Abstract
Background
It is important, for drug-resistance surveillance, to identify human immunodeficiency virus type 1 (HIV-1) strains that have undergone antiretroviral drug selection.
Methods
We compared the prevalence of protease and reverse-transcriptase (RT) mutations in HIV-1 sequences from persons with and without previous treatment with protease inhibitors (PIs), nucleoside RT inhibitors (NRTIs), and nonnucleoside RT inhibitors (NNRTIs). Treatment-associated mutations in protease isolates from 5867 persons and RT isolates from 6247 persons were categorized by whether they were polymorphic (prevalence, >0.5%) in untreated individuals and whether they were established drug-resistance mutations. New methods were introduced to minimize misclassification from transmitted resistance, population stratification, sequencing artifacts, and multiple hypothesis testing.
Results
Some 36 established and 24 additional nonpolymorphic protease mutations at 34 positions were related to PI treatment, 21 established and 22 additional nonpolymorphic RT mutations at 24 positions with NRTI treatment, and 15 established and 11 additional nonpolymorphic RT mutations at 15 positions with NNRTI treatment. In addition, 11 PI-associated and 1 NRTI-associated established mutations were polymorphic in viruses from untreated persons.
Conclusions
Established drug-resistance mutations encompass only a subset of treatment-associated mutations; some of these are polymorphic in untreated persons. In contrast, nonpolymorphic treatment-associated mutations may be more sensitive and specific markers of transmitted HIV-1 drug resistance.
HIV-1 mutations that emerge during in vitro passage experiments, reduce in vitro drug susceptibility in site-directed mutants, or occur in large numbers of clinical isolates have been widely recognized as being related to drug resistance [1, 2]. These established mutations, however, have 2 limitations for drug-resistance surveillance. First, most of them are identified during pre-clinical and early clinical drug development. In contrast, treatment-related mutations that are uncommon, emerge after prolonged treatment, or act primarily in combination with other mutations often remain unrecognized. Second, several of the established mutations are polymorphic and occur commonly in untreated patients.
We previously reported that mutations at 23 protease positions not recognized as drug-resistance positions were significantly more likely to occur in persons receiving protease inhibitors (PIs) than in PI-naive persons [3] and that mutations at 9 reverse-transcriptase (RT) positions not recognized as drug-resistance positions were significantly more likely to occur in persons receiving nucleoside RT inhibitors (NRTIs) than in NRTI-naive persons [4]. However, in those previous studies, we grouped together different mutations at the same position and did not examine the relationship between specific amino acids and treatment. In the present study, we examine associations between specific mutations and treatments in HIV-1 protease and RT in a data set that is more than twice as large as those used for the previous studies. Mutations are categorized according to their strength of association with treatment and the extent of polymorphism in untreated viruses.
PATIENTS AND METHODS
Patients, viruses, and mutations
We analyzed HIV-1 subtype B sequences from persons with well-characterized histories of antiretroviral treatment. Sequences were obtained from the Stanford HIV RT and Protease Sequence Database [5] and included sequences from published studies and previously unpublished sequences generated at Stanford University from patients living in northern California (GenBank accession numbers AY796421–AY798497 and AY800656–AY802758). For viruses undergoing sequencing at Stanford University, treatment histories were obtained from patient charts and pharmacy records as part of an institutional review board—approved collaboration. For data from published studies, we supplemented treatment histories by requesting information from the study authors. Data from patients for whom there was uncertainty about whether a particular drug class was received were not analyzed.
Protease positions 1–99 and RT positions 1–240 were analyzed. Mutations were defined as differences from the consensus B amino acid reference sequence (see Appendix). This sequence contains the most common amino acid at each position in early alignments of published protease and RT sequences from untreated persons [5, 6]. Whereas recent alignments had a plurality of prolines at position 63 of protease, early alignments had a plurality of leucines; this, by convention, has become the consensus at that position.
For patients who provided >1 sample, only the latest sample obtained while they were receiving treatment was analyzed. Only sequences determined by dideoxyterminator sequencing were included. Mutations present as part of a mixture based on the sequencing electropherogram or the GenBank sequence were excluded from analysis. We obtained 96% of protease and 93% of RT sequences using direct polymerase chain reaction (PCR; “population-based”) sequencing. The term “isolate” was used to describe the viral material that was sequenced whether the sequencing was done by use of population-based or clonal sequencing.
Established drug-resistance positions and mutations were defined by use of 2 reviews on HIV-1 drug resistance and the International AIDS Society—USA expert panel mutation list [1, 2, 7]. On the basis of this list of mutations, protease positions 10, 20, 24, 30, 32, 33, 36, 46, 47, 48, 50, 53, 54, 63, 71, 73, 77, 82, 84, 88, and 90 were categorized as established PI-resistance positions. RT positions 41, 44, 62, 65, 67, 69, 70, 74, 75, 77, 115, 116, 118, 151, 184, 210, 215, and 219 were categorized as established NRTI-resistance positions. RT positions 100, 103, 106, 108, 181, 188, 190, 225, 230, and 236 were categorized as established nonnucleoside RT inhibitor (NNRTI)—resistance positions.
Nonpolymorphic mutations were defined as mutations present in ⩽0.5% of viruses from untreated persons. This number was chosen to distinguish mutations likely to reflect natural variation in protease and RT from more-rare mutations that would be more likely to reflect transmitted drug-resistant viruses, sequencing errors, or rare nonviable virus variants.
Analysis
To identify PI-related mutations, we compared the prevalence of protease mutations in PI-treated versus PI-naive persons. To identify NRTI-related mutations, we compared the prevalence of RT mutations in NRTI-treated (but NNRTI-naive) versus RTI-naive persons. To identify NNRTI-related mutations, we compared the prevalence of RT mutations in NRTI- and NNRTI-treated versus NRTI-treated (but NNRTI-naive) persons. For RT mutations related to both NRTI and NNRTI treatment, we examined the relative contributions of NRTI and NNRTI treatment using a generalized linear model that accounted for 3 levels of NRTI treatment (1–2, 3–4, and >4 NRTIs). For each comparison, only mutations significantly related to treatment and >2 times more common in treated persons are described. Because of marked imbalances in the number of persons who received different drugs in the same class and because most persons received multiple NRTIs and PIs, we did not correlate mutations with individual drugs.
