SUMMARY
Transmission of highly pathogenic avian influenza across species into human populations can result in lethal infections and increases the potential of a pandemic outbreak. A major determinant of species tropism is the influenza polymerase subunit PB2. We show here that a dominant inhibitory activity in human cells potently and selectively restricts the function of polymerase containing an avian-like PB2 with glutamic acid at residue 627. This inhibitory activity dramatically reduces virus production by polymerases containing PB2 K627E without reducing their relative infectivity. Restricted polymerases fail to assemble into ribonucleoprotein complexes, resulting in decreased genome transcription, replication, and virus production. However, both wild-type and mutant PB2 bind to viral nucleoprotein when expressed alone, suggesting that species-specific conformational alterations within the polymerase trimer disrupt ribonucleoprotein assembly. Understanding the molecular basis for influenza virus host cell specificity and pathogenesis will enable rational strategies to treat and prevent future avian influenza outbreaks in humans.
INTRODUCTION
Recurring outbreaks of highly pathogenic H5N1 avian influenza virus in domestic and wild bird populations pose a serious threat to public health. Outbreaks decimate bird populations and can result in severe and often fatal avian influenza virus infections in humans (Chen et al., 2006b; Chen et al., 2005; Hatta et al., 2001; Maines et al., 2005). Because transmission of influenza virus from bird to human populations has the potential to establish new pandemics, understanding species tropism and virulence of these viruses is critical to prevention and treatment efforts (Subbarao et al., 2006).
Along with its central role in viral replication and gene expression, the influenza virus RNA-dependent RNA polymerase is a major determinant of viral tropism and pathogenicity. The influenza polymerase is a ~250 kD heterotrimer composed of three polypeptides: PB1, PB2, and PA. The polymerase associates with viral RNA and the viral nucleoprotein (NP) to form ribonucleoprotein (RNP) complexes that mediate transcription via a “cap-snatching” technique that utilizes short host-derived 7mG-capped RNAs as primers for synthesis of influenza mRNAs (Supplemental Figure 1A) (Elton et al., 2005). RNPs also direct replication of the minus-sense viral RNA genome (vRNA) through a plus-sense complementary RNA (cRNA) intermediate. Following replication, vRNA segments are packaged as vRNPs into newly budding virions. The PB2 subunit has long been identified as a host-range determinant (Almond, 1977); replication in human and avian systems depends on the identity of amino acid 627 (Subbarao et al., 1993). Glutamic acid predominates at position 627 in avian PB2, whereas lysine is present almost exclusively at this position in human isolates (Chen et al., 2006a). The presence of a lysine at residue 627 enhances viral replication and pathogenicity in mammalian model systems (Hatta et al., 2001; Munster et al., 2007; Salomon et al., 2006) and is associated with lethality in humans infected with H5N1 and H7N7 avian viruses (Fouchier et al., 2004; Gao et al., 1999). Conversely, the presence of a glutamic acid at this position severely attenuates replication efficiency and pathogenicity in mammalian systems (Hatta et al., 2001; Shinya et al., 2004).
Amino acid 627 lies outside of the identified functional domains and subunit-binding sites in PB2 and the molecular basis for restricted polymerase function has not been established. By analyzing the function of PB2 variants in transient heterokaryons formed between human and avian cells, we show here that a dominant inhibitory activity in human cells potently and selectively restricts the function of avian-like PB2 K627E. This inhibitor dramatically reduces viral production by polymerases containing PB2 K627E without reducing the relative infectivity of the virions that are produced. The restricted polymerase fails to both bind NP and assemble into vRNP complexes in human cells, resulting in decreased genome transcription, replication, and virus production. However, both WT and mutant PB2 bind to NP when expressed alone, suggesting that it is the conformation of the polymerase trimer that is altered by changes at PB2 residue 627. The blockade is relieved during late stages of viral infection, allowing assembly of PB2 K627E-containing vRNP and virus budding. Our data indicate that the blockade targets the polymerase at an early stage during replication. Thus, adaptation of avian PB2 to replication in human cells involves escape from species-specific restriction(s) of polymerase function. Exploiting this inhibitory activity may provide for new prevention strategies.
RESULTS
Dominant restriction of avian-like influenza polymerase in human cells
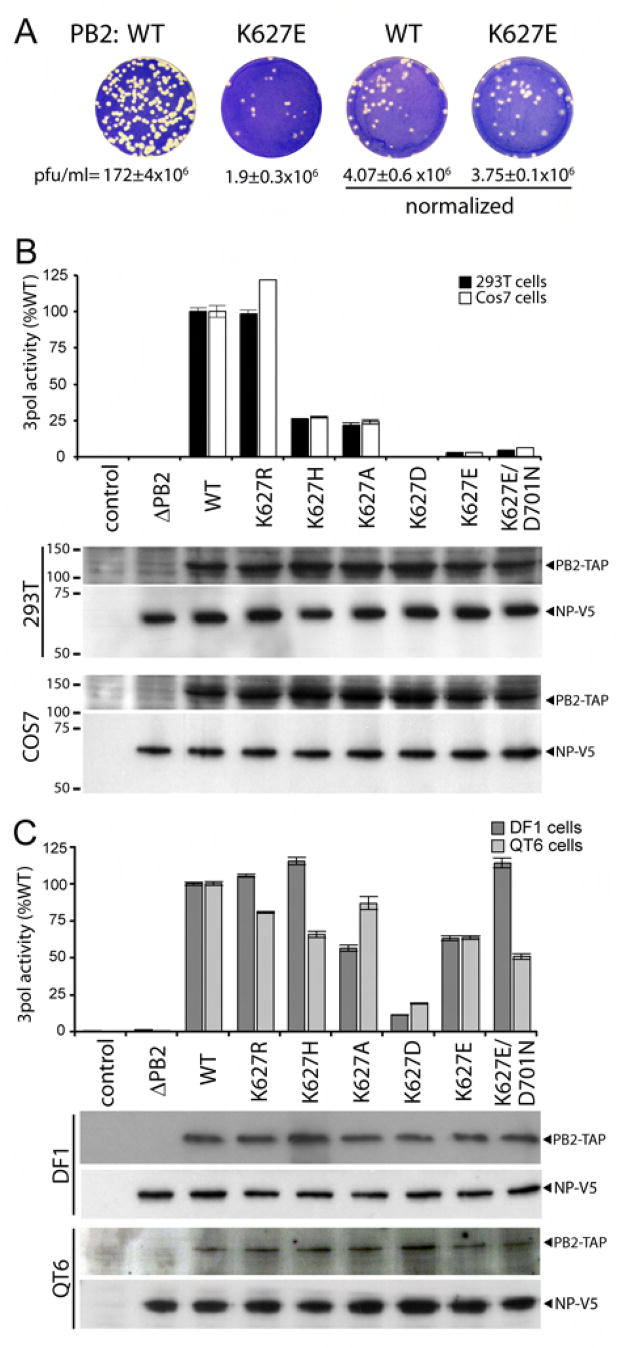
The avian-signature glutamic acid at position 627 in PB2 has been associated with reduced virus replication in mammalian cells, mice, ferrets, and humans (Clements et al., 1992; Fouchier et al., 2004; Gao et al., 1999; Hatta et al., 2001; Munster et al., 2007; Salomon et al., 2006; Shinya et al., 2004; Subbarao et al., 1993). To determine whether this reduction results from decreased virion production, viral infectivity or a combination of the two, we measured the infectivity of normalized amounts of recombinant virus. Recombinant A/WSN/33 virus was produced in 293T cells in the presence of PB2 containing a lysine at position 627 (WT) or the PB2 mutant containing glutamic acid at this position (K627E). For simplicity, we refer to PB2 K627E as “avian-like,” recognizing that additional amino acid differences might exist between PB2 from A/WSN/33 and various avian strains. The use of a plasmid-based reverse genetics system permitted analysis of individual residues in isogenic virus backgrounds and initially ensured equivalent expression of WT and K627E