Summary
Fine-tuning of light signaling is crucial to plant development. Following light-triggered nuclear translocation, the photoreceptor phytochrome A (phyA) regulates gene expression under continuous far-red light, and is rapidly destabilized upon red light irradiation by E3 ubiquitin ligases including COP1. Here we provide evidence that the light signaling repressors SPA proteins contribute to COP1-mediated phyA degradation and that a COP1/SPA1 protein complex is tightly associated with phyA ubiquitination activity. Furthermore, a phosphorylated phyA form accumulates in the nucleus and preferentially associates with the COP1/SPA1 complex. In contrast, underphosphorylated phyA predominantly associates with the phyA-signaling intermediates FHY3 and FHY1. However, COP1 associates with underphosphorylated phyA in the absence of FHY3 or FHY1, suggesting that phyA associations with FHY3 and FHY1 protect underphosphorylated phyA from being recognized by the COP1/SPA complex. We propose that light-induced phyA phosphorylation acts as a switch controlling differential interactions of the photoreceptor with signal propagation or attenuation machineries.
Keywords: phytochrome A, phosphorylation, ubiquitination, COP1, SPA proteins, light signaling
Introduction
Plants monitor light by a set of photoreceptors such as phytochromes (phys) to optimize development. Phys are dimeric chromoproteins that undergo photoconversion between the red light (600–700 nm; R) absorbing Pr form and the far red light (700–750 nm; FR) absorbing, biologically active Pfr form (Chen et al., 2004). Light activation of phys induces their nuclear translocation (Nagatani, 2004; Nagy and Schafer, 2002), thereby reprogramming gene expression through direct interaction with several transcription factors of the bHLH family, including PIF3 and PIF3-like proteins (Al-Sady et al., 2006; Bauer et al., 2004; Hiltbrunner et al., 2005; Khanna et al., 2004; Martinez-Garcia et al., 2000). In Arabidopsis, phyA is the primary photoreceptor for responses to continuous far-red light (FRc), termed FR high irradiance response (FR-HIR), and is rapidly degraded upon exposure to red or white light (Chen et al., 2004; Clough and Vierstra, 1997). Genetic and genomic evidence points to a pivotal role of two sets of nuclear factors in phyA signaling: FHY1 and FHL that promote phyA nuclear accumulation (Desnos et al., 2001; Zhou et al., 2005), and FHY3 and FAR1 that act as transcription factors essential for FHY1/FHL gene expression (Hudson et al., 1999; Lin et al., 2007; Wang and Deng, 2002; Wang et al., 2002). These findings indicate that critical phyA signaling events occur within the nucleus (Chen et al., 2004; Nagatani, 2004; Nagy and Schafer, 2002).
In the R-induced phyA destabilization, the photomorphogenic repressor protein COP1 acts as an E3 ubiquitin ligase that directly binds and targets phyA for degradation (Seo et al., 2004). COP1 also targets several photomorphogenesis-promoting transcription factors for degradation (Jang et al., 2005; Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2004; Seo et al., 2003; Yang et al., 2005). COP1 contains a RING-finger domain, a coiled-coil domain, and a WD40 domain that directly recognizes its substrate proteins (Holm et al., 2001). COP1 is present as part of large protein complex(es) that also contains the nuclear-resided light signaling repressor SPA1 (Hoecker et al., 1999; Saijo et al., 2003). SPA1 is a member of the WD40 domain-containing SPA protein family, and was shown to modulate the E3 ligase activity of COP1 in vitro (Saijo et al., 2003; Seo et al., 2003). Genetic and biochemical data suggest that all four SPA proteins (SPA1–SPA4) work together with COP1 in repressing light signaling (Hoecker, 2005; Laubinger et al., 2004). However, the precise mechanisms by which SPA proteins support COP1-mediated substrate ubiquitination remain unclear.
Reversible phosphorylation has also been described as a signal-attenuating mechanism for phyA: phosphorylated phyA shows low Pfr stability in vivo and low affinity in vitro for signaling intermediates (Kim et al., 2004; Ryu et al., 2005). Previous studies have identified phosphorylation sites on oat phyA (Lapko et al., 1999; Yeh and Lagarias, 1998), of which distinct phosphorylation sites seem to affect the stability and protein-protein interactions of the photoreceptor (Kim et al., 2004; Ryu et al., 2005). However, it remains to be elucidated how phyA phosphorylation status is linked to signal propagation or attenuation in vivo. Our data suggest that phosphorylation-dependent sorting of phyA into different protein complexes provides a molecular basis for balanced control of plant development in changing light environment.