To distinguish mutations developing in multiple individuals from those developing in a smaller number of founder viruses, we reconstructed the ancestral sequences at each node of a phylogenetic tree for all sequences and counted the number of times that each mutation was predicted to have developed. Mutations for which founder viruses accounted for ⩾50% of occurrences were not considered to be treatment-related mutations. Neighbor-joining trees were created by use of the HKY85 model with γ-distribution (PAUP* version 4.0b10; Sinauer) for each gene. Ancestral sequences were reconstructed by use of MESQUITE (version 1.02; available at: http://www.mesquiteproject.org).
Two steps were taken to reduce the influence of transmitted drug resistance: isolates from persons with primary HIV-1 infection in studies published after the year 2000 were not included, and isolates from untreated persons that had ⩾2 established nonpolymorphic drug-related mutations were excluded. Isolates from untreated persons that contained a single established nonpolymorphic drug-resistance mutation were excluded from analyses of mutations other than that mutation.
Holm’s method [8] was used to control the familywise error rate for multiple hypothesis testing. For each drug class, we examined the P values for each χ2 test that compared the prevalence in treated and untreated persons (or the NNRTI coefficient in the general linear model) of mutations that occurred in HIV-1 isolates from at least 2 treated persons. The P values were ranked in descending order. Starting from the smallest P (rank r = n, where n is the number of hypotheses), we compared each P of rank r with a significance cutoff of 0.01/r as long as Pr⩽ 0.01/r. All P values of Pr…Pn were considered to be statistically significant.
Mutations at established drug-resistance positions [1, 2, 7] that were significantly related to treatment (P < .05) before but not after adjustment for multiple comparisons were classified as treatment-related mutations, because these positions had already been related to resistance—usually in phenotypic and clinical studies—and therefore required only confirmation in our analysis.
RESULTS
Virus isolates
A total of 8426 protease isolates from 5867 persons with well-characterized PI treatment histories were analyzed; 1547 (26%) of these persons were from northern California, and their samples underwent sequencing at Stanford University. Protease sequences from 479 persons from northern California had been previously unpublished. In addition, 8786 RT isolates from 6247 persons with well-characterized RT inhibitor treatment histories were analyzed; 1563 (25%) of these persons were from northern California, and their samples underwent sequencing at Stanford University. RT sequences from 500 persons from northern California had been previously unpublished. When multiple isolates from a single person were available, we analyzed only the latest obtained during treatment.
Treatment histories
A total of 2689 persons were PI naive; 3178 persons received ⩾1 PI. Among the PI-experienced persons, 1492 received 1 PI and 1686 received >1 PI. Among the persons who received 1 PI, 614 received indinavir, 479 received nelfinavir, 220 received saquinavir, 119 received ritonavir, 26 received amprenavir, 18 received lopinavir, and 16 received atazanavir.
A total of 2123 persons were NRTI naive; 2551 persons were NRTI experienced but NNRTI naive. Of these 2551 persons, 381 received 1 NRTI and 2170 received >1 NRTI. Among the persons who received 1 NRTI, 264 received zidovudine, 43 received abacavir, 39 received stavudine, 25 received didanosine, 8 received lamivudine, and 2 received zalcitabine.
Of the 1573 persons who received ⩾1 NNRTI, 1245 received 1 NNRTI and 328 received >1 NNRTI. Among the persons who received 1 NNRTI, 619 received nevirapine, 521 received efavirenz, and 105 received delavirdine.
PI-related mutations
Of 1881 (99 positions × 19 nonconsensus amino acids) possible protease mutations, 265 (14.0%) at 81 positions occurred in isolates from ⩾2 persons who received a PI. After adjustment for 265 comparisons, 63 mutations at 38 positions were related to PI treatment (familywise error rate, ⩽0.01). An additional 10 uncommon mutations at 9 established PI-resistance positions were related to PI treatment only before correction for multiple comparisons (P = .001–.02). Therefore, 73 protease mutations—including 53 at 21 established drug-resistance positions and 20 at 17 additional positions—were significantly related to PI treatment.
Of the 73 PI-related mutations, 60 at 34 positions were nonpolymorphic (table 1). These included 36 of 46 established mutations (L10F/R, K20I/T, L24I, D30N, V32I, L33F, M46I/L, I47V/A, G48V, I50V/L, F53L, I54V/M/L/T/A/S, G73S/T/C/A, V82A/T/F/S, I84V/A/C, N88D/S, and L90M), 6 additional mutations at established drug-resistance positions (K20V, M46V, G48M, F53Y, A71I, and N88T), and 18 mutations at additional positions (V11I, L23I, E34Q, E35G, K43T, K55R, Q58E, I66F, C67F, T74S/P/A, L76V, P79A, I85V, L89V, Q92R, and C95F).
Table 1.