PB2 proteins independent of their functionality. Plaque assays were performed on canine (MDCK) cells which display only a minor dependence on PB2 amino acid 627 for the lab-adapted strain A/WSN/33 (data not shown) and recent H5N1 isolates (Hatta et al., 2007; Labadie et al., 2007; Massin et al., 2005; Shinya et al., 2004), whereas other isolates containing a glutamic acid at position 627 do not replicate efficiently in MDCK cells (Subbarao et al., 1993). The PB2 K627E mutation reduced the infectious plaque assay titer to only ~1% compared to that of WT, similar to previous observations (Figure 1A). This decreased infectious titer was associated with a concomitant decrease in virion production. PB2 K627E viral supernatants contained 4.6% of the hemagglutination activity possessed by WT (data not shown). Both WT and K627E PB2 displayed similar infectious titers when the viral inoculum was normalized by hemagglutination titer (Figure 1A). Thus, the PB2 K627E mutation reduces virus replication primarily by reducing the production of viral particles, whereas the particles that are produced are as infectious as WT.
Figure 1. PB2 amino acid 627 decreases virus production by regulating polymerase activity in human cells.
(A) Recombinant A/WSN/33 virus was produced in 293T cells expressing WT or K627E PB2. Viral titers (plaque forming units (pfu)/ml) were measured by plaque assays on MDCK cells (n=2–3+/− standard deviation) before and after normalizing inoculums based on hemaggluttination units. (B) Viral polymerase activity was determined in primate cells transfected with expression plasmids for PB1, PA, NP, the indicated PB2-TAP, and a vRNA-luciferase reporter plasmid. Control cells were transfected with reporter plasmid alone. Luciferase activity was determined in cell extracts. PB2-TAP and NP-V5 were detected by western blot. Molecular weights are shown in kilodaltons. (C) Polymerase activity assays were performed in the avian cells as in (B), except the vRNA-luciferase reporter was driven by a chicken pol I promoter. For all activity assays, n=3 +/− standard deviation.
To further understand the defects associated with a polymerase containing PB2 K627E, we utilized a cell-based polymerase activity assay to determine the species-specific amino acid requirements at this position (Supplemental Figure 1B). Human 293T cells were transfected with vectors expressing PB1, PA, NP and wild-type or mutant TAP-tagged PB2 (PB2-TAP) cloned from the mouse-adapted lab strain A/WSN/33 based on a human influenza isolate. Wild-type PB2, in the context of human isolates, contains a lysine at position 627. The presence of a C-terminal TAP tag on PB2 does not significantly alter function (Engelhardt et al., 2005; Fodor and Smith, 2004). Cells were co-transfected with a reporter vector expressing minus-sense luciferase RNA flanked by neuraminidase viral RNA sequences (Regan et al., 2006). The vRNA-luciferase reporter is replicated and transcribed only in the presence of functional influenza polymerase. Polymerase reconstituted with WT PB2 produced significant amounts of luciferase as measured by a luciferase activity assay, whereas control cells transfected as above but with reporter alone or with polymerase expression plasmids lacking PB2 (ΔPB2) produced only background levels of luciferase (<0.5%) (Figure 1B). Mutation of PB2 to the avian-like K627E severely impaired luciferase production; luciferase activity was reduced to only ~2.5% WT activity, in agreement with previous reports demonstrating reduced function of polymerase containing a glutamic acid at PB2 residue 627 in human cells (Labadie et al., 2007; Naffakh et al., 2000; Salomon et al., 2006). The conservative PB2 K627R mutation functioned at levels similar to WT. K627H and K627A mutations reduced polymerase activity, although these mutants still retained ~25% WT activity. The K627D mutation demonstrated the most profound defect, with polymerase activity indistinguishable from background. Western blot analysis detected similar levels of NP and WT or mutant PB2 expression, suggesting that differences in polymerase function were not the result of differential protein expression, stability, or transfection efficiency. Similar results were observed when experiments were repeated in the African green monkey cells Cos7 (Figure 1B) and human HeLa cells (Supplemental Figure 2), indicating generally decreased function of avian-like PB2 K627E in primate cells. In addition, similar results were observed whether polymerase activity assays were performed at 33, 37, or 41°C (data not shown). These data show that a basic residue at position 627 is critical for production of viral mRNA and that acidic residues at this position severely compromise transcriptional activity in primate cells.
To gain further insight into the mechanism of restriction, we investigated polymerase activity at the cellular level. The luciferase-based assays measure polymerase activity in bulk, making it unclear if the polymerase activity associated with PB2 K627E is equally impaired throughout the entire cell population or if the reduction in activity as compared to wild type results from a stochastic event where only a subset of cells restrict polymerase function. To differentiate these two possibilities, we measured polymerase activity utilizing a vNS-GFP reporter plasmid (Supplemental Figure 3). GFP was produced by WT polymerase less than 20 hr post-transfection and was saturated within 48 hr. By contrast, in cells expressing PB2 K627E GFP appeared with delayed kinetics at lower levels throughout the entire population. RFP was expressed equivalently in both transfections and GFP was present in a similar number of cells at 70 hr post-transfection for WT and PB2 K627E indicating equivalent transfection efficiencies. These data suggest that the reduction in polymerase activity does not occur randomly within a population, but rather is an intrinsic property of all cells in this culture.
