurboGFP™ Control Transduction particles (SHC003V) and Non-target shRNA Control Transduction particles (SHC002V) (Sigma) in 96-well plates for optimal growth conditions, viral dosage, puromycin selection concentration and assay times prior to screening. On day 1, SKOV-3TR cells were seeded at 1.6x104 cells/well in triplicate, on 96-well plates and labeled with plate A, B or C, and incubated for 24 hr at 37°C (Fig. 1). On day 2, 8 μg/ml of hexadimethrine bromide were added to each well, and infected using 8 μl of lentiviral particles encoding shRNA against different drug resistance associated genes to appropriate wells, and incubated for 24 hr at 37°C. On day 3, plate A was kept as the control without the addition of puromycin or paclitaxel, the shRNA viral particle-containing medium was removed and replaced it with fresh medium. For plate B, viral particle-containing medium was removed and replaced with fresh medium containing 5 μg/ml of puromycin for selection of transduced cells. For plate C, viral particle-containing medium was removed and replaced with fresh medium containing 5 μg/ml of puromycin and 0.1 μM of paclitaxel. From day 4 to day 10, fresh medium was replaced with necessity as described above and evaluated for cytotoxicity in any wells under the microscope. On Day 10, the number of viable cells was determined via CellTiter 96®AQueous One Solution Cell Cytotoxicity Assay (Promega, Madison, WI)
Figure 1.
Flow chart of the functional lentiviral shRNA screen measuring the effect of drug resistant gene knockdown on paclitaxel sensitivity. The drug resistance associated 132-lentiviral shRNA library was established. For the screen, 1.6x104 SKOV-3TR cells per well in 96-well plates were infected with lentiviral particles encoding shRNA against different drug resistance associated genes. The infected cells were subsequently selected with puromycin following a protocol provided by the manufacturer. The number of viable cells was analyzed using the CellTiter 96®AQueous One Solution Cell Cytotoxicity Assay and visualized under the microscope.
Determination of the Results and Analysis
The general format was designed to include evaluation of each target gene through shRNA lentiviral-mediated gene knockdown. The experiments were conducted in two control plates and one experimental plate to permit plate-to-plate comparisons (Fig. 1). The control plates (plate A and plate B) were used to evaluate transduction efficiency, and to exclude target genes that were lethal during knockdown in the absence of chemotherapy. The latter control (plate B) was necessary because the goal of this study was to identify genes that are important in acquired drug resistance and not cell survival. Therefore, one plate (plate A) was given only lentivirus to confirm that the shRNA is not lethal in the absence of puromycin and paclitaxel (a positive result is cell survival); A second plate (plate B) was exposed to puromycin and lentivirus to confirm efficiency of infection (once again a positive result is cell survival). The third plate (plate C) was given both lentiviral shRNA and 0.1 μM paclitaxel. This is typically a sublethal dose of paclitaxel for the SKOV-3TR cell line and a positive result would be cell death at 4–10 days. shRNA targeted genes that are associated with cells surviving in plate A and B and dying in plate C were identified as “hits” and selected for further study. Each of the 132 genes is represented by three to five different shRNA lentiviral particle constructs targeting different sites in each gene. Because these shRNA are complimentary to different regions of the mRNA, it reduces the risk of “off target effects.” To further minimize the possibility of “off target” hits, we only focused on genes identified as functional drug resistance genes by two or more shRNA targeting the same gene. All experiments used cells infected with a MISSION® non-target vector control virus (Sigma) as a negative control for expression changes. Once hits were identified, we validated that the targeted genes indeed have been knocked down by RT-PCR and Northern analysis.
RNA Extraction
RNA was collected from SKOV-3 and SKOV-3TR. Total RNA was isolated using TRIzol® Reagent (Invitrogen) according to the manufacturer’s instructions. Additionally, RNA quality was determined via ethidium bromide staining following agarose/formaldehyde gel electrophoresis.
RT-PCR
For the PRP-4 gene, the results from the shRNA screening were verified using RT-PCR and Northern blot analysis. RT-PCR was performed using the sense and antisense primers to human PRP-4: sense primer 5′-ATAAGAATGCGGCCCGCGGAAGTTCAAGATGGCCGCCG-3′ to introduce a Not I site as underlined, and antisense primer: 5′-GGTGGATCCCACACACTCAAACCCTTGGAG-3′ ( GenBank # NM_003913 ) to introduce a BamH I site as underlined. The introduced restriction enzyme sites were designed for following confirmatory functional studies. TRIzol extracted total RNA was DNase-treated to remove contaminating genomic DNA according to the manufacturer’s protocol (Invitrogen). RT-PCRs were performed using the Titan One Tube RT-PCR systems (Roche, Indianapolis, IN) following the manufacturer’s protocol.
Northern Analysis
PRP-4 RT-PCR products were cloned into the pCR®2.1 vector using a TA Cloning® kit (Invitrogen) and the cloned fragment was confirmed by sequencing. The cDNA inserts were cut using Not I and BamH I (Promega) and purified using the QIAEXII Gel Extraction Kit (Qiagen Inc, Chatsworth, CA). PRP-4 probes were labeled with 32P-dCTP using the Megaprime DNA Labeling System. (Amersham Pharmacia Biotech, Piscataway, NJ). Total RNA was extracted using the TRIzol reagents as described above. RNA was separated by electrophoresis in 1.2% agarose/formaldehyde gels (approximately 5μg total RNA per lane), transferred to Hybond N-plus nylon membranes (Amersham Biosciences) and UV cross-linked. A one hour prehybridization step was performed in Rapid-hyb buffer (Amersham Biosciences), followed by a 2 hour hybridization of the 32P-labeled PRP-4 probe in the same buffer. The blots were washed twice at room temperature with 2X SSC-0.1%SDS for 15 min and twice at 65°C with 0.2X SSC-0.1% SDS for 15 min. Blots were exposed to autoradiography X-ray film with an intensifying screen. Finally, to confirm the amounts of RNA loaded in each lane, blots were hybridized with a β-actin probe. Relative quantification was performed using densitometry.