Protease inhibitor (PI)—related mutations and extent of polymorphism in HIV-1 isolates from PI-naive persons
| Mutations | Nonpolymorphica |
Polymorphica |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos | AA | Naive, % | Treatment, % | Pb | Pos | AA | Naive, % | Treatment, % | Pb | |
| Established PI resistance | 10 | F | 0.2 | 5.4 | <.001 | 10 | I | 7.8 | 35 | <.001 |
| R | 0 | 0.7 | <.001 | V | 2.1 | 4.5 | <.001 | |||
| 20 | Ic | 0.1 | 5.8 | <.001 | 20 | R | 1.4 | 8.7 | <.001 | |
| T | 0.1 | 2.3 | <.001 | M | 0.5 | 2.2 | <.001 | |||
| 24 | I | 0 | 5.3 | <.001 | 33 | I | 0.6 | 1.3 | .006d | |
| 30 | N | 0 | 11.6 | <.001 | 36 | I | 12.2 | 26.8 | <.001 | |
| 32 | I | 0 | 4.6 | <.001 | V | 0.5 | 2 | <.001 | ||
| 33 | Fc | 0.2 | 5.3 | <.001 | 63e | P | 55.2 | 73.9 | <.001 | |
| 46 | I | 0.1 | 19.7 | <.001 | 71 | T | 4.7 | 10 | <.001 | |
| Lc | 0.2 | 9.3 | <.001 | V | 2.2 | 29.9 | <.001 | |||
| 47 | A | 0 | 0.2 | .02d | 77e | I | 25 | 32.4 | <.001 | |
| V | 0.1 | 2.2 | <.001 | |||||||
| 48 | V | 0 | 4.1 | <.001 | ||||||
| 50 | L | 0 | 0.5 | <.001 | ||||||
| V | 0 | 1 | <.001 | |||||||
| 53 | L | 0.1 | 3.7 | <.001 | ||||||
| 54 | A | 0 | 0.6 | <.001 | ||||||
| L | 0 | 2.1 | <.001 | |||||||
| M | 0 | 1.2 | <.001 | |||||||
| T | 0 | 1 | <.001 | |||||||
| V | 0 | 19.1 | <.001 | |||||||
| S | 0 | 0.2 | .02d | |||||||
| 73 | A | 0 | 0.3 | .004d | ||||||
| C | 0 | 1.1 | <.001 | |||||||
| S | 0 | 8.2 | <.001 | |||||||
| T | 0 | 1.8 | <.001 | |||||||
| 82 | A | 0 | 23 | <.001 | ||||||
| F | 0 | 1.4 | <.001 | |||||||
| S | 0 | 0.4 | .001d | |||||||
| T | 0 | 2.8 | <.001 | |||||||
| 84 | A | 0 | 0.3 | .007d | ||||||
| C | 0 | 0.4 | .002d | |||||||
| V | 0 | 12.6 | <.001 | |||||||
| 88 | D | 0 | 8 | <.001 | ||||||
| S | 0 | 2 | <.001 | |||||||
| 90 | M | 0.2 | 34.2 | <.001 | ||||||
| At established PI-resistance positions | 20 | V | 0 | 0.4 | <.001 | |||||
| 46 | V | 0 | 0.7 | <.001 | ||||||
| 48 | M | 0 | 0.3 | .004d | ||||||
| 53 | Y | 0 | 0.3 | .004d | ||||||
| 71 | I | 0.1 | 2.2 | <.001 | ||||||
| 88 | T | 0 | 0.3 | .007d | ||||||
| At positions not deemed to be established PI-resistance positions |
11 | Ic | 0.2 | 1 | <.001 | 72 | L | 0.5 | 1.4 | <.001 |
| 23 | I | 0 | 0.7 | <.001 | 92 | K | 0.6 | 1.9 | <.001 | |
| 34 | Q | 0 | 0.8 | <.001 | ||||||
| 35 | Gc | 0 | 0.7 | <.001 | ||||||
| 43 | T | 0.1 | 2.9 | <.001 | ||||||
| 55 | R | 0.1 | 3 | <.001 | ||||||
| 58 | E | 0.1 | 3.6 | <.001 | ||||||
| 66 | F | 0.1 | 1 | <.001 | ||||||
| 67 | F | 0 | 0.7 | <.001 | ||||||
| 74 | Ac | 0.2 | 1 | <.001 | ||||||
| P | 0 | 1 | <.001 | |||||||
| Sc | 0.1 | 4.3 | <.001 | |||||||
| 76 | V | 0 | 1.5 | <.001 | ||||||
| 79 | A | 0 | 0.5 | <.001 | ||||||
| 85 | V | 0 | 3.1 | <.001 | ||||||
| 89 | V | 0 | 1.3 | <.001 | ||||||
| 92 | R | 0.2 | 0.8 | <.001 | ||||||
| 95 | F | 0 | 1.2 | <.001 | ||||||
NOTE. Established positions and mutations were categorized by use of 2 recently published reviews on HIV-1 drug resistance and recent updates to the International AIDS Society—USA expert panel mutation list [1, 2, 7]. AA, amino acid; Pos, position.
Nonpolymorphic mutations occurred in ⩽0.5% of sequences from untreated persons.
Holm’s method with a familywise error rate of ⩽0.01 was used to identify results that were statistically significant in multiple comparisons [8].
K20I is the consensus amino acid in subtypes G and CRF02_AG. V11I, L33F, E35G, M46L, and P74A/S were polymorphic in ⩾1 non-B subtypes (range, 1%–4%).
This mutation was related to PI treatment only before correction for multiple comparisons.
This mutation was <2 times more prevalent in viruses from treated persons. An exception was made for this mutation because it did not meet this criterion, which was applied to all other mutations, as outlined in Patients and Methods.
The polymorphic mutations included 11 mutations at 7 established positions and 2 at additional positions (table 1). Two mutations at PI-resistance positions were not related to treatment: L33V (1% in treated vs. 2% in untreated persons) and V82I (1.3% in treated vs. 1.4% in untreated persons).
Among the 73 PI-related mutations, 59, 63, and 69 were related to PI treatment (P<.05) in persons who received 1, 2, or ⩾3 PIs, respectively. Among sequences with only 1 nonpolymorphic PI-related mutation, 90% were established drug-resistance mutations, and 10% were one of the additional treatment-related mutations. Among sequences with >1 nonpolymorphic PI-related mutation, 58.4% had only established drug-resistance mutations, 41.4% had a combination of established and additional mutations, and 0.1% had only additional mutations (figure 1).
Figure 1.