To determine if the decreased transcription associated with mutations at position 627 was the result of a general polymerase defect or a host-specific restriction, we performed polymerase activity assays in avian cells. Western blotting showed similar levels of NP and WT or mutant PB2 for each assay, indicating equivalent transfection efficiency (Figure 1C). Polymerase activity of all PB2 mutants restricted in human cells was significantly enhanced in both chicken DF1 cells and quail QT6 cells (Figure 1C). Specifically, PB2 K627E increased from ~2.5% WT activity in primate cells to over 70% WT activity in avian cells. PB2 K627D transcription was also increased, but this mutant still displayed only modest activity. Together, these data demonstrate that a variety of mutations are tolerated at PB2 residue 627 for the production of functional polymerase in avian cells. Thus, the predominance of glutamic acid at position 627 in avian influenza populations (Chen et al., 2006a) is not due solely to a requirement for polymerase activity, but might result from additional selective pressures to maintain glutamic acid at this position. Furthermore, the activity of these mutants in avian cells suggests that their decreased activity in primate cells does not reflect an intrinsic defect in polymerase function, but rather represents a species-specific restriction.
Recent clinical specimens of H5N1 isolated from lethal human infections possess the avian-signature glutamic acid at position 627 in PB2, but a small fraction also have an asparagine at position 701, versus the aspartic acid typical of human strains (de Jong et al., 2006). It is possible that the D701N change might compensate for the decreased polymerase activity associated with glutamic acid at position 627 in humans. However, the polymerase activity of the double-mutant PB2 K627E/D701N remained significantly impaired in 293T and Cos7 cells, with activity similar to PB2 K627E (Figure 1B). The double mutation did not enhance activity in quail QT6 cells, whereas the paired mutations completely restored activity to WT levels in chicken DF1 cells (Figure 1C). These results suggest that in some hosts these changes might represent compensatory mutations, but in primate cells the conversion of position 701 to asparagine does not compensate for the defects caused by PB2 K627E mutation in the context of the A/WSN/33 polymerase.
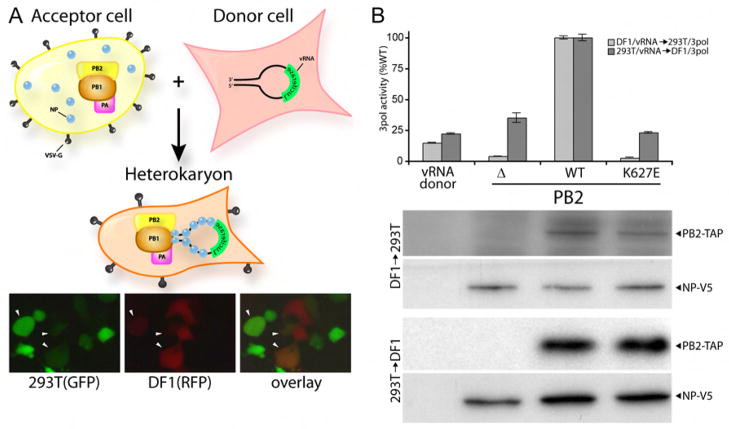
The species-restricted functionality of PB2 K627E suggests at least two possibilities: permissive avian cells might contain a factor or activity that compensates for an inherent defect associated with PB2 K627E, or non-permissive primate cells contain an activity that selectively impairs these polymerases. We tested these two possibilities with a transient heterokaryon assay in which human and avian cell hybrids were formed by the fusogenic VSV-G protein (Figure 2A). If avian cells contain a compensatory activity, DF1 cells expressing vRNA-luciferase are predicted to rescue the activity of polymerase containing PB2 K627E expressed in restrictive human cells upon fusion. Conversely, if human cells contain an inhibitory activity, fusion of 293T cells expressing vRNA-luciferase to polymerase-expressing avian cells is predicted to impair the otherwise functional PB2 K627E. Critically, replication complexes only form in cells that have undergone fusion. Fusion efficiency was monitored by the appearance of dual-fluorescent cells in parallel experiments where donor and acceptor cells expressed RFP and GFP, respectively (Figure 2A). Fusion of donor DF1 cells to 293T cells expressing polymerase reconstituted polymerase activity for WT PB2, but not PB2 K627E (Figure 2B). Activity of the PB2 K627E polymerase was similar to the activity observed in the absence of PB2, which was less than that observed in donor DF1 cells alone. No polymerase activity was observed in unfused cells (data not shown). These data suggest that avian cells do not contain an activity or factor that compensates for mutations at PB2 residue 627. Fusion of donor 293T cells to DF1 cells recovered polymerase activity for WT polymerase, but severely impaired the function of polymerase containing PB2 K627E (Figure 2B). PB2 K627E polymerase activity was reduced to background levels, compared to the ~70% activity displayed by this mutant when reporter assays were performed in unfused DF1 and QT6 cells (Figure 1C). Similar results were observed when heterokaryon formation was mediated by polyethylene glycol (Supplemental Figure 4). Western blotting of lysates from mixed cell cultures shows equivalent levels of WT and mutant PB2-TAP and NP, indicating that the lack of polymerase activity did not result from changes in protein stability upon heterokaryon formation (Figure 2B). Together, these data provide strong evidence that human cells contain an activity that selectively inhibits polymerase with the avian-signature PB2 K627E.
Figure 2. A dominant inhibitory activity restricts PB2 function in human cells.
(A) Experimental strategy for formation of heterokaryons by expression of the fusogenic VSV-G protein. Heterokaryon formation between 293T cells expressing GFP and DF1 cells expressing RFP was monitored by the detection of dual-fluorescent cells (arrowheads). (B) Polymerase activity in heterokaryons expressing NP, PB1, PA, and no PB2, WT, or K627E PB2-TAP was determined by luciferase assay. Protein expression was monitored by western blot. n=3 +/− standard deviation.
Polymerases containing mutant PB2 assemble and localize to the nucleus
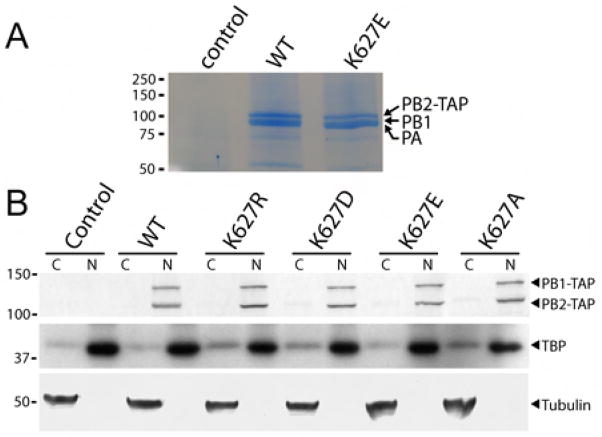
The heterokaryon assays suggest the presence of an activity in primate cells that restricts the function of the avian-like PB2 K627E polymerase, but do not indicate the specific molecular defects associated with this inhibition. One possible explanation is a failure to assemble the trimeric polymerase complex. To test this possibility, we purified WT and K627E PB2-TAP polymerase complexes from 293T cell nuclear extracts. Both WT and mutant PB2-TAP co-precipitated approximately stoichiometric amounts of PB1 and PA (Figure 3A and data not shown). PB1 and PA migrate at similar positions in this gel system and have been identified by mass spectrometry in similar experiments (data not shown). Thus, PB2 K627E does not prevent assembly of the trimeric viral polymerase.