Cytotoxicity Assay
Chemotherapy drug cytotoxicity was assessed in vitro using the MTT assay as previously described (19). Briefly, 2×103 cells per well were plated in 96-well plates in culture medium (RPMI 1640 supplemented with 10% fetal bovine serum and penicillin/streptomycin) containing increasing concentrations of chemotherapy drug such as paclitaxel. After 7 days of culture, 10 μl of MTT (5 mg/ml in PBS, obtained from Sigma) was added to each well and the plates were incubated for 4 h. The resulting formazan product was dissolved with acid-isopropanol and the absorbance at a wavelength of 490 nm (A490) was read on a SPECTRAmax® Microplate Spectrophotometer (Molecular Devices, Sunnyvale, CA). The absorbance values were normalized by assigning the value of the control line in the medium without drug to 1.0 and the value of the no cell control to 0. Experiments were performed in triplicate. Dose response curves were fitted with use of GraphPad PRISM® 4 software (GraphPad Software, San Diego, CA).
pIRESPRP-4 Expression Vector Construction
Clonetech’s (Palo Atlo, CA) mammalian expression vector pIRESneo was used for functional expression study. The expression cassette of pIRESneo contains the human cytomegalovirus major immediate early promoter/enhancer (pCMV) followed by a multiple cloning site (MCS) and a synthetic intron known to enhance the stability of the mRNA. A 3094 base pair cDNA fragment containing the full ORF of human PRP-4 was amplified by RT-PCR from the RNA of SKOV-3TR, a paclitaxel resistant cell line that highly overexpresses PRP-4. The resulting PRP-4 RT-PCR product was cloned into pCR®2.1 vector using Invitrogen’s Original TA Cloning Kit. After sequence confirmation, PRP-4 was cut from the pCR®2.1 vector, purified, subcloned into the MCS of expression vector pIRESneo, and subsequently sequenced to confirm the correct ORF. Expression of PRP-4 cDNA was under the control of the pCMV.
Transfection and Production of Stable Cell Lines
Transfections were performed using LipofectAmine Plus reagents (Invitrogen) as follows: approximately 5x105 SKOV-3 cells were plated into 90 millimeter tissue culture dishes and cultured overnight. Prior to transfection, the growth medium was replaced with serum free RPMI 1640 and cultured for three hours. LipofectAmine reagent containing 5 μg of pIRESempty, pIRESPRP-4 was combined with Plus reagent and applied to the cells. After culture for four hours, the media was replaced with RPMI 1640 containing 10% fetal bovine serum. G418 sulfate (Invitrogen) selection (300μg/ml) was started at 24 hours post transfection. The selection medium was changed every 2 days.
Western Blotting
The human PRP-4 antibody was generously provided by Dr. Regis Giet (University Rennes, France) (20). The Pgp antibody C219 was purchased from Signet (Dedham, MA). The mouse monoclonal antibody to human actin was purchased from Sigma-Aldrich. PRP-4 and Pgp proteins were analyzed in total cell lysates. Total cell lysates were prepared, and Western blot analysis was performed as previously described. Briefly, the cells were lysed in 1X RIPA lysis buffer (Upstate Biotechnology, Charlottesville, VA) and protein concentration was determined by the DC Protein Assay (Bio-Rad). Twenty-five micrograms of total protein were resolved on NuPage™ 4–12% Bis-Tris Gels (Invitrogen) and immunoblotted with specific antibodies. Primary antibodies were incubated in TBS (pH 7.4) with 0.1% Tween-20 with gentle agitation overnight at 4°C. Horseradish peroxidase (HRP)-conjugated secondary antibodies (Bio-Rad) were incubated in TBS (pH 7.4) with 5% nonfat milk (Bio-Rad) and 0.1% Tween-20, at a 1:2000 dilution for one hour at room temperature with gentle agitation. Positive immunoreactions were detected by using SuperSingal® West Pico Chemiluminescent Substrate (Pierce Biotechnology, Inc. Rockford, IL).
Results
Lentiviral shRNA Screen for Mediators of Paclitaxel Sensitivitiy in Ovarian Cancer cells
To investigate the potential functional role of previously identified drug resistance associated genes in ovarian cancer, we identified 291 human gene transcripts listed in the NCBI database that have been found to be over-expressed in at least two out three resistant cell lines (SKOV-3TR, OVCAR8TR and MCF-7TR). Based on the list of 291 genes, we constructed a 132- gene lentiviral shRNA library. This was submitted as part of the Sigma MISSION® shRNA collection. This library contains 504 siRNA templates targeting 132 drug resistance associated genes. The library was cloned into a lentiviral-based pLKO.1-puro shRNA expression vector. Up to five shRNA sequences were individually cloned into pLKO.1-puro for broad coverage of each target gene. The hairpin structure includes an intramolecular 20–21 bp stem and 6 base loop that is recognized and cleaved by Dicer upon expression via the U6 (pol III) promoter in the host cell. The resulting siRNA duplex then continues in the RNAi pathway by association with RISC. The puromycin resistance marker is present to allow for stable selection in mammalian cells.