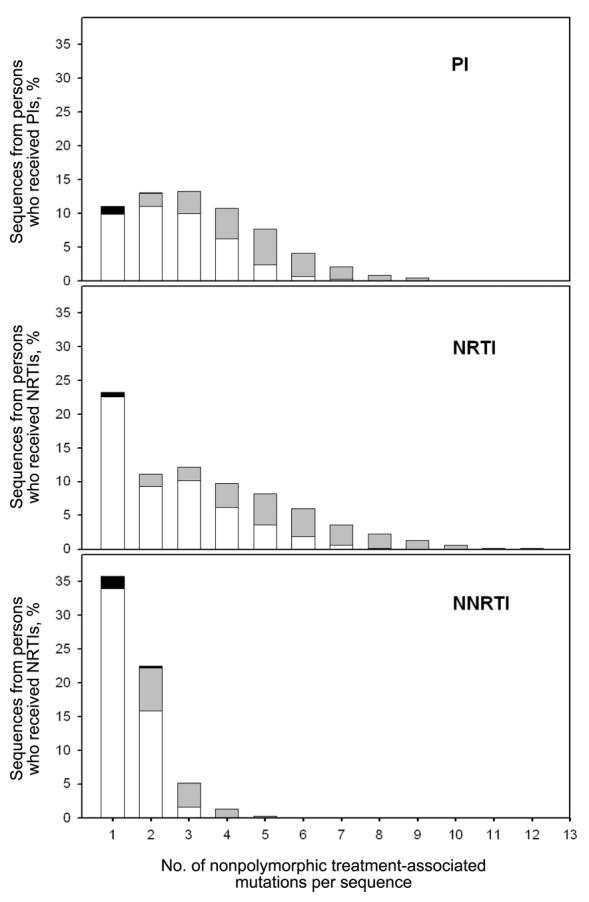
Classification of nonpolymorphic mutations in sequences from treated persons. The x-axis segregates sequences according to the no. of nonpolymorphic mutations per sequence. Histograms show the proportion of sequences containing only established drug-resistance mutations (white rectangles), only additional treatment-related mutations (black rectangles), or a combination of established and additional mutations (gray rectangles). The y-axis shows the proportion that each type of sequence contributes to the total no. of sequences from treated persons. The proportions of sequences from treated persons lacking nonpolymorphic treatment-related mutations are not shown. NNRTI, nonnucleoside reverse-transcriptase inhibitor; NRTI, nucleoside reverse-transcriptase inhibitor; PI, protease inhibitor.
NRTI-related mutations
Of 4560 (240 positions × 19 nonconsensus amino acids) possible RT mutations, 383 (8.4%) at 150 positions (between positions 1 and 240) occurred in isolates from ⩾2 persons who received an NRTI. After adjustment for 383 comparisons, 40 mutations at 28 positions were significantly related to NRTI treatment (familywise error rate, ⩽0.01). An additional 7 uncommon mutations at 6 established NRTI-resistance positions were related to NRTI treatment only before correction for multiple comparisons (P = .002–.01). Therefore, 47 RT mutations, including 34 at 18 established drug-resistance positions and 13 at 10 additional positions, were significantly related to NRTI treatment.
Of the 47 NRTI-related mutations, 43 at 24 positions were nonpolymorphic (table 2). These included 21 of 23 established mutations (M41L, E44D, A62V, K65R, D67N, T69D/ins, K70R, L74V, V75I, F77L, Y115F, F116Y, Q151M, M184V/I, L210W, T215Y/F, and K219Q/E), 12 additional mutations at established drug-resistance positions (E44A, D67G/E, T69N/S, L74I, V75M/T, T215I/V, and K219N/R), and 10 mutations at additional positions (K43N/Q/E, A98G, E203K, H208Y, D218E, K223Q, and L228H/R).
Table 2.
Nucleoside reverse-transcriptase inhibitor (NRTI)—related mutations according to previous classifications and extent of polymorphism in HIV-1 isolates from untreated persons.
| Mutations | Nonpolymorphica |
Polymorphica |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos | AA | Naive, % | Treatment, % | Pb | Pos | AA | Naive, % | Treatment, % | Pb | |
| Established NRTI resistance | 41 | L | 0.5 | 33.5 | <.001 | 118 | I | 2.2 | 11.4 | <.001 |
| 44 | Dc | 0.3 | 5 | <.001 | ||||||
| 62 | V | 0.1 | 2 | <.001 | ||||||
| 65 | R | 0.1 | 1 | <.001 | ||||||
| 67 | N | 0 | 24.9 | <.001 | ||||||
| 69 | D | 0.1 | 5.3 | <.001 | ||||||
| Ins | 0 | 0.5 | .003d | |||||||
| 70 | R | 0.2 | 22.6 | <.001 | ||||||
| 74 | V | 0 | 2.8 | <.001 | ||||||
| 75 | I | 0 | 1.7 | <.001 | ||||||
| 77 | L | 0.1 | 2 | <.001 | ||||||
| 115 | F | 0 | 1 | <.001 | ||||||
| 116 | Y | 0 | 1.9 | <.001 | ||||||
| 151 | M | 0 | 2.6 | <.001 | ||||||
| 184 | I | 0.1 | 0.5 | .01d | ||||||
| V | 0.2 | 47.5 | <.001 | |||||||
| 210 | W | 0 | 19.1 | <.001 | ||||||
| 215 | F | 0.1 | 7.2 | <.001 | ||||||
| Y | 0.1 | 34.7 | <.001 | |||||||
| 219 | E | 0.1 | 2.1 | <.001 | ||||||
| Q | 0.2 | 13 | <.001 | |||||||
| At established NRTI-resistance positions | 44 | A | 0 | 0.7 | <.001 | |||||
| 67 | G | 0 | 1 | <.001 | ||||||
| E | 0 | 0.5 | .004d | |||||||
| 69 | N | 0.5 | 4.5 | <.001 | ||||||
| S | 0.5 | 1.6 | <.001 | |||||||
| 74 | I | 0.1 | 0.7 | .001d | ||||||
| 75 | M | 0 | 1 | <.001 | ||||||
| T | 0 | 0.7 | <.001 | |||||||
| 215e | I | 0 | 0.4 | .006d | ||||||
| V | 0.1 | 0.7 | .002d | |||||||
| 219 | R | 0.2 | 1 | .004d | ||||||
| N | 0.1 | 0.8 | <.001 | |||||||
| At positions not deemed to be established NRTI-resistance positions |
43 | Ec | 0.2 | 3.7 | <.001 | 39 | A | 2.2 | 6.6 | <.001 |
| Q | 0.1 | 2.8 | <.001 | 90 | I | 1.1 | 2.7 | <.001 | ||
| N | 0.1 | 1 | <.001 | 104 | N | 0.6 | 2 | .001 | ||
| 98 | Gc | 0.1 | 1.5 | <.001 | ||||||
| 203 | K | 0 | 1.9 | <.001 | ||||||
| 208 | Y | 0.1 | 3.6 | <.001 | ||||||
| 218 | E | 0 | 3.2 | <.001 | ||||||
| 223 | Q | 0 | 1 | <.001 | ||||||
| 228 | H | 0.1 | 3.4 | <.001 | ||||||
| R | 0 | 1.4 | <.001 | |||||||
NOTE. Established drug-resistance positions and mutations were categorized by use of 2 recently published reviews on HIV-1 drug resistance and recent updates to the International AIDS Society—USA expert panel mutation list [1, 2, 7]. AA, amino acid; Pos, position.