Results and Discussion
In an attempt to purify the presumed COP1-SPA1 complex(es), we employed a modified tandem affinity purification (TAP)-based procedure (Saijo et al., 2003) to isolate SPA1-associated proteins in Arabidopsis (Figure 1A). As expected, both COP1 and SPA1 proteins co-purified throughout the TAP procedure (Figure 1A). Gel filtration analysis confirmed co-fractionation of both proteins in the purified samples (Figure S1A in the Supplemental Research Data). Mass spectrometry analysis of an ~72 kDa band specifically associated with TAPSPA1 showed a 100% match to COP1 (Figure 1A). In contrast, phyA was not co-purified throughout the TAP procedure, indicating that phyA is not an integral component of the COP1/SPA1 complex(es) (Figure S1B), despite evident co-precipitation of TAPSPA1 with phyA (Figure S1C).
Figure 1. The COP1/SPA1 complex associates with an E3 activity for phyA ubiquitination.
(A) TAPSPA1 co-purified proteins from seedlings following the purification scheme (right) were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and silver-stained. Immunoblots with anti-myc (for TAPSPA1) and anti-COP1 antibodies are presented (middle). The box shows COP1 peptide fragments identified by mass spectrometry analysis. GFP, TAPGFP; SPA1, TAPSPA1; IgG, Immunoglobulin G; 3C–P, 3C protease. (B and C) phyA ubiquitination by TSC in vitro. Purified TSC (B) and an analytical scale TSC fraction (C) as well as their corresponding TAPGFP controls were assayed for E3 activity on GSTphyA. Ubiquitinated products were detected by immunoblotting (WB) with anti-phyA and ubiquitin (Ub) antibodies (B) or with anti-phyA and Flag antibodies (C). phyA was recovered by immunoprecipitation (IP) with anti-GST antibodies (C). The open and closed arrowheads indicate positions of apparently mono-ubiquitinated and unmodified phyA, respectively.
We then examined the E3 activity of isolated TAPSPA1 and COP1 complex (TSC) toward phyA in vitro, using recombinant Arabidopsis phyA fused with glutathione S-transferase (GSTphyA) as the substrate. Neither the photoconversion nor absence of the chromophore influenced in vitro phyA ubiquitination by recombinant COP1 (Seo et al., 2004). We detected a size shift consistent with mono-ubiquitination of phyA when incubated with TSC sample but not with the TAPGFP control sample in the presence of recombinant E1 and E2 (Figure 1B). At present, the biological significance of phyA mono-ubiquitination is unknown. We further tested whether the COP1/SPA1 complex mediates phyA poly-ubiquitination that generally leads to subsequent proteasomal degradation (Jabben et al., 1989; Pickart, 2001). To facilitate specific detection of ubiquitinated phyA, we developed a procedure using Flag-tagged ubiquitin (Ub) and partially purified TSC at an analytical scale. We recovered GSTphyA from the E3 assay reactions and then performed immunoblot analysis with anti-Flag antibody. We detected the attachment of Flag-Ub to GSTphyA in both high molecular weight (poly-ubiquitinated) as well as apparent mono-ubiquitinated GSTphyA forms, specifically when incubated with the TSC sample (Figure 1C). Co-purification and hence tight association of the COP1/SPA1 complex with robust E3 activity for phyA supports a notion that the COP1/SPA1 complex(es) represents the core biochemical entity of an E3 ligase responsible for phyA ubiquitination.