Initial screening conditions utilized the paclitaxel-resistant cell line SKOV-3TR (Fig. 1). We found a 5-day incubation with 5 μg/ml of puromycin was effective for generating complete cell death of untransfected cells. Infection of SKOV-3TR cells with MISSON® TurboGFP™ Control lentivirus and subsequent selection with puromycin confirmed high transduction efficiency. Likewise, transduction with a MISSON® Non-target lentivirus containing a ‘non-target shRNA’ control that fails to target any known genes was non-toxic to SKOV-3TR. After optimizing the transduction efficiency and puromycin selection concentration, we infected SKOV-3TR cells with a preselected lentiviral shRNA library that targeted each of the 132 drug resistance associated genes. After 4 to 10 days, the specific gene knockdown associated with 0.1 μM paclitaxel-induced cell death was identified. Overall, 6 gene “hits” were identified where knockdown could reverse paclitaxel resistance; of these, MDR1, survivin, and MDM2 (murine double minute 2) were already known to be associated with drug resistance or apoptosis resistance while PRP-4, THOC1, and Krit1 (Krev interaction trapped 1) are novel genes with unknown function in drug resistance (Table 1). Significantly, three of five PRP-4 knockdown constructs resulted in the reversal of paclitaxel sensitivity in SKOV-3TR (Table 2, Fig. 2). Levels of knockdown ranged from approximately 60% to 90%. These data suggest that an shRNA screen is a practical platform for genome-scale screening of drug resistant genes in drug resistant cancer cells and demonstrate that shRNA knockdown of specific genes can synergize drug sensitivities. These results also demonstrate the feasibility of screening with large collections of lentiviral vectors of drug resistance associated genes after genome-wide microarray studies to identify drug enhancers and suppressors.
Table 1.
The list of positive hits from the lentiviral shRNA screen in SKOV-3TR
| Gene name | Symbol | Functions | Positive hits |
|---|---|---|---|
| Pre-mRNA Processing factor 4 | PRP-4 | pre-mRNA splicing, Interacts with Clk1 C-terminus | 3 out 5 |
| Survivin | EPR-1 | Inhibit apoptosis, regulate mitosis | 2 out 4 |
| Multidrug resistance 1 | MDR1 | efflux pump responsible for decreased drug accumulation | 2 out 5 |
| Nuclear matrix protein p84 | THOC1 | protein transcription and RNA export | 2 out 5 |
| Oncoprotein mdm2 | MDM2 | Inhibit cell cycle arrest and apoptosis | 2 out 4 |
| Krev interaction trapped 1 | Krit1 | unknown | 2 out 5 |
Table 2.
MISSION™ TRC PRP-4 shRNA Target Set
| TRC number | Sequence | Target region | Reverse resistance |
|---|---|---|---|
|
TRCN0000000719
(TRC 719) |
CCGGCCCTATCAACTGTCTTATGTACTCGA
GTACATAAGACAGTTGATAGGGTTTTT |
3UTR | Yes |
|
|
|||
|
TRCN0000000720(
TRC 720) |
CCGGGCTGCTGATGTTAAAGAGTATCTCG
AGATACTCTTTAACATCAGCAGCTTTTT |
CDS | No |
|
|
|||
|
3TRCN0000000721(
TRC 721) |
CCGGCTCAAGATCAAGCAAGGAAATCTCG
AGATTTCCTTGCTTGATCTTGAGTTTTT |
CDS | No |
|
|
|||
|
TRCN0000000722(
TRC 722) |
CCGGGAATGAAAGTTGAGCAGGAATCTCG
AGATTCCTGCTCAACTTTCATTCTTTTT |
CDS | Yes |
|
|
|||
|
TRCN0000000723(
TRC 723) |
CCGGAGCAAGTCAAAGGAGAGGAAACTCG
AGTTTCCTCTCCTTTGACTTGCTTTTTT |
CDS | Yes |
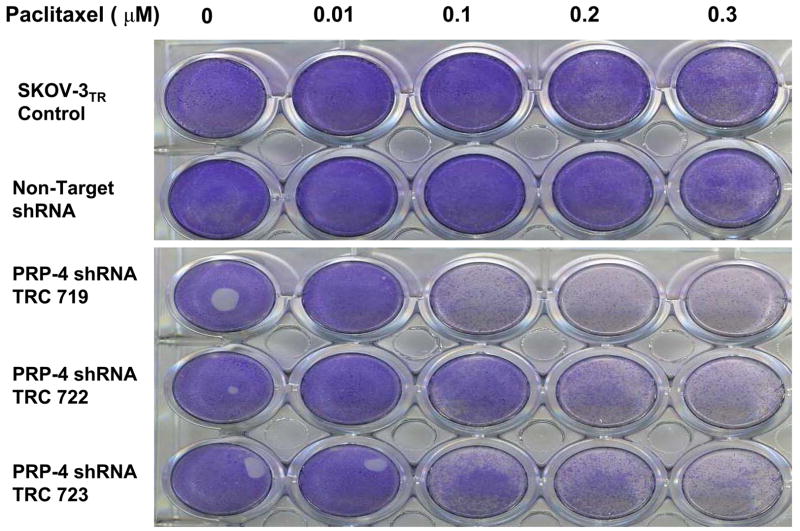
Figure 2.