Nonpolymorphic mutations occurred in ⩽0.5% of sequences from untreated persons.
Holm’s method [8] with a familywise error rate of ⩽0.01 was used to identify results that were statistically significant in the presence of multiple comparisons.
K43E is the consensus amino acid for subtype CRF01_AE. E44D and A98G were polymorphic in ⩾1 non-B subtype (range, 1%– 3%).
These 7 rare nonpolymorphic mutations at established NRTI-related positions were related to NRTI treatment only before correction for multiple comparisons.
T215S/C/D/E occurred in 0.1%–0.5% of NRTI-naive persons and 0.1%–0.8% of NRTI-treated persons and were therefore not significantly related to NRTI treatment, although, as noted in Discussion, these mutations would be expected to be reliable indicators of antiretroviral drug selection.
V118I occurred in 2.2% of untreated and 11.4% of NRTI-treated persons and was the only polymorphic, established NRTI-related mutation. Four T215 revertants (T215S/C/D/E) were not significantly related to treatment, because they occurred in NRTI-naive persons nearly as often as they did in NRTI-treated persons.
Among the 47 NRTI-related mutations, 24, 38, and 47 were related to NRTI treatment (P<.05) in persons who received 1, 2, or ⩾3 NRTIs, respectively. Among sequences with only 1 nonpolymorphic NRTI-related mutation, 97% were established drug-resistance mutations and 3% were one of the additional treatment-related mutations. Among sequences with >1 nonpolymorphic NRTI-related mutation, 57.6% had only established drug-resistance mutations, and 42.4% had a combination of established and additional mutations (figure 1).
NNRTI-related mutations
Of 4560 (240 positions × 19 nonconsensus amino acids) possible RT mutations, 348 (7.6%) at 131 positions (between positions 1 and 240) occurred in isolates from ⩾2 persons who received an NNRTI. After adjustment for 348 comparisons, 32 mutations at 21 positions were related to NNRTI treatment (familywise error rate, ⩽0.01). An additional 5 uncommon mutations at 4 established NNRTI-resistance positions were related to NNRTI treatment only before correction for multiple comparisons (P = .001–.03). Therefore, 37 RT mutations, including 19 at 10 established drug-resistance positions and 18 at 12 additional positions were significantly related to NNRTI treatment.
Of the 37 NNRTI-related mutations, 26 mutations at 15 positions were nonpolymorphic (table 3). These included 15 established mutations (L100I, K103N, V106A/M, V108I, Y181C/I, Y188L/H/C, G190A/S, P225H, M230L, and P236L), 4 additional mutations at established drug-resistance positions (K103S, Y181V, and G190E/Q), and 7 mutations at additional positions (K101P/E/N, E138Q, H221Y, F227L, and K238T).
Table 3.
Nonnucleoside reverse-transcriptase inhibitor (NNRTI)—related mutations according to previous classifications and extent of polymorphism in HIV-1 isolates from untreated persons.
| Mutations | Nonpolymorphica |
Polymorphica |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos | AA | Naive, % | NRTI alone, % |
NRTI and NNRTI, % |
Pb | Pos | AA | Naive, % | NRTI alone, % |
NRTI and NNRTI, % |
Pb | |
| Established NNRTI resistance | 100 | I | 0 | 0 | 4.5 | <.001 | ||||||
| 103 | N | 0.3 | 0.4 | 47 | <.001 | |||||||
| 106 | A | 0 | 0 | 2.2 | <.001 | |||||||
| M | 0 | 0 | 0.6 | <.001 | ||||||||
| 108 | I | 0.3 | 0.4 | 5.4 | <.001 | |||||||
| 181 | C | 0 | 0.2 | 22.1 | <.001 | |||||||
| I | 0 | 0 | 0.7 | <.001 | ||||||||
| 188 | C | 0 | 0 | 0.3 | .03c | |||||||
| H | 0 | 0 | 0.4 | .004c | ||||||||
| L | 0 | 0.1 | 3.8 | <.001 | ||||||||
| 190 | A | 0 | 0.1 | 12.7 | <.001 | |||||||
| S | 0 | 0.1 | 3.3 | <.001 | ||||||||
| 225 | H | 0 | 0 | 3 | <.001 | |||||||
| 230 | L | 0 | 0 | 1.2 | <.001 | |||||||
| 236 | L | 0.1 | 0.1 | 0.9 | .001c | |||||||
| At established NNRTI-resistance positions | 103 | S | 0 | 0 | 1 | <.001 | ||||||
| 181 | V | 0 | 0 | 0.4 | .003c | |||||||
| 190 | Q | 0 | 0 | 0.3 | .03c | |||||||
| E | 0 | 0 | 0.6 | <.001 | ||||||||
| At positions not deemed to be established NNRTI-resistance positions | 101 | E | 0.2 | 0.3 | 5.6 | <.001 | 65 | R | 0.1 | 1 | 2.9 | <.001 |
| N | 0 | 0 | 0.5 | <.001 | 74 | I | 0.1 | 0.7 | 3.3 | <.001 | ||
| P | 0 | 0 | 1.2 | <.001 | V | 0 | 2.8 | 9.3 | <.001 | |||
| 138 | Q | 0 | 0 | 0.7 | <.001 | 75 | M | 0 | 1 | 2.9 | <.001 | |
| 221 | Y | 0 | 0.3 | 4.6 | <.001 | 98 | G | 0.1 | 1.5 | 4.1 | <.001 | |
| 227 | Ld | 0 | 0 | 1.6 | <.001 | 101 | Q | 0.4 | 0.9 | 2.8 | <.001 | |
| 238 | T | 0.1 | 0.1 | 2.4 | <.001 | 179 | D | 1.3 | 0.6 | 1.8 | <.001 | |
| I | 2.4 | 3 | 5.6 | <.001 | ||||||||
| 219 | N | 0.1 | 0.8 | 3.3 | <.001 | |||||||
| 228 | H | 0.1 | 3.4 | 7.5 | <.001 | |||||||
| R | 0 | 1.4 | 4.2 | <.001 | ||||||||
NOTE. Established drug-resistance positions and mutations were categorized by use of 2 recently published reviews on HIV-1 drug resistance and recent updates to the International AIDS Society—USA expert panel mutation list [1, 2, 7]. AA, amino acid; Pos, position.