Figure 3. Avian-like PB2 mutants form trimeric polymerase complexes in the nucleus.
(A) Influenza polymerase was TAP-tag purified from nuclear extracts of 293T cells expressing PB1, PA, and PB2-TAP. Proteins were detected by Coomassie staining. (B) Subcellular localization of PB1 and PB2 were determined by western blotting of nuclear (N) and cytoplasmic (C) extracts prepared from 293T cells transfected with expression vectors for PB1-TAP, PA, and the indicated PB2-TAP. The identity of each fraction was confirmed by western blotting for tubulin, a cytoplasmic protein, and TATA-binding protein (TBP), a nuclear protein. Molecular weight in kilodaltons is shown.
Another possible explanation is alteration of the subcellular localization of mutant polymerase. Influenza polymerase-catalyzed transcription and viral replication occurs within the nucleus of infected cells (Herz et al., 1981; Jackson et al., 1982). The polymerase subunits rely on cellular proteins for nuclear import, and changes in the C-terminus of PB2 have been associated with species-specific defects in subcellular localization (Gabriel et al., 2008; Naito et al., 2007). Therefore, we assayed the subcellular distribution of various polymerase complexes. Western blotting of nuclear and cytoplasmic fractions of 293T cells shows that all PB2 variants localized almost exclusively in the nucleus (Figure 3B). PB1 co-localized with the PB2 variants, consistent with the ability of mutant PB2 to associate with PB1 and PA as demonstrated above. Control western blots for proteins found primarily in the cytoplasm, tubulin, and the nucleus, TATA-binding protein, confirm the identity of the subcellular fractions and demonstrate minimal cross-contamination (Figure 3B). These data indicate that mutation of PB2 residue 627 does not prevent polymerase assembly or proper subcellular localization.
PB2 K627E mutation impairs assembly of ribonucleoprotein complexes without disrupting the NP binding site
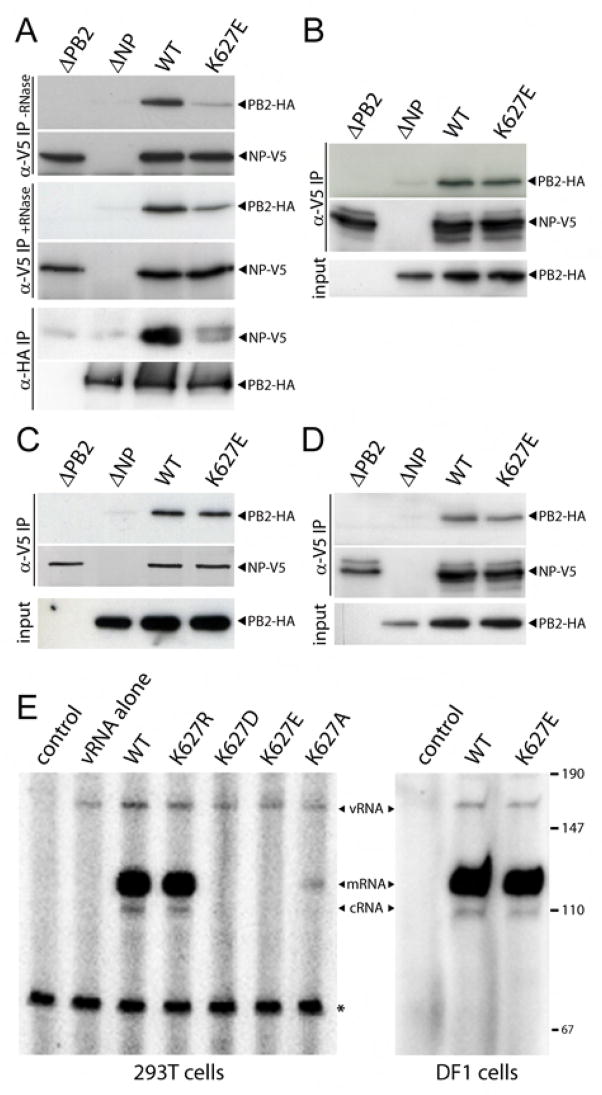
Assembly of the vRNP, a complex consisting of vRNA coated by NP and the polymerase, is essential to both viral replication and transcription. NP is a major structural protein that binds both vRNA and cRNA and interacts with the PB1 and PB2 subunits to facilitate polymerization (Biswas et al., 1998; Medcalf et al., 1999). PB2 from a primary avian isolate with a glutamic acid at position 627 was found to immunoprecipitate lower amounts of NP in human cells relative to that observed for a mutant containing lysine at this position (Labadie et al., 2007). We confirmed that the inhibitory activity in human cells correlates with decreased RNP assembly; WT polymerase was specifically co-precipitated by NP whereas precipitation of PB2 K627E polymerase was significantly decreased (Figure 4A). Treatment of cell lysates with RNase, and the presumptive disassembly of vRNPs, did not restore interaction between NP and PB2 K627E (Figure 4A), suggesting that the binding defect is largely independent of vRNA. The low amounts of PB2 K627E precipitated by NP might account for the low infectious titers and activity of this polymerase detected in 293T cells (Figure 1). Similar results were observed in reciprocal immunoprecipitations, where significantly less NP was precipitated by polymerase containing PB2 K627E compared to WT (Figure 4A). However, WT and PB2 K627E polymerase were equally precipitated from lysates prepared from transfected avian DF1 cells (Figure 4B). The chaperone protein Hsp90 has been identified as a cellular cofactor in polymerase assembly (Naito et al., 2007). WT and mutant PB2 both co-precipitated Hsp90 from 293T cell extracts, indicating that assembly defects are not simply the result of a failure to bind Hsp90 (Supplemental Figure 5).
Figure 4. PB2 K627E mutation prevents assembly of vRNP without disrupting the NP binding site.