Effect of PRP-4 knockdown on paclitaxel sensitivitiy in SKOV-3TR cells. Clonogenic assay transduction of PRP-4 lentiviral shRNA vector confers paclitaxel sensitivity to SKOV-3TR cells. After transfection with the PRP-4 lentiviral shRNA, puromycin selected clones were plated and exposed to varying concentrations of paclitaxel for 6 days. The cells were fixed and stained with crystal violet to visualize the viable cells, with darker staining representing the more viable cells.
Confirmation of Identified PRP-4 Knockdown and the Reversal of Paclitaxel Resistance
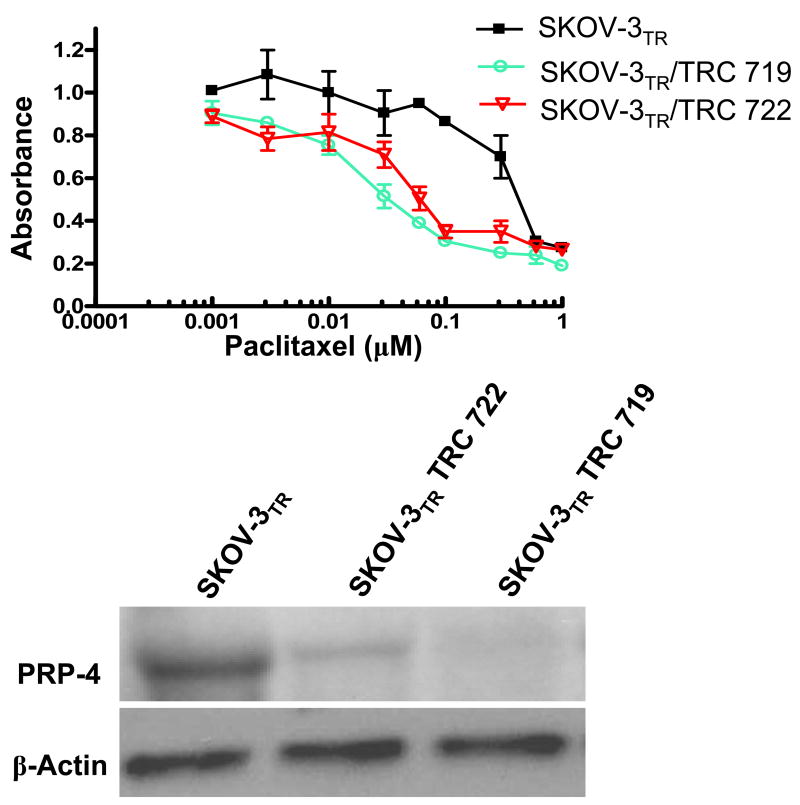
The insertion of viral DNA into a host chromosome often leads to disruption of host genes. Therefore, to validate that the reversal of paclitaxel resistant phenotype of the positive cells is indeed caused directly by the shRNA knockdown of the PRP-4 gene instead of nonspecific defects induced by random viral DNA insertion, we reinfected the SKOV-3TR cells with each positive lentivirus and assayed for cell death with paclitaxel. We used a clonogenic assay in our initial assay to determine paclitaxel sensitivitiy (Fig. 2.). For confirmation, we used an alternative liquid culture cell MTT cyototoxic assay to demonstrate that the reversal phenotypes we detected were not assay specific (Fig. 3 A). Using our established viral infection protocol, we found if viral infection and knockdown of the target gene in SKOV-3TR cells does not alter the paclitaxel sensitivities, the infected cell will grow well in 0.1 μM paclitaxel-containing medium. On the other hand, if viral infection and knockdown of specific genes (such as MDR1, survivin or PRP-4) does lead to drug sensitivity, the SKOV-3 TR cells will die in medium containing 0.1 μM paclitaxel. We confirmed that three out of five PRP-4 shRNA target knockdowns significantly reversed paclitaxel resistance in SKOV-3TR cells (Fig. 2 and Fig. 3 A). The two out of five constructs that did not reduce palitaxel resistance could not significantly block PRP-4 expression (Table 2). These results suggest that by treating the SKOV-3TR cells with low doses of paclitaxel (0.1 μM) combined with PRP-4 shRNA, apoptosis or cell killing induced by paclitaxel could be enhanced. Northern blot analysis demonstrated that PRP-4 was significantly depleted in the PRP-4 shRNA infected and puromycin selected cells. Both RT-PCR (data not shown) and Northern blot analysis demonstrated that PRP-4 gene expression was indeed knocked down in shRNA infected clones (Fig. 3 B).
Figure 3.
Confirmation of PRP-4 depletion in clones and effect on paclitaxel sensitivitiy. (A) MTT assay shows validation of PRP-4 knockdown in the reversal of paclitaxel resistance. Data shows two of three positive target sites of PRP-4 by shRNA. (B). Duplicate sample sets of total RNA isolated from the SKOV-3 TR, SKOV-3TR/TRC719, and SKOV-3TR/TRC722 cell lines analyzed in the MTT assay were subjected to Northern blot analysis using cDNA probe directed against PRP-4 (upper panel) and β-actin (lower panel)
Evaluation of PRP-4 Knockdown and Drug Sensitivities in OVCAR8TR cells
After identification of PRP-4 as a new regulator of chemoresistance in SKOV-3TR, the effect of PRP-4 inhibition on drug sensitivities was further examined in human ovarian cancer cell drug resistant cell line OVCAR8TR. Inhibition of PRP-4 expression by shRNA in OVCAR8TR combined with sublethal doses of paclitaxel (0.1 μM), doxorubicin (0.5 μM), vincristine (0.1 μM), or cisplatin (1 μM), demonstrated increase drug sensitivity to paclitaxel, doxorubicin, vincristine but not to cisplatin (Supplementary materials Fig. 1). It is noteworthy that both SKOV-3TR and OVCAR8TR were showed not resistance to cisplatin as previously reported.