Nonpolymorphic mutations occurred in ⩽0.5% of sequences from untreated persons or those who received NRTIs but not NNRTIs.
To identify NNRTI-related mutations, the prevalence of mutations in persons treated with both NRTIs and NNRTIs was compared with their prevalence in persons treated with NRTIs alone. For mutations related to both NRTIs and NNRTIs, the relative contributions of NRTI and NNRTI treatment was assessed by use of a generalized linear model that accounted for 3 different levels of NRTI treatment (1–2 NRTIs, 3–4 NRTIs, and >4 NRTIs). Holm’s method with a familywise error rate of ⩽0.01 was used to identify results that were statistically significant in the presence of multiple comparisons [8].
These 5 rare nonpolymorphic mutations at established NNRTI-related positions were related to NNRTI treatment only before correction for multiple comparisons.
F227L occurred at a prevalence of 2% in subtype F.
The polymorphic mutations included A98G, K101Q, and V179D/I and 7 NRTI-related mutations (table 3). The 7 NRTI mutations occurred 3–8 times more commonly in persons who received NRTIs and NNRTIs than in those who received only NRTIs; these remained significantly related to NNRTI treatment in a multivariate model that controlled for the number of NRTIs received.
Several mutations at established drug-resistance positions were polymorphic and/or were weakly or not at all related to NNRTI treatment: A98S (8.4% in treated vs. 5.8% in untreated persons), 101R (1% in treated vs. 0.5% in untreated persons), 103R (1.9% in treated vs. 2.2% in untreated persons), 106I (1.4% in treated vs. 1.2% in untreated persons), 179I (5.6% in treated vs. 3.1% in untreated persons), and 238R (0.2% in treated vs. 0.2% in untreated persons).
Among the 37 NNRTI-related mutations, 36 and 32 were related to NNRTI treatment (P<.05) in persons who received 1 or ⩾2 NNRTIs, respectively. Among sequences with only 1 nonpolymorphic NNRTI-related mutation, 94.7% were established drug-resistance mutations, and 5% were one of the additional treatment-related mutations. Among sequences with >1 nonpolymorphic NNRTI-related mutation, 59.8% had only established drug-resistance mutations, 39.2% had a combination of established and additional mutations, and 0.9% had only additional mutations (figure 1).
DISCUSSION
Of 73 PI-related mutations, 60 (82%)–36 established drug-resistance and 24 additional mutations—at 34 positions were nonpolymorphic. Of 47 NRTI-related mutations, 43 (91%)–21 established drug-resistance and 22 additional mutations—at 24 positions were nonpolymorphic. Of 37 NNRTI-related mutations, 26 (70%)–15 established drug-resistance and 11 additional mutations—at 15 positions were nonpolymorphic. The additional treatment-related mutations occurred less frequently than the established mutations and were more likely to occur in combination with other treatment-related mutations, which possibly explains the delay in their recognition. Among treated persons, 10%, 3%, and 5% of sequences with a single nonpolymorphic treatment-related mutation had one of these additional PI-, NRTI-, or NNRTI-related mutations, respectively, which suggests that, in these persons, one of the additional mutations was the only indication of previous selective drug pressure.
Nonpolymorphic treatment-related mutations are particularly important for drug-resistance surveillance, but their accurate identification is challenging. Misclassification can result from the following causes: (1) transmission of drug-resistant viruses, (2) differences in specific HIV-1 variants among different human populations (population stratification), (3) variability in sequence quality, and (4) the requirement for multiple statistical comparisons. The identification of NNRTI-related mutations is further complicated by the fact that NNRTIs are nearly always administered in combination with NRTIs.
Effect of transmitted resistance
The transmission of drug-resistant HIV-1 strains weakens cross-sectional analyses that are designed to identify treatment-related mutations, because some isolates from untreated persons will also have treatment-related mutations. Therefore, we adopted 2 approaches to reduce the influence of transmitted drug resistance. First, we excluded isolates from untreated persons with primary HIV-1 infection published in studies after the year 2000. Second, we excluded isolates from untreated persons that contained ⩾2 established nonpolymorphic drug-related mutations. The second approach was predicated on the strong likelihood that the presence of ⩾2 drug-resistance mutations at conserved sites in untreated persons does not reflect natural sequence variation but is, instead, most consistent with previous selective drug pressure.
Nonetheless, it is likely that some of the untreated isolates in the study were transmitted from persons who received antiretroviral therapy. Many of the nonpolymorphic treatment-related mutations occurred in untreated persons at a frequency of 0.1%–0.2%; a few—including the RT mutations M41L, E44D, T69S/N, K103N, and V108I—occurred at a frequency of 0.3%–0.5%. Whereas some of these may represent naturally occurring rare variants, others are likely to represent transmitted resistance, which suggests that any cutoff between polymorphic and nonpolymorphic mutations is slightly ambiguous. Although most T215 revertants were not significantly related to treatment, previous studies of primary HIV-1 infection have demonstrated that these mutations indicate transmission from a person treated with NRTIs [9] and should be considered treatment-related mutations.