Cell lysates prepared from 293T (A) or DF1 (B) cells transfected with expression vectors for PB1, PA, NP, WT or K627E PB2-HA, and the vRNA-luciferase reporter were subject to immunoprecipitation and western blotting. Where indicated, lysates were treated with RNase. (C) and (D) Immunoprecipitations and western blots were performed on lysates from 293T (C) and DF1 (D) cells expressing NP and WT or K627E PB2. The levels and ratio of PB2 and NP were the same as in (A) and (B). (E) Primer extension assays were used to detect RNAs produced in 293T cells or DF1 cells expressing viral polymerases containing WT or mutant PB2 in the presence of a vRNA-reporter. Products are indicated and the size of a DNA standard is shown in nucleotides. A non-specific band (*) serves as an internal loading control.
PB2 interacts with NP independent of PB1 and PA, possibly via a C-terminal binding region that might encompass amino acid 627 (Biswas et al., 1998; Medcalf et al., 1999; Poole et al., 2004). To further probe the binding between NP and PB2 K627E, we expressed only these two proteins in 293T and DF1 cells. To our surprise, and in contrast to previous results (Labadie et al., 2007), WT and PB2 K627E were immunoprecipitated at similar levels when expressed in either human or avian cells (Figure 4C and D). Thus, PB2 K627E is competent to bind NP when expressed alone, but not in the context of the trimeric holoenzyme. Furthermore, these data suggest that changes at amino acid 627 do not disrupt the NP binding site(s) on PB2. Consequently, these results suggest that the species-restricted activity of PB2 K627E is the result of a defect(s) in polymerase assembly or conformation that precludes binding to NP within the vRNP complex.
To directly assess the consequences of defective vRNP assembly on enzyme activity, we measured the products of vRNA replication (vRNA and cRNA) and transcription (mRNA). Primer extension assays were performed on total RNA extracted from 293T cells expressing the polymerase, NP, and the vRNA-luciferase reporter. Both plus-sense and minus-sense viral RNAs were detected in the presence of WT polymerase, but not in untransfected cells (Supplemental Figure 6). Viral mRNA is the predominant species, as previously reported (Fodor and Smith, 2004). Similar results were obtained when probes were used alone or in combination; therefore, tandem primer extension analysis was performed to analyze viral RNAs. Primer extension demonstrates that both WT and PB2 K627R polymerase synthesized significant amounts of mRNA, cRNA, and increased levels of vRNA, quantified at 2.3- and 1.9-fold, respectively, over that of the reporter vector alone (Figure 4E). This is consistent with the high levels of luciferase detected in the polymerase activity assays in human cells (Figure 1B). By contrast, only low levels of mRNA were detected in the presence of PB2 K627A, and no synthesis of any viral RNAs was detected above background in the presence of PB2 K627D or K627E (Figure 4E). vRNA, cRNA and mRNA were detected in DF1 cells expressing WT or PB2 K627E polymerase (Figure 4E), indicating that WT and mutant PB2 support both replication and transcription of viral RNAs in avian cells. Quantification shows that PB2 K627E produces ~80% of the mRNA product compared to WT (data not shown), in agreement with results from the polymerase activity assays (Figure 1C). These data demonstrate that species-restricting mutations in PB2 prevent both replication and transcription of vRNA in human, but not avian, cells. mRNA is produced shortly after infection (Glass et al., 1975), and thus it appears that inhibition of binding of PB2 K627E polymerase to NP disrupts one of the earliest steps of viral infection.
Recovery of mutant PB2 function during virus assembly
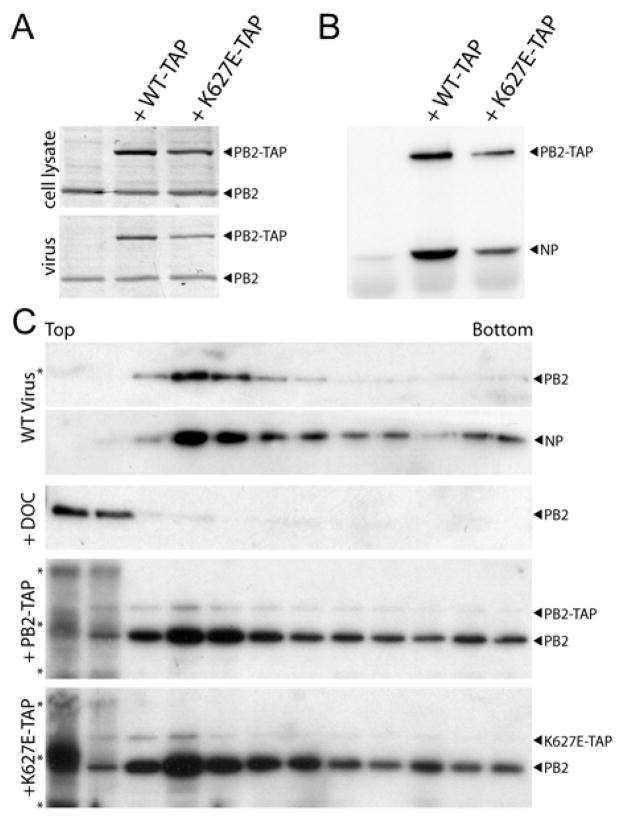
Because the block to PB2 K627E activity in primate cells occurs during the early stages of viral gene expression and genome replication, we next tested whether co-expressing WT PB2 can bypass an early blockade and rescue activity of PB2 K627E during later stages of virus production. Recombinant virus was produced in 293T cells by transfection of a genomic expression vector and plasmids encoding NP, PB1, PA, PB2, and an equivalent amount of WT or mutant PB2-TAP where indicated. Western blot analysis of cell lysates indicated that PB2 and PB2-TAP were expressed in virus-producing cells (Figure 5A). Both WT and K627E PB2-TAP were detected in cell-free virions harvested at 24 hr (Figure 5A), 48 hr, and 72 hr post-transfection (data not shown). Thus, even though our data indicate that PB2 K627E is defective at early stages of virus replication, it is still packaged into virions at levels similar to WT PB2-TAP when untagged PB2 is co-expressed, raising the possibility that the defects associated with K627E might be absent during late stages of infection or alleviated by WT PB2.
Figure 5. Defective assembly of mutant polymerase into vRNP is overcome during late stages of infection.
(A) Recombinant virus was produced in 293T cells transfected with pTM-pol I-WSN-All and expression vectors for NP, PB1, PA, PB2, and WT or K627E PB2-TAP. PB2 and PB2-TAP were detected by western blot of cell lysate or cell-free virions with anti-PB2 serum. (B) PB2-TAP was precipitated with rabbit IgG-agarose from virus-producing cell lysates. Co-precipitating NP was detected by anti-NP western blot. (C) Purified virus was lysed and separated on a linear 30–60% glycerol gradient. Where indicated, 1 % deoxycholate (DOC) was present during viral lysis. Fractions were collected and PB2 and NP were detected by western blot. Non-specific bands at the top of the gradient are indicated (*).