Effect of PRP-4 Transfection on in vitro Growth of Transfected Cells
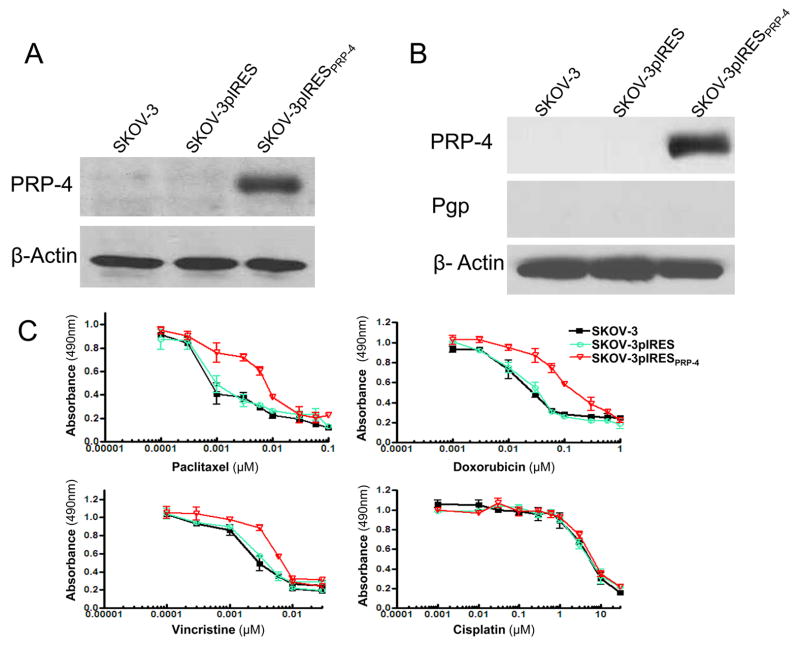
To further confirm the role of PRP-4 in drug resistance of ovarian cancer, we evaluated the effect of transfected overexpression of PRP-4 in drug sensitive SKOV-3. Transfection of PRP-4 into SKOV-3 with subsequent cloning selection of transfectants demonstrated overexpression of the respective gene and protein (Fig. 4 A, B). Analysis of these same transfectants demonstrates no change in MDR1 gene or Pgp protein expression as compared with the parental line or empty vector transfectant control (Fig. 4 B). MTT assay demonstrates a modest increase in paclitaxel, resistance in transfected cells. Cloned is relatively resistant to paclitaxel, doxorubicin, vincristine but not to cisplatin (Fig. SKOV-3pIRESPRP-4 4.C).
Figure 4.
Exogenous expression of PRP-4 confers paclitaxel resistance to the ovarian cancer cell line SKOV-3. (A) Confirmation of PRP-4 gene knockdown by Northern blot. Duplicate sample sets of total RNA isolated from the SKOV-3, SKOV-3pIRES, and SKOV-3pIRESPRP-4 cell lines analyzed in the MTT assay were subjected to Northern blot analysis using cDNA probe directed against PRP-4 (upper panel) and β-actin (lower panel). (B) Confirmation of PRP-4 protein knockdown by Western blot. Total protein was isolated from SKOV-3, SKOV-3pIRES, and SKOV-3pIRESPRP-4 cell lines and analyzed by Western blotting with anti PRP-4, Pgp or anti-actin antibodies. (C) The relative cytotoxicity of paclitaxel, doxorubicin, vincristine and cisplatin in SKOV-3 derived cell lines (SKOV-3pIRESPRP-4) stably transfected with a pIRESPRP-4 expression vector and in the parental (SKOV-3) and empty vector (SKOV-3pIRES) controls were assessed using the MTT assay. All samples were analyzed in triplicate.
Discussion
Multi-drug resistance often leads to eventual chemotherapy failure in patients with advanced ovarian cancer. There are many putative mechanisms for this process of inducing chemoresistance. MDR1 has been implicated in many examples of multi-drug resistance. Additionally, prior studies have reported that non-MDR1 transporter proteins cause multi-drug resistance in human cancer cell lines (21–23). We previously reported paclitaxel resistant cell lines are associated with overexpression of a broad range of transcriptional changes, including gene families involved in cell growth/maintenance, cell structure, signal transduction, and inflammatory response (4, 8). Furthermore, transfection of HER2/neu, c-H-ras, Bcl-2, IL-6 or MAGE into drug-sensitive cell lines has conferred drug-resistance in vitro (24–28). These findings illustrate that multiple mechanisms can be selected to cause drug resistance in ovarian cancer cell lines, and they may all contribute partially to drug resistance in ovarian cancer.