Population stratification
To distinguish mutations developing in multiple individuals from those developing in a smaller number of founder viruses, we reconstructed the ancestral sequences at each node of a phylogenetic tree for all sequences and counted the number of times that each mutation developed along the tree. Mutations for which founder viruses accounted for ⩾50% of occurrences were not considered to be treatment-related mutations. This led to the exclusion of 1 protease mutation (T96S), which was reported primarily in 1 study of treated persons from 1 location [10].
One-fourth of the persons in the study were from northern California. Although we previously reported that patterns of drug-resistance mutations in subtype B isolates from heavily treated persons in northern California are similar to those in other parts of the world [11], it is possible that this large proportion of sequences from 1 region may have influenced our results. Drug-resistance surveillance efforts will benefit if investigators from different geographic regions also release primary sequence data on persons with well-characterized anti-retroviral treatment histories.
Sequence quality
All but a few sequences in the study were obtained by use of direct PCR or population-based sequencing of plasma virus. Thus, the presence of mixtures of >1 amino acid at the same position was expected. Indeed, 1 study of protease and RT sequences from heavily treated persons reported that electrophoretic mixtures occurred at 1% of all nucleotides and 5% of nucleotides at drug-resistance positions and that most electrophoretic mixtures reflected mixed virus populations rather than technical artifacts [12]. Nonetheless, when data from multiple different studies are combined, mutations present as part of a mixture are more likely than pure mutants to represent technical artifacts. For example, in the current data set (∼17,000 sequences), stop codons (n = 314) and active site mutations (protease positions 25–27 and RT positions 110, 185, and 186; n = 71) were 3–6 times more likely to be part of a mixture than they were to be in their pure form.
Multiple hypothesis testing
Holm’s method [8] was used to control for multiple comparisons, which reduced the risk of type I errors. This method is a sequential Bonferroni-type procedure that is appropriate for situations in which multiple statistically significant associations are expected. Because 265 protease, 383 RT, and 348 RT mutations, respectively, were present in ⩾2 persons who received PIs, NRTIs, and NNRTIs, we controlled for these numbers of comparisons using a familywise error rate of ⩽0.01.
Moreover, other research results support the supposition that many of the additional mutations that we identified contribute to HIV-1 drug resistance. For example, the substrate cleft mutation L23I decreases susceptibility to nelfinavir [13]; C95F increases the energy requirements related to inhibitor binding [14]; and E34Q, K43T, G48M, I54S, Q58E, T74S, L76V, and L89I have been related to decreased susceptibility to lopinavir [15–17].
Among the additional NRTI-related mutations, T69N reduces susceptibility to several NRTIs [18]; V75T reduces susceptibility to stavudine [19, 20]; H208Y is highly correlated with M41L, L210W, and T215Y [4]; and mutations at positions 43, 203, 218, and 228 have been related to poor virologic response to salvage treatment that contains tenofovir [21].
Among the additional NNRTI-related mutations, A98G, K101E, and V179D each cause >2-fold decreased susceptibility to ⩾1 NNRTI [20, 22, 23]; F227L, especially in combination with V106A, reduces susceptibility to nevirapine [24]; and K101P, K103S, and K238T reduce susceptibility to each NNRTI [25, 26]. Certain mutations at position 138 have also been shown to decrease HIV-1 susceptibility to an experimental class of NNRTIs [27].
Confounding effect of NRTI treatment on the identification of NNRTI-related mutations
Because NNRTI-treated persons are more likely to have also received NRTIs, it is not surprising that several mutations appeared to be selected by both NRTIs and NNRTIs, despite our regression model that attempted to control for the extent of NRTI treatment. The NNRTI-related mutation A98G occurred in 0.1% of untreated persons, 1.5% of those who received NRTIs, and 4.1% of those who received NRTIs and NNRTIs. Conversely, 7 NRTI-related mutations (K65R, L74V/I, V75M, K219N, and L228R/H) occurred 3–8 times more commonly in NNRTI-treated persons and were significantly related to NNRTI treatment even after we controlled for the extent of NRTI treatment. Three of these mutations (L74V/I and V75M) have previously been related to NNRTI resistance, and a structural mechanism for that association has been proposed [28]. For the remaining NRTI mutations, the association may reflect drug usage patterns. For example, the incidence of K65R has increased concurrently with the increased use of regimens that contain both tenofovir and NNRTIs.
Conclusion
We identified 60 nonpolymorphic PI-related mutations, 43 nonpolymorphic NRTI-related mutations, and 26 nonpolymorphic NNRTI-related mutations that are indicators of antiretroviral selective drug pressure in subtype B isolates. Eleven of these 129 mutations were polymorphic in ⩾1 non-B subtype, which indicates the need for similar studies of each of the other common viral subtypes [29]. The nonpolymorphic treatment-related mutations identified in the present study—or a carefully chosen subset—would be useful for drug-resistance surveillance.
Acknowledgments
Financial support: National Institutes of Allergy and Infectious Diseases (grant AI46148-01 to S.-Y.R. and R.W.S.); California University-Wide AIDS Research Program (to R.W.S., W.J.F., A.R.Z., and L.H.).
APPENDIX
Protease consensus B reference sequence: PQITLWQRPLVTIKIGGQLKEALLDTGADDTVLEEMNLPGRWKPKMIGGIGGFIKVRQYDQILIEICGHKAIGTVLVGPTPVNIIGRNLLTQIGCTLNF.
RT consensus B reference sequence (positions 1–240): PISPIETVPVKLKPGMDGPKVKQWPLTEEKIKALVEICTEMEKEGKISKIGPENPYNTPVFAIKKKDSTKWRKLVDFRELNKRTQDFWEVQLGIPHPAGLKKKKSVTVLDVGDAYFSVPLDKDFRKYTAFTIPSINNETPGIRYQYNVLPQGWKGSPAIFQSSMTKILEPFRKQNPDIVIYQYMDDLYVGSDLEIGQHRTKIEELRQHLLRWGFTTPDKKHQKEPPFLWMGYELHPDKWT.