To further test this hypothesis, we assayed binding between PB2-TAP and NP in virus-producing cells (Figure 5B). WT and K627E PB2-TAP co-precipitated NP at similar relative amounts from cell lysate, providing further evidence that the K627E mutant is functional during late stages of virus production. This is in contrast to the extremely low amounts of NP co-precipitated when PB2 K627E polymerase was expressed without WT PB2 (Figure 4A). To eliminate the possibility that NP co-precipitation was due to PB2-TAP that was not assembled into a trimeric polymerase complex or that virion-associated PB2-TAP was non-specifically packaged, we analyzed the incorporation of PB2-TAP into vRNPs within virions. Glycerol gradient centrifugation was performed using vRNPs isolated from cell-free virions. Free, non-specifically incorporated polymerase is predicted to remain at the top of the gradient, whereas polymerase associated with vRNPs migrates into the gradient (Klumpp et al., 2001). Similar amounts of lysed viral cores were separated on a linear 30–60% glycerol gradient and fractions were analyzed by western blot (Figure 5C). WT PB2 migrated into the gradient and cosedimented with fractions containing the majority of NP. As expected, free PB2 released from vRNPs by treatment with 1% deoxycholate remained at the top of the gradient. By contrast, glycerol gradients of viruses containing WT PB2 in combination with WT or K627E PB2-TAP demonstrate that the PB2-TAP variants migrated into the gradient with a profile similar to that of untagged PB2. Blotting of these gradients reveals that although less PB2-TAP was packaged into virions compared to untagged PB2, PB2-TAP still sedimented with vRNPs. These results indicate that WT and K627E PB2-TAP polymerases are incorporated into virions as vRNPs. This is consistent with our earlier observation that virions containing a PB2 K627E polymerase were equally infectious compared to those with a WT polymerase once normalized to hemagglutinin content, even though the overall infectious viral titers were reduced 100-fold (Figure 1). Together, these data suggest that the inhibitory activity targeting PB2 K627E is absent during virus assembly and budding.
DISCUSSION
Cross-species transmission of influenza from birds to humans requires the adaptation of viral genes to the new host. Several species-specific determinants of viral replication and pathogenicity have been identified, including adaptations in HA that enhance processing and govern receptor specificity, NS1 sequences that deregulate innate anti-viral responses, and a point mutant in PB2 that increases nuclear localization in human cells (Gabriel et al., 2008; Govorkova et al., 2005; Hatta et al., 2001; Jackson et al., 2008; Krug, 2006). We have shown here that changes within the PB2 subunit of the influenza virus polymerase are necessary to overcome an inhibitory activity or factor in human cells that selectively restricts the function of an avian-like polymerase. This inhibitory activity prevents assembly of vRNPs, thereby disrupting genomic replication and the production of viral mRNA, a critical step early in virus replication. This potent block early in infection suggests that an inhibitory factor is present in mammalian cells prior to infection, perhaps constitutively expressed. However, the inhibitory effect is relieved during later stages of the viral life cycle. Virions that were produced by PB2 K627E polymerase were as infectious as WT, suggesting that residual polymerase activity was sufficient to make new virions and that the processes of vRNP export, virion assembly and budding are unaffected by the species-specific restriction. The ability of WT PB2 to rescue the function of PB2 K627E during virus production also indicates that the blockade does not target mutant PB2 during late stages of replication. Thus, transmission of virus from birds to humans is restricted in part by an innate activity in human cells that disrupts polymerase function early in infection.
Several host co-factors have been identified that interact with the polymerase and nucleoprotein (Deng et al., 2006; Engelhardt et al., 2005; Gabriel et al., 2008; Huarte et al., 2001; Kawaguchi and Nagata, 2007; Mayer et al., 2007; Momose et al., 2001; Momose et al., 2002; Naito et al., 2007; Tarendeau et al., 2007). However, few interact directly with PB2 and of those that do, binding is independent of changes at position 627 (Supplemental Figure 5 and (Gabriel et al., 2008)). Inhibitory activity may be mediated directly by an anti-viral factor exclusive to primates that specifically targets PB2, or possibly a more fortuitous blockade by a cellular activity affecting viral proteins. It also remains possible that this activity results from evolutionarily divergent forms of a protein factor common to avian and primate hosts. The change in restriction during the viral life cycle may result from alterations in cellular protein function, possibly due to repression of host-protein production during viral infection (Krug et al., 1989), or changes to viral proteins that influence polymerase activity and susceptibility to the inhibitor. It is of great interest to determine if the inhibitory activity is due to binding by a host factor, binding by a small molecule, modification by a host factor, or some indirect effect on vRNP assembly.
Adaptation is a multigenic process and other mutations within the influenza replication machinery also enhance replication in mammalian hosts. Whereas position 627 in PB2 is a key regulator of species-specific polymerase activity, replication and pathogenicity is not always strictly correlated with the presence of lysine at PB2 627 (Gabriel et al., 2005; Gao et al., 1999; Govorkova et al., 2005). For example, recent infections in humans with avian H5N1 display a ~60% case fatality rate, despite many of these viruses retaining a glutamic acid at residue 627. Other changes may partially overcome defects associated with a glutamic acid at PB2 residue 627; the PB2 D701N and NP N319K mutations enhanced polymerase activity and lethality in the mouse-adapted H7N7 isolate SC35M in mammalian hosts even though this virus contains a glutamic acid at residue 627 in PB2 (Gabriel et al., 2005). The same PB2 D701N mutation was observed in lethal infections with H5N1 viruses that posses PB2 with glutamic acid at residue 627 (de Jong et al., 2006). However, the PB2 D701N mutation did not rescue polymerase activity of a PB2 K627E polymerase derived from the A/WSN/33 strain (Figure 1). Furthermore, introducing a lysine at amino acid 627 in PB2 of the parental SC35 virus increased polymerase activity 20-fold, compared to the more modest 6-fold increase associated with PB2 D701N (Gabriel et al., 2005). The varied effects of mutations in different viral strains and cells highlight the complex interplay between the subunits of the viral replication machinery and between the replication machinery and the host.