In this study, our goal was to use a lentiviral shRNA-mediated genetic screen to identify genes that are important in paclitaxel resistance in ovarian cancer cell lines. We extend previous studies of cDNA microarray analysis using paclitaxel-resistance associated genes. We observed that knockdown of PRP-4 and five additional genes (MDR1, survivin, THOC1, MDM2 and Krit1) are associated with significant reversal of paclitaxel-resistance. We further validated the functional role of PRP-4 in paclitaxel resistance. Previous studies demonstrated that up-regulated expression of PRP-4 is broadly associated with multidrug resistance, and increased PRP-4 transcript levels are detected in paclitaxel resistant cell lines. We confirm these observations in the ovarian cancer cell line SKOV-3TR and, importantly, we demonstrate that overexpression of the PRP-4 protein in a paclitaxel sensitive ovarian cancer cell line led to a modest increase in paclitaxel, doxorubicin, vincristine resistance. This data implies an important role for PRP-4 in the development of the chemotherapy drug resistance.
The PRP-4 gene encodes a 150-kD serine-threonine protein kinase that has been implicated in the regulation of mRNA splicing, and mutations in PRP-4 lead to the accumulation of pre-mRNAs (29). PRP-4 is also involved in mitosis; the expression of a dominant truncated PRP-4 protein could induce mitotic aberrations, suggesting a dual PRP-4 function in RNA splicing and mitosis (30, 31). The catalytic domain of PRP-4 shows significant similarity to the JNK/stress activated protein kinase type of MAPK including the TPY motif, suggesting that PRP-4 may play an important role in cell differentiation (32). PRP4 has been reported to mediate cellular signaling (33). The precise cellular function of PRP-4 in cancer cells remains unclear, although recent studies in Hela cells have found that PRP-4 expression is important for chromosome alignment (20). Several lines of evidence support that PRP-4 belongs to the family of spindle assembly checkpoint regulatory genes (20, 33). Our results also demonstrate the important role of PRP-4 kinase in regulating cell survival and drug sensitivity. The roles of PRP-4 in mRNA splicing and mitosis do not have clear connections with its apparent role in the development of the multidrug resistance. However, the PRP-4 gene is expressed in a wide spectrum of both normal and neoplastic tissues (33), and the determination of PRP-4 function in cancer cells may clarify its association with acquired drug resistance. The precise definition of its role awaits further investigation.
Enhanced paclitaxel sensitivitiy and drug induced apoptosis was not limited to PRP-4 gene knockdown; we also observed increased paclitaxel sensitivity and cell apoptosis with shRNA that was directed against the drug resistant target MDR1, anti-apoptotic protein MDM2 and survivin, which is associated with tumor cell survival and anti-apoptosis. All three genes have been associated with resistance to chemotherapy and poor prognosis (Table 1). For example, overexpression of MDR1 has been associated with resistance to paclitaxel and adriamycin in several cancer cell lines (34, 35). Knockdown of MDR1 expression by RNAi could partially reverse drug resistance (11). Overexpression of survivin has been associated with drug resistance and poor prognosis in breast and ovarian cancers (36, 37). MDM2 gene encodes a protein that inhibits p53-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain (38). Overexpression of MDM2 in several types of human cancers is associated with a poor prognosis (39, 40). Because of its functions in cell cycle and apoptosis, MDM2 may also play a role in the development of drug resistance in tumor cells. It has been reported that overexpression of MDM2 resulted in expression of the MDR1 gene and its protein Pgp in human glioblastoma cells and resulting in decreased sensitivity to etoposide (VP-16) and doxorubicin (41). More recent reports showed that MDM2 bound to TopoII resulted in decreased cellular enzyme content. Knockdown of MDM2 by RNAi stabilized TopoIIalpha and decreased resistance to TopoII-targeting drugs (etoposide, mitoxantrone) (15). Another study found that Curcumin, a dietary component that has anticancer, chemosensitization, and radiosensitization effects down-regulates MDM2. Curcumin also inhibited the growth of these cells and enhanced the cytotoxic effects of gemcitabine (16, 42). The human THOC1 gene, also known as hHpr1 or p84, encodes a protein that was originally recognized as a nuclear matrix component that binds the retinoblastoma tumor suppressor protein (43). Changes in the nuclear matrix and resulting alterations in nuclear structure have been recognized to correlate with tumor progression (42). Indeed, overexpression of THOC1 has recently been documented in human breast cancer with THOC1 levels correlating with tumor size and metastases (44).
In conclusion, we identified PRP-4 and several other genes that, when knocked down, could partially reverse drug-resistance in the ovarian cancer cell line SKOV-3TR and OVCAR8TR. Significantly, overexpression of the PRP-4 protein in a drug-sensitive cell line was sufficient to increase paclitaxel resistance, suggesting that PRP-4 may directly participate in the development of clinical drug resistance. The apparent synergy that we observed between PRP-4 repression and paclitaxel sensitivity (Table 1), in comparison with other known drug resistance target genes such as MDR1, MDM2 and survivin, suggest that shRNA chemosensitizer screens could find molecular components closely related to the mode of action of a given drug. All of these genes, in addition to PRP-4, may be developed as new targets in improving cancer treatment using combination chemotherapy. A combination of low-dosage paclitaxel with MDR1, survivin, MDM2 or PRP-4 inhibitors could decrease the apoptotic threshold in drug resistant cells and may prove to be an effective anti-cancer strategy in vivo.
Supplementary Material
Acknowledgments
This project was supported by the Grant from the Ovarian Cancer Research Foundation (OCRF) and the Ovarian Cancer SPORE at DF/HCC. Dr. Duan is supported, in part, through a grant from the National Cancer Institute, NIH (Nanotechnology Platform Partnership), R01-CA119617. Support has also been provided by the Gaetagno and Wechsler funds. We thank Dr. Regis Giet for providing the PRP-4 antibodies. In addition, we would like to acknowledge Dr. Michael Seiden at Fox Chase Cancer Center for providing useful advice during the drafting of this manuscript.