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Hirsch MS, Brun-Vezinet F, Clotet B, et al. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society—USA Panel. Clin Infect Dis. 2003;37:113–28. doi: 10.1086/375597. [DOI] [PubMed] [Google Scholar]
- 2.Clavel F, Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–35. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 3.Wu TD, Schiffer CA, Gonzales MJ, et al. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J Virol. 2003;77:4836–47. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzales MJ, Wu TD, Taylor J, et al. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS. 2003;17:791–9. doi: 10.1097/01.aids.0000050860.71999.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuiken CL, Foley B, Hahn BH, et al. Human retroviruses and AIDS: a compilation and analysis of nucleic and amino acid sequences. Los Alamos National Laboratory; Los Alamos, NM: 1999. [Google Scholar]
- 7.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: 2004. Top HIV Med. 2004;12:119–24. [PubMed] [Google Scholar]
- 8.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 9.Yerly S, Rakik A, De Loes SK, et al. Switch to unusual amino acids at codon 215 of the human immunodeficiency virus type 1 reverse transcriptase gene in seroconvertors infected with zidovudine-resistant variants. J Virol. 1998;72:3520–3. doi: 10.1128/jvi.72.5.3520-3523.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masciotra S, Livellara B, Belloso W, et al. Evidence of a high frequency of HIV-1 subtype F infections in a heterosexual population in Buenos Aires, Argentina. AIDS Res Hum Retroviruses. 2000;16:1007–14. doi: 10.1089/08892220050058425. [DOI] [PubMed] [Google Scholar]
- 11.Gonzales MJ, Belitskaya I, Dupnik KM, Rhee SY, Shafer RW. Protease and reverse transcriptase mutation patterns in HIV type 1 isolates from heavily treated persons: comparison of isolates from northern California with isolates from other regions. AIDS Res Hum Retroviruses. 2003;19:909–15. doi: 10.1089/088922203322493085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shafer RW, Hertogs K, Zolopa AR, et al. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J Clin Microbiol. 2001;39:1522–9. doi: 10.1128/JCM.39.4.1522-1529.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston E, Winters MA, Rhee SY, Merigan TC, Schiffer CA, Shafer RW. Association of a novel human immunodeficiency virus type 1 protease substrate cleft mutation, L23I, with protease inhibitor therapy and in vitro drug resistance. Antimicrob Agents Chemother. 2004;48:4864–8. doi: 10.1128/AAC.48.12.4864-4868.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prashar V, Hosur MV. 1.8A X-ray structure of C95M/C1095F double mutant of tethered HIV-1 protease dimer complexed with acetyl pepstatin. Biochem Biophys Res Commun. 2004;323:1229–35. doi: 10.1016/j.bbrc.2004.08.226. [DOI] [PubMed] [Google Scholar]
- 15.Parkin NT, Chappey C, Petropoulos CJ. Improving lopinavir genotype algorithm through phenotype correlations: novel mutation patterns and amprenavir cross-resistance. AIDS. 2003;17:955–61. doi: 10.1097/00002030-200305020-00003. [DOI] [PubMed] [Google Scholar]
- 16.Loutfy MR, Raboud JM, Walmsley SL, et al. Predictive value of HIV-1 protease genotype and virtual phenotype on the virological response to lopinavir/ritonavir-containing salvage regimens. Antivir Ther. 2004;9:595–602. [PubMed] [Google Scholar]
- 17.Mueller S, Daeumer M, Kaiser R, Walter H, Colonno R, Korn K. Susceptibility to saquinavir and atazanavir in highly protease inhibitor (PI) resistant HIV-1 is caused by lopinavir-induced drug resistance mutation L76V. Antivir Ther. 2004;9:S44. [Google Scholar]
- 18.Winters MA, Merigan TC. Variants other than aspartic acid at codon 69 of the human immunodeficiency virus type 1 reverse transcriptase gene affect susceptibility to nucleoside analogs. Antimicrob Agents Chemother. 2001;45:2276–9. doi: 10.1128/AAC.45.8.2276-2279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacey SF, Larder BA. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob Agents Chemother. 1994;38:1428–32. doi: 10.1128/aac.38.6.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masquelier B, Tamalet C, Montes B, et al. Genotypic determinants of the virological response to tenofovir disoproxil fumarate in nucleoside reverse transcriptase inhibitor-experienced patients. Antivir Ther. 2004;9:315–23. [PubMed] [Google Scholar]
- 22.Byrnes VW, Emini EA, Schleif WA, et al. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reserve transcriptase inhibitors. Antimicrob Agents Chemother. 1994;38:1404–7. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacheler L, Jeffrey S, Hanna G, et al. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol. 2001;75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balzarini J, Pelemans H, Esnouf R, De Clercq E. A novel mutation (F227L) arises in the reverse transcriptase of human immunodeficiency virus type 1 on dose-escalating treatment of HIV type 1-infected cell cultures with the nonnucleoside reverse transcriptase inhibitor thiocarboxanilide UC-781. AIDS Res Hum Retroviruses. 1998;14:255–60. doi: 10.1089/aid.1998.14.255. [DOI] [PubMed] [Google Scholar]
- 25.Harrigan PR, Mo T, Wynhoven B, et al. Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitor and clinical correlates. AIDS. 2005;19:549–54. doi: 10.1097/01.aids.0000163930.68907.37. [DOI] [PubMed] [Google Scholar]
- 26.Petropoulos C, Chappey C, Parkin NT. High-level resistance to HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the absence of known resistance mutations [abstract H-451]. Program and abstracts of the 43rd Interscience Conference on Antimicrobial Agents and Chemotherapy (Chicago); Washington, DC. 2003; American Society for Microbiology; p. 307. [Google Scholar]
- 27.Balzarini J, Karlsson A, Sardana VV, Emini EA, Camarasa MJ, De Clercq E. Human immunodeficiency virus 1 (HIV-1)-specific reverse transcriptase (RT) inhibitors may suppress the replication of specific drug-resistant (E138K) RT HIV-1 mutants or select for highly resistant (Y181C→C181I) RT HIV-1 mutants. Proc Natl Acad Sci USA. 1994;91:6599–603. doi: 10.1073/pnas.91.14.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleim JP, Rosner M, Winkler I, et al. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor- specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:325–37. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]