PB2 K627E binds to NP in human cells when expressed alone, but not when assembled into the trimeric polymerase (Figure 4), raising the possibility that the glutamic acid to lysine change might induce conformational alterations within the holoenzyme with species-specific consequences. Structural models of the influenza polymerase generated by electron microscopy suggest large-scale conformational rearrangements within the polymerase that occur upon vRNP formation, specifically changes including PB2 and regions that bind NP (Area et al., 2004; Torreira et al., 2007). In this scenario, trimeric polymerases containing PB2 K627E might adopt a conformation that precludes the conformational changes required for NP binding and vRNP assembly in mammalian cells. This is consistent with the fact that residue 627 lies outside of all known protein binding sites in PB2. This alternative conformation could be influenced by species-specific binding partners or post-translational modifications. For example, studies have shown that the enhancement of polymerase activity and virus lethality associated with the PB2 D701N mutant results from changes in conformational flexibility that increase binding to the nuclear import adaptor importin α1 in a species-specific fashion (Gabriel et al., 2008; Tarendeau et al., 2007). A better understanding of the conformational changes the polymerase undergoes upon vRNP formation might inform on the assembly defect manifest by the PB2 K627E mutation.
The potent block to avian-like influenza polymerase during early stages of viral replication in human cells provides a strong selective pressure for PB2 mutations. Indeed, virtually all strains of influenza that have circulated in humans contain PB2 with a lysine at amino acid 627 (Chen et al., 2006a). A single nucleotide change in avian PB2 vRNA results in an adaptive mutation that introduces a lysine at amino acid 627 and restores polymerase activity in mammalian cell culture (Figure 1), viral replication in animal models (Hatta et al., 2001; Munster et al., 2007; Salomon et al., 2006; Subbarao et al., 1993), and is often associated with increased replication and lethality in humans (Clements et al., 1992; Hatta et al., 2001; Munster et al., 2007; Salomon et al., 2006). Escape from this species-restricting factor is also a hallmark of the pandemic 1918 strain (Taubenberger et al., 2005). These findings indicate that mutations in PB2 play a key role in the evolution of influenza as it is transmitted from birds to humans. But, it remains to be determined whether the vRNP assembly defects displayed in human cells by PB2 with a glutamic acid at residue 627 from the A/WSN/33 strain (Figure 4), or the primary avian isolate A/Mallard/Marquenterre/MZ237/83 (Labadie et al., 2007), are generally applicable to other primary isolates, especially lethal H5N1 isolates that retain a glutamic acid at this residue (Gao et al., 1999). Regardless, it is alarming to note that many of the viruses isolated during a recent outbreak of H5N1 virus infection in wild birds contained PB2 with a lysine at position 627 (Chen et al., 2006b). Thus, viruses with adaptive mutations for replication in humans are circulating in migratory waterfowl populations.
In summary, we demonstrate that an inhibitory activity in human cells restricts the function of influenza polymerase containing an avian-like PB2 by preventing assembly of polymerase into the vRNP. This assembly defect reduces virion production by blocking replication and transcription of viral RNAs, and likely represents a major barrier to cross-species transmission of influenza from birds to humans. Determining the mechanisms of restriction will provide crucial insight into virus-host interactions and factors influencing the infectivity and relative pathogenicity of viruses newly transferred into humans, and may ultimately identify new targets for therapeutic intervention.
METHODS
Reagents
The plasmids pCDNA-PA, pCDNA-PB1, pCDNA-PB1-TAP, pCDNA-PB2, and pCDNA-PB2-TAP derived from influenza A/WSN/33 have been described (Engelhardt et al., 2005). pCDNA-NP-V5 and pCMV4-PB2-HA were constructed with cDNA derived from A/WSN/33. pCAGGS-WSN-NP 0/12 and the vRNA expression vector pTM-polI-WSN-All were gifts of Y. Kawaoka (Neumann et al., 2005). pTM-ΔPB2 was created by excising the PB2 expression cassette from pTM-polI-WSN-All. pBD-PB2 and pBD-PB2 K627E are bi-directional expression constructs for production of PB2 (A/WSN/33) vRNA and mRNA based on pHW2000 (Hoffmann et al., 2000). The vRNA-luciferase reporter plasmid pHH21-vNA-luc contains the luciferase coding sequence in the anti-sense orientation flanked by the 5′ and 3′ untranslated regions of the vNA gene segment from the A/WSN/33 isolate and was previously described (Regan et al., 2006). The vRNA-luciferase reporter plasmid used in avian cells, pgHH21-vNA-luc, was created by replacing the human pol I promoter of pHH21-vNA-luc with a minimal chicken pol I promoter (Massin et al., 2005) constructed de novo from oligonucleotides. pVSV-G was purchased from Clontech. PB2 mutants were created by PCR-based strategies and confirmed by sequencing.
Immunoreagents include anti-protein A (Sigma), anti-NP (ATCC hybridoma HB-65), anti-V5 (Bethyl Laboratories), anti-HA (Roche), anti-TBP (BioDesign), anti-tubulin (Sigma), protein A-agarose (Sigma), and rabbit-IgG agarose (Sigma). PB2 antiserum was created by immunization of rabbits with His6-PB2(1–156) purified from E. coli (Josman LLC).
All cells were maintained in Dulbecco’s modified Eagles medium supplemented with 10% FBS. Cells were transfected with FuGene6 (Roche) or calcium phosphate. Luciferase assays were performed according to the manufacturer’s instructions (Promega). Cytoplasmic and nuclear fractions were isolated with the NE-PER kit (Pierce). Immunoprecipitations were performed with cell lysates prepared in 50 mM Tris pH 7.4, 50 mM NH4Cl, 15% glycerol, and 0.5% NP40. Purification of TAP-tagged polymerase was as described (Engelhardt et al., 2005).
Primer extension
RNA was extracted from 293T or DF1 cells 48 hr after transfection with pCDNA-PB1, pCDNA-PA, pCDNA-NP-V5, WT or mutant pCDNA-PB2 and pHH21-vNA-luc or pgHH21-vNA-luc. Primer extension assays were performed using radiolabeled probes directed against luciferase (Fodor and Smith, 2004). Products were resolved on 8% polyacrylamide gels containing 7 M urea and detected and quantified by phosphorimaging.
Virus production
Recombinant virus was produced in 293T transfected with pTM-ΔPB2, pCAGGS-WSN-NP 0/12, pCDNA-PA, pCDNA-PB1, and pBD-PB2 or pBD-PB2 K627E. Virus was harvest 48 hr post-transfection. Hemagglutination activity was measured using a 0.5% solution of turkey red blood cells and the infectious titer was determined on MDCK cells. Recombinant virus was also produced in the presence of PB2-TAP by transfecting 293T cells with pTM-polI-WSN-All, pCAGGS-WSN-NP 0/12, pCDNA-PA, pCDNA-PB1, pCDNA-PB2, and pCDNA-PB2-TAP WT or K627E. Virus was harvested 24 hr later and vRNPs were fractionated on a linear 30–60% glycerol gradient as described (Klumpp et al., 2001).