Abbreviations
- shRNA
short hairpin RNA
- PRP-4
pre-mRNA Processing factor-4
- MDR1
multidrug resistance gene 1
Footnotes
Financial support: Ovarian Cancer Research Foundation (OCRF) and the Ovarian Cancer SPORE at DF/HCC.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.See HT, Kavanagh JJ, Hu W, Bast RC. Targeted therapy for epithelial ovarian cancer: current status and future prospects. Int J Gynecol Cancer. 2003;13:701–34. doi: 10.1111/j.1525-1438.2003.13601.x. [DOI] [PubMed] [Google Scholar]
- 2.Yusuf RZ, Duan Z, Lamendola DE, Penson RT, Seiden MV. Paclitaxel resistance: molecular mechanisms and pharmacologic manipulation. Curr Cancer Drug Targets. 2003;3:1–19. doi: 10.2174/1568009033333754. [DOI] [PubMed] [Google Scholar]
- 3.Duan Z, Lamendola DE, Yusuf RZ, Penson RT, Preffer FI, Seiden MV. Overexpression of human phosphoglycerate kinase 1 (PGK1) induces a multidrug resistance phenotype. Anticancer Res. 2002;22:1933–41. [PubMed] [Google Scholar]
- 4.Lamendola DE, Duan Z, Yusuf RZ, Seiden MV. Molecular description of evolving paclitaxel resistance in the SKOV-3 human ovarian carcinoma cell line. Cancer Res. 2003;63:2200–5. [PubMed] [Google Scholar]
- 5.Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–95. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barre B, Vigneron A, Perkins N, Roninson IB, Gamelin E, Coqueret O. The STAT3 oncogene as a predictive marker of drug resistance. Trends Mol Med. 2007;3:4–11. doi: 10.1016/j.molmed.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 7.van Vlerken LE, Duan Z, Seiden MV, Amiji MM. Modulation of intracellular ceramide using polymeric nanoparticles to overcome multidrug resistance in cancer. Cancer Res. 2007;67:4843–50. doi: 10.1158/0008-5472.CAN-06-1648. [DOI] [PubMed] [Google Scholar]
- 8.Duan Z, Lamendola DE, Duan Y, Yusuf RZ, Seiden MV. Description of paclitaxel resistance-associated genes in ovarian and breast cancer cell lines. Cancer Chemother Pharmacol. 2005;55:277–85. doi: 10.1007/s00280-004-0878-y. [DOI] [PubMed] [Google Scholar]
- 9.Duan Z, Brakora KA, Seiden MV. MM-TRAG (MGC4175), a novel intracellular mitochondrial protein, is associated with the taxol- and doxorubicin-resistant phenotype in human cancer cell lines. Gene. 2004;340:53–9. doi: 10.1016/j.gene.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Bosher JM, Labouesse M. RNA interference: genetic wand and genetic watchdog. Nat Cell Biol. 2000;2:E31–6. doi: 10.1038/35000102. [DOI] [PubMed] [Google Scholar]
- 11.Duan Z, Brakora KA, Seiden MV. Inhibition of ABCB1 (MDR1) and ABCB4 (MDR3) expression by small interfering RNA and reversal of paclitaxel resistance in human ovarian cancer cells. Mol Cancer Ther. 2004;3:833–8. [PubMed] [Google Scholar]
- 12.Cullen BR. Induction of stable RNA interference in mammalian cells. Gene Ther. 2006;13:503–8. doi: 10.1038/sj.gt.3302656. [DOI] [PubMed] [Google Scholar]
- 13.Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Dann CT. New technology for an old favorite: lentiviral transgenesis and RNAi in rats. Transgenic Res. 2007;16:571–80. doi: 10.1007/s11248-007-9121-z. [DOI] [PubMed] [Google Scholar]
- 15.Nayak MS, Yang JM, Hait WN. Effect of a single nucleotide polymorphism in the murine double minute 2 promoter (SNP309) on the sensitivity to topoisomerase II-targeting drugs. Cancer Res. 2007;67:5831–9. doi: 10.1158/0008-5472.CAN-06-4533. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Zhang Z, Hill DL, Wang H, Zhang R. Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 2007;67:1988–96. doi: 10.1158/0008-5472.CAN-06-3066. [DOI] [PubMed] [Google Scholar]
- 17.Zaffaroni N, Daidone MG. Survivin expression and resistance to anticancer treatments: perspectives for new therapeutic interventions. Drug Resist Updat. 2002;5:65–72. doi: 10.1016/s1368-7646(02)00049-3. [DOI] [PubMed] [Google Scholar]
- 18.Duan Z, Feller AJ, Penson RT, Chabner BA, Seiden MV. Discovery of differentially expressed genes associated with paclitaxel resistance using cDNA array technology: analysis of interleukin (IL) 6, IL-8, and monocyte chemotactic protein 1 in the paclitaxel-resistant phenotype. Clin Cancer Res. 1999;5:3445–53. [PubMed] [Google Scholar]
- 19.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–42. [PubMed] [Google Scholar]
- 20.Montembault E, Dutertre S, Prigent C, Giet R. PRP4 is a spindle assembly checkpoint protein required for MPS1, MAD1, and MAD2 localization to the kinetochores. J Cell Biol. 2007;179:601–9. doi: 10.1083/jcb.200703133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haimeur A, Conseil G, Deeley RG, Cole SP. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab. 2004;5:21–53. doi: 10.2174/1389200043489199. [DOI] [PubMed] [Google Scholar]