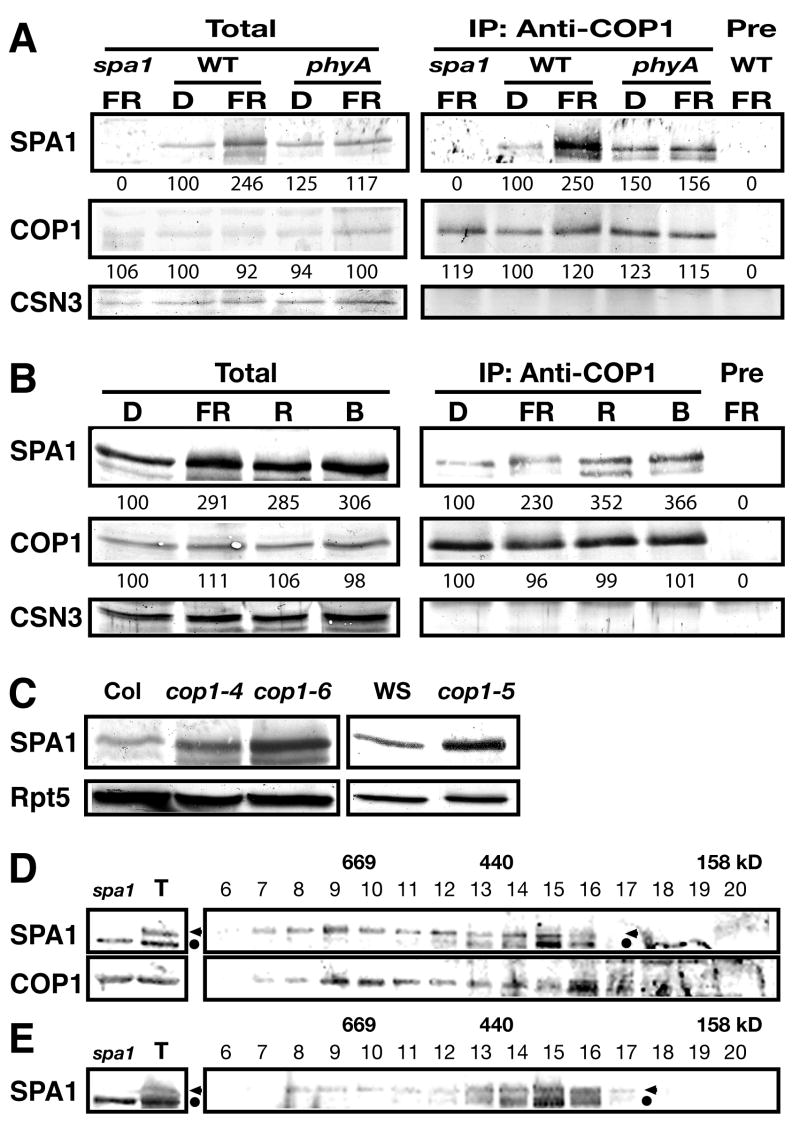
We next characterized the endogenous COP1/SPA1 complex. SPA1 abundance is elevated under FRc in the wild type but not in a phyA-101 null mutant, indicating that phyA is responsible for the increase in SPA1 accumulation under FRc (Figure 2A). As previously reported (Fittinghoff et al., 2006), SPA1 is more abundant under various light conditions compared to darkness (Figure 2B). We then verified endogenous COP1-SPA1 interaction by co-immunoprecipitation (co-IP) analysis of the two proteins (Figure 2A). Although essentially the same amounts of COP1 were precipitated with COP1-specific antibodies, the yield of SPA1 in COP1 co-IP was slightly higher under all light conditions examined than under darkness (Figures 2A and B), likely reflecting the elevation in total SPA1 abundance under light conditions. A phyA-induced SPA1 accumulation followed by enhanced COP1-SPA1 association may serve to desensitize phyA-signaling under FRc. In addition, the results support the previous finding that SPA1 also functions under light conditions other than FRc (Fittinghoff et al., 2006). Moreover, SPA1 accumulates to higher levels in the hyper-photomorphogenic cop1 mutant backgrounds than in a wild type background (Figure 2C). Furthermore, gel filtration analysis of endogenous COP1 and SPA1 showed their co-fractionation (Figure 2D). The size of endogenous SPA1 peak fractions (Fractions 8–10 in Figure 2D) was essentially the same as that of purified TSC (Figure S1A). However, the SPA1 peak fractions were slightly shifted toward smaller size in the cop1–5 null mutant (Figure 2E), supporting the idea that SPA1 acts as part of the COP1 complex(es).
Figure 2. Endogenous COP1/SPA1 complex characterization.
(A) phyA-induced SPA1 accumulation and SPA1-COP1 interaction. COP1 co-immunoprecipitates were analyzed by immunoblotting with the indicated antibodies on the left. Pre, control immunoprecipitation (IP) with the pre-immune sera. D, continuous darkness; FR, continuous far-red light. All genotypes used are in the RLD background. (B) COP1 co-IP of SPA1 from the wild type RLD under various light conditions. R, continuous red light; B, continuous blue light. Numbers under lanes in (A) and (B) indicate relative band intensities that were quantified and normalized for each panel. (C) Immunoblotting of dark-grown seedling lysates. CSN3 (A and B), and Rpt5 (C) were monitored as loading controls. WS, the Wassilewskija ecotype. (D and E) Gel filtration profiles of SPA1 and COP1 in the dark-grown wild type RLD (D) and a cop1–5 null mutant (E). T and spa1 represent total lysate inputs and those from spa1–3 used for gel filtration, respectively. The fraction numbers and molecular weight are indicated above. The arrowhead and dot indicate SPA1 and a cross-reacting band recognized by anti-SPA1 antibodies, respectively.
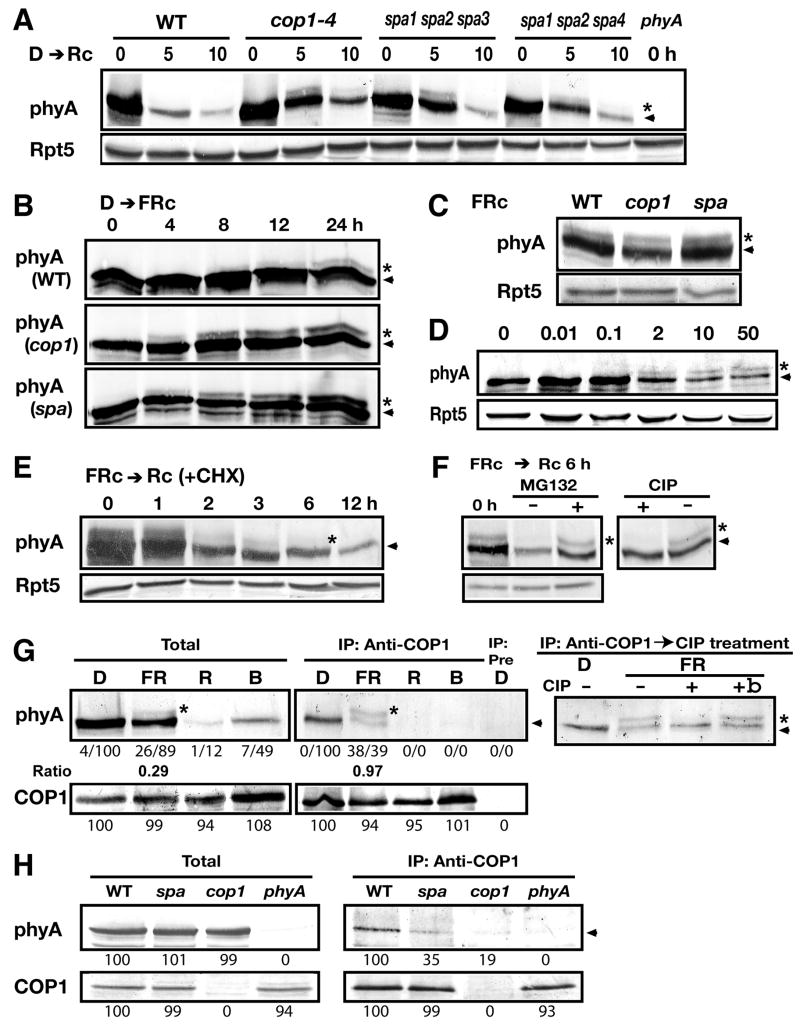
To examine functional significance of the COP1/SPA protein complex in phyA degradation, we monitored phyA degradation kinetics in the spa1 spa2 spa3 and spa1 spa2 spa4 triple mutants, considering genetic redundancy among the SPA genes (Laubinger et al., 2004). Both spa triple mutants exhibited a delay in Rc-induced phyA degradation similar to cop1–4 (Figure 3A). Thus, SPA proteins indeed contribute to phyA degradation. All SPA proteins may act as part of the COP1 E3 complex(es), as demonstrated herein with the COP1/SPA1 complex. The contribution of each SPA protein may be different, as the spa1 spa2 spa3 mutant shows a more severe cop1–4 mutant-like phenotype than the spa1 spa2 spa4 mutant in phyA-dependent responses (Figure S2; Laubinger et al., 2004). However, it is notable that light-induced phyA degradation still occurs in the null cop1–5 allele, revealing the presence of a COP1-independent phyA-degradation pathway (Figure S3A). Interestingly, no significant defect was observed for pulsed R-induced phyA degradation in the cop1 mutants tested (Figure S3B), suggesting that the COP1-dependent and independent pathways contribute differentially to the light-induced phyA elimination under distinct light conditions.
Figure 3. The COP1/SPA complex(es) preferentially associate with phosphorylated phyA in vivo.
(A–B) Five-day-old dark-grown seedlings exposed to Rc (A) or FRc (B) for the indicated times and FRc-grown seedlings (C and D) were subjected to immunoblot analyses. The cop1–4 (cop1), spa1spa2spa3 (spa), spa1spa2spa4, phyA-201 (phyA) mutants, and wild type (WT) were used. Rpt5 was monitored as a loading control in (A and C–F). The numbers above the panel indicate FRc fluence rate (μmol/m2s) in (D). The asterisk and arrowhead respectively represent positions of the large and unmodified phyA forms in (A–E). (E) Immunoblot analysis of FRc-grown seedlings exposed to Rc for the indicated times in the presence of 100 μM cycloheximide. (F) FRc-grown seedlings pre-treated with or without 50 μM MG132 were transferred to Rc for 6 h. Lysates of the MG132-treated plants were incubated with or without calf intestinal alkaline phosphatase (CIP). The asterisk and arrowhead represent the phosphorylated and underphosphorylated phyA forms, respectively, in (F–H). (G) Five-day-old seedlings were subjected to COP1 co-IP analysis under the indicated continuous light conditions (D, darkness; FR, far-red light; R, red light; B blue light). Pre represents a mock IP control with the pre-immune sera. Co-immunoprecipitated phyA with COP1 was incubated with CIP (+), boiled inactive CIP (+b), or without CIP (−). (H) COP1-phyA co-IP was reduced in the cop1–4 and spa triple mutant under darkness. Numbers below immunoblots indicate relative band intensities normalized for each panel. The ratio of the phosphorylated to underphosphorylated form is shown for phyA.
We noticed that a slow-migrating (or large) phyA form accumulates in the cop1 and spa triple mutants upon continuous R (Rc) but not in the wild type (Figure 3A). The large phyA band was not detectable in darkness despite high phyA abundance (Figure 3A) as well as during pulsed R-induced degradation (Figure S3B). It was reported that low-rate of phyA degradation occurs upon FRc (Clough and Vierstra, 1997). We found that a large phyA form accumulates when dark-grown seedlings were exposed to prolonged FRc (for approximately 8 hours in the wild type, Figure 3B). Noticeably, the large phyA form emerges significantly earlier and is more abundant at early time points (4–8 h) in the cop1 and spa triple mutants than in the wild type (Figure 3B). We were unable to detect this phyA form in 2–60 minutes upon Rc or FRc (data not shown), a time period during which a phyA-GFP fusion is re-localized to the early-type nuclear speckles (Bauer et al., 2004). These data suggest that both prolonged Rc and FRc facilitate the large phyA formation, and that this modified phyA form may be a preferred target for COP1-mediated degradation. However, COP1 and SPA proteins seem to have little effect on the overall phyA steady-state levels under extended FRc (5 days), as no marked differences in phyA levels were observed among the wild type, cop1 and spa triple mutants (Figure 3C). In addition, we observed that high fluence rates of FRc promote the formation of the large phyA species (Figure 3D).
We then examined whether both phyA forms are destabilized upon Rc. When transferring FRc-grown seedlings to Rc in the presence of the protein synthesis inhibitor cycloheximide, the amount of both phyA forms declines rapidly (Figure 3E). Pretreatment of plants with the proteasome inhibitor MG132 stabilized both phyA forms (Figure 3F). Subsequent incubation with protein phosphatase specifically diminished the large phyA form (Figure 3F). Thus, we conclude that the large and small phyA forms represent phosphorylated and underphosphorylated phyA, respectively. The results also suggest that Rc-induced elimination of both phyA forms is dependent on the 26S proteasome pathway.
To determine which form of phyA might be the preferred substrate for COP1-mediated degradation, we performed co-IP analysis for endogenous proteins (Figure 3G). Interestingly, the large phyA band was significantly enriched through the co-IP with COP1 under FRc, compared to the underphosphorylated phyA form (Figure 3G). Subsequent protein phosphatase treatment confirmed that the enriched large phyA form by COP1 co-IP procedure is phosphorylated phyA (Figure 3G). This suggests that COP1 preferentially associates with the phosphorylated phyA form, hereafter designated COP1-associated phosphorylated phyA (cap-phyA), under FRc. Both cap-phyA and underphosphorylated phyA are expected to be predominantly in the Pr state under FRc, with a pool of phyA photo-cycling between Pfr and Pr forms (Mancinelli, 1994; Shinomura et al., 2000). It is unclear whether the two photochemical forms differ in the relevant phosphorylation status. Given the requirement of prolonged exposure to FRc at a high fluence rate for cap-phyA accumulation (Figures 3B, 3D and S3C), cap-phyA-mediated modulation of phyA signaling may be more pronounced under prolonged than pulsed FR. In dark-grown seedlings, COP1 co-IP recovers only underphosphorylated phyA, consistent with the lack of cap-phyA detection from total protein extracts (Figure 3G). We were unable to observe robust co-IP between COP1 and cap-phyA upon a brief Rc treatment of dark-grown seedlings, when active phyA ubiquitination and its immediate degradation are expected to occur.
To test whether SPA proteins help COP1 to bind phyA, we compared COP1-phyA association between the spa1spa2spa3 triple mutant and wild type plants. To facilitate the comparison, we performed co-IP analysis with dark-grown seedlings from which high amount of phyA is readily recovered in COP1 co-IP (Figure 3G). phyA levels are essentially the same in the wild type as in the spa triple mutant (Figure 3H). However, the yield of phyA in COP1 co-IP was reduced in spa triple mutant compared to the wild type, whereas similar amounts of COP1 were recovered (Figure 3H). Thus, this difference may reflect a decrease in the amount of phyA associated with COP1 in the absence of 3 out of 4 SPA proteins. Our data suggest that SPA proteins likely contribute to the COP1 E3-substrate recognition, a critical step in ubiquitination (Pickart, 2001). Furthermore, SPA1 co-IP analysis indicates that SPA1-phyA association is dependent on COP1 (Figure S4). Together, our data suggest that functional COP1/SPA complex(es) may be required for effective recognition of phyA.
In light of the notion that the majority of COP1/SPA proteins and phyA is distributed in different subcellular compartments (nuclear and cytosolic, respectively) under darkness, we suspected a possibility that their interaction might occur during our IP procedure. To localize both phyA forms, we performed biochemical fractionation of the endogenous proteins from the wild type seedlings. COP1 and phyA were detected in the nuclear fraction under all light conditions tested. Notably, we also detected a low level of phyA in the nuclear fraction of dark-grown seedlings (Figure 4A). phyA nuclear translocation is known to be mediated by both the very low fluence response and HIR. Thus, the phyA pool detected in the nuclear fraction of dark-grown seedlings was likely generated by brief exposure to white light before seedling germination in our procedure. Strikingly, cap-phyA was found to accumulate at high levels in the nucleus of FRc-grown seedlings, but not of seedlings exposed to red light.
Figure 4. The nuclear-resident phyA-signaling intermediates FHY3 and FHY1 preferentially associate with underphosphorylated phyA in vivo.
(A) Immunoblot analysis of purified nuclear (N) and nuclei-depleted soluble (S) fractions from 5-day-old wild type seedlings grown under the indicated light conditions: D, continuous darkness; FR, FRc; D–R, D followed by Rc for 10 min. An approximately 12-fold quantity (in protein content) of the nuclear fraction compared to the nuclear-depleted fraction on a per-tissue amount basis was loaded. Fraction markers used were histone H3 (Histone), cytosolic HSP90 and phosphoenolpyruvate carboxylase (PEPC). Specificity of anti-FHY3 antibodies was verified (right). (B) Co-IP of phyA with the indicated antibodies using FRc-grown seedlings. Respective pre-immune serum was used as an IP control (Pre). The Rpt5 immunoblots are shown to verify co-IP specificity. (C) R-induced phyA-FHY3 dissociation. Design of experiments (top left): Total protein lysates of the FRc-grown phyA-overexpressing seedlings were irradiated with 5 min of FR light pulse (FR) or 5 min of FR light pulse immediately followed by 5 min of R light pulse (FR/R), and then subjected to co-IP analysis with anti-FHY3 and anti-COP1 antibodies (bottom). Immunoblot analysis of lysates of FRc-grown seedlings kept under Rc for 3 h ensured phyA stability during the procedure (top right). (D) COP1 co-IP analysis of FRc-grown seedlings including wild types (Col-0 and No-0), fhy3-1 far1–2, and fhy1-1. Relative band intensities normalized for each panel and the ratio of cap-phyA to underphosphorylated phyA are shown. The asterisk and arrowhead