Heterokaryon formation
Donor cells were transfected with pHH21-vNA-luc (293T cells) or pgHH21-vNA-luc (DF1 cells). Separately, acceptor cells were transfected with pCDNA-PB1, -PA, -PB2, -NP-V5, and pVSV-G. Transfected cells were incubated overnight and washed extensively. Donor cells were trypsinized and mixed with acceptor cells at a 1:1 ratio. Heterokaryon formation was monitored visually with donor and acceptor cells expressing RFP and GFP, respectively. Cells were harvested ~30 hr after mixing and subject to luciferase assays and western blot.
Supplementary Material
Acknowledgments
We thank C. Anderson, A. Fisher, and M.Yasukawa for assistance, P. Sarnow and M. Botchan for helpful discussions, and E. Fodor, T. Parslow, and Y. Kawaoka for reagents. A.M. is funded by a Kirschstein NRSA. J.A.D. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almond JW. A single gene determines the host range of influenza virus. Nature. 1977;270:617–618. doi: 10.1038/270617a0. [DOI] [PubMed] [Google Scholar]
- Area E, Martin-Benito J, Gastaminza P, Torreira E, Valpuesta JM, Carrascosa JL, Ortin J. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc Natl Acad Sci U S A. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Boutz PL, Nayak DP. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J Virol. 1998;72:5493–5501. doi: 10.1128/jvi.72.7.5493-5501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, et al. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis. 2006a;12:1353–1360. doi: 10.3201/eid1209.060276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Qi Q, Tian G, Fan S, Zhao H, et al. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J Virol. 2006b;80:5976–5983. doi: 10.1128/JVI.00110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJ, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JS, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–192. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Clements ML, Subbarao EK, Fries LF, Karron RA, London WT, Murphy BR. Use of single-gene reassortant viruses to study the role of avian influenza A virus genes in attenuation of wild-type human influenza A virus for squirrel monkeys and adult human volunteers. J Clin Microbiol. 1992;30:655–662. doi: 10.1128/jcm.30.3.655-662.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J Virol. 2006;80:11911–11919. doi: 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D, Digard P, Tiley L, Ortin J. Structure and function of the influenza virus RNP. In: Kawaoka Y, editor. Influenza Virology: Current Topics. Norfolk UK: Caister; 2005. p. 1036. [Google Scholar]
- Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J Virol. 2005;79:5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J Virol. 2004;78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Herwig A, Klenk HD. Interaction of Polymerase Subunit PB2 and NP with Importin alpha1 Is a Determinant of Host Range of Influenza A Virus. PLoS Pathog. 2008;4:e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass SE, McGeoch D, Barry RD. Characterization of the mRNA of influenza virus. J Virol. 1975;16:1435–1443. doi: 10.1128/jvi.16.6.1435-1443.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorkova EA, Rehg JE, Krauss S, Yen HL, Guan Y, Peiris M, Nguyen TD, Hanh TH, Puthavathana P, Long HT, et al. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J Virol. 2005;79:2191–2198. doi: 10.1128/JVI.79.4.2191-2198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- Hatta M, Hatta Y, Kim JH, Watanabe S, Shinya K, Nguyen T, Lien PS, Le QM, Kawaoka Y. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 2007;3:1374–1379. doi: 10.1371/journal.ppat.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz C, Stavnezer E, Krug R, Gurney T., Jr Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell. 1981;26:391–400. doi: 10.1016/0092-8674(81)90208-7. [DOI] [PubMed] [Google Scholar]
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Sanz-Ezquerro JJ, Roncal F, Ortin J, Nieto A. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J Virol. 2001;75:8597–8604. doi: 10.1128/JVI.75.18.8597-8604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: The NS1 protein four C-terminal residues modulate pathogenicity. 2008:0800482105. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Caton AJ, McCready SJ, Cook PR. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature. 1982;296:366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi A, Nagata K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. Embo J. 2007;26:4566–4575. doi: 10.1038/sj.emboj.7601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp K, Hooker L, Handa B. Influenza virus endoribonuclease. Methods Enzymol. 2001;342:451–466. doi: 10.1016/s0076-6879(01)42566-3. [DOI] [PubMed] [Google Scholar]
- Krug R, Alonso-Caplen F, Julkunen I, Katze MG. Expression and Replication of the Influenza Virus Genome. In: Krug R, editor. The Influenza Viruses. New York: Plenum Press; 1989. pp. 89–152. [Google Scholar]
- Krug RM. Virology. Clues to the virulence of H5N1 viruses in humans. Science. 2006;311:1562–1563. doi: 10.1126/science.1125998. [DOI] [PubMed] [Google Scholar]
- Labadie K, Dos Santos Afonso E, Rameix-Welti MA, van der Werf S, Naffakh N. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology. 2007;362:271–282. doi: 10.1016/j.virol.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massin P, Rodrigues P, Marasescu M, van der Werf S, Naffakh N. Cloning of the chicken RNA polymerase I promoter and use for reverse genetics of influenza A viruses in avian cells. J Virol. 2005;79:13811–13816. doi: 10.1128/JVI.79.21.13811-13816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res. 2007;6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medcalf L, Poole E, Elton D, Digard P. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J Virol. 1999;73:7349–7356. doi: 10.1128/jvi.73.9.7349-7356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Basler CF, O’Neill RE, Iwamatsu A, Palese P, Nagata K. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Kuiken T, Fouchier RA. The Molecular Basis of the Pathogenicity of the Dutch Highly Pathogenic Human Influenza A H7N7 Viruses. J Infect Dis. 2007;196:258–265. doi: 10.1086/518792. [DOI] [PubMed] [Google Scholar]
- Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J Gen Virol. 2000;81:1283–1291. doi: 10.1099/0022-1317-81-5-1283. [DOI] [PubMed] [Google Scholar]
- Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Fujii K, Kino Y, Kawaoka Y. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc Natl Acad Sci U S A. 2005;102:16825–16829. doi: 10.1073/pnas.0505587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E, Elton D, Medcalf L, Digard P. Functional domains of the influenza A virus PB2 protein: identification of NP- and PB1-binding sites. Virology. 2004;321:120–133. doi: 10.1016/j.virol.2003.12.022. [DOI] [PubMed] [Google Scholar]
- Regan JF, Liang Y, Parslow TG. Defective assembly of influenza A virus due to a mutation in the polymerase subunit PA. J Virol. 2006;80:252–261. doi: 10.1128/JVI.80.1.252-261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K, Swayne DE, Olsen CW. Epidemiology and control of human and animal influenza. In: Kawaoka Y, editor. Influenza Virology: Current Topics. Wymondham Caister Academic Press; 2006. pp. 229–280. [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- Torreira E, Schoehn G, Fernandez Y, Jorba N, Ruigrok RW, Cusack S, Ortin J, Llorca O. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res. 2007;35:3774–3783. doi: 10.1093/nar/gkm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.