- 22.Litman T, Druley TE, Stein WD, Bates SE. From MDR to MXR: new understanding of multidrug resistance systems, their properties and clinical significance. Cell Mol Life Sci. 2001;58:931–59. doi: 10.1007/PL00000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buys TP, Chari R, Lee EH, et al. Genetic changes in the evolution of multidrug resistance for cultured human ovarian cancer cells. Genes Chromosomes Cancer. 2007;46:1069–79. doi: 10.1002/gcc.20492. [DOI] [PubMed] [Google Scholar]
- 24.Benz CC, Scott GK, Sarup JC, et al. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24:85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 25.Isonishi S, Hom DK, Thiebaut FB, et al. Expression of the c-Ha-ras oncogene in mouse NIH 3T3 cells induces resistance to cisplatin. Cancer Res. 1991;51:5903–9. [PubMed] [Google Scholar]
- 26.Miyashita T, Reed JC. bcl-2 gene transfer increases relative resistance of S49.1 and WEHI7.2 lymphoid cells to cell death and DNA fragmentation induced by glucocorticoids and multiple chemotherapeutic drugs. Cancer Res. 1992;52:5407–11. [PubMed] [Google Scholar]
- 27.Duan Z, Lamendola DE, Penson RT, Kronish KM, Seiden MV. Overexpression of IL-6 but not IL-8 increases paclitaxel resistance of U-2OS human osteosarcoma cells. Cytokine. 2002;17:234–42. doi: 10.1006/cyto.2001.1008. [DOI] [PubMed] [Google Scholar]
- 28.Duan Z, Duan Y, Lamendola DE, et al. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin-resistant human cancer cell lines. Clin Cancer Res. 2003;9:2778–85. [PubMed] [Google Scholar]
- 29.Rosenberg GH, Alahari SK, Kaufer NF. prp4 from Schizosaccharomyces pombe, a mutant deficient in pre-mRNA splicing isolated using genes containing artificial introns. Mol Gen Genet. 1991;226:305–9. doi: 10.1007/BF00273617. [DOI] [PubMed] [Google Scholar]
- 30.Kiger AA, Baum B, Jones S, et al. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross T, Lutzelberger M, Weigmann H, Klingenhoff A, Shenoy S, Kaufer NF. Functional analysis of the fission yeast Prp4 protein kinase involved in pre-mRNA splicing and isolation of a putative mammalian homologue. Nucleic Acids Res. 1997;25:1028–35. doi: 10.1093/nar/25.5.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyata Y, Nishida E. Distantly related cousins of MAP kinase: biochemical properties and possible physiological functions. Biochem Biophys Res Commun. 1999;266:291–5. doi: 10.1006/bbrc.1999.1705. [DOI] [PubMed] [Google Scholar]
- 33.Kojima T, Zama T, Wada K, Onogi H, Hagiwara M. Cloning of human PRP4 reveals interaction with Clk1. J Biol Chem. 2001;276:32247–56. doi: 10.1074/jbc.M103790200. [DOI] [PubMed] [Google Scholar]
- 34.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 35.Tang-Wai DF, Kajiji S, DiCapua F, de Graaf D, Roninson IB, Gros P. Human (MDR1) and mouse (mdr1, mdr3) P-glycoproteins can be distinguished by their respective drug resistance profiles and sensitivity to modulators. Biochemistry. 1995;34:32–9. doi: 10.1021/bi00001a005. [DOI] [PubMed] [Google Scholar]
- 36.Kleinberg L, Florenes VA, Nesland JM, Davidson B. Survivin, a member of the inhibitors of apoptosis family, is down-regulated in breast carcinoma effusions. Am J Clin Pathol. 2007;128:389–97. doi: 10.1309/E899BG1282M5D505. [DOI] [PubMed] [Google Scholar]
- 37.Cohen C, Lohmann CM, Cotsonis G, Lawson D, Santoianni R. Survivin expression in ovarian carcinoma: correlation with apoptotic markers and prognosis. Mod Pathol. 2003;16:574–83. doi: 10.1097/01.MP.0000073868.31297.B0. [DOI] [PubMed] [Google Scholar]
- 38.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladanyi M, Cha C, Lewis R, Jhanwar SC, Huvos AG, Healey JH. MDM2 gene amplification in metastatic osteosarcoma. Cancer Res. 1993;53:16–8. [PubMed] [Google Scholar]
- 40.Cordon-Cardo C, Latres E, Drobnjak M, et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–9. [PubMed] [Google Scholar]
- 41.Kondo S, Kondo Y, Hara H, et al. mdm2 gene mediates the expression of mdr1 gene and P-glycoprotein in a human glioblastoma cell line. Br J Cancer. 1996;74:1263–8. doi: 10.1038/bjc.1996.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat Rev Cancer. 2004;4:677–87. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 43.Durfee T, Mancini MA, Jones D, Elledge SJ, Lee WH. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J Cell Biol. 1994;127:609–22. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Lin AW, Zhang X, Wang Y, Wang X, Goodrich DW. Cancer cells and normal cells differ in their requirements for Thoc1. Cancer Res. 2007;67:6657–64. doi: 10.1158/0008-5472.CAN-06-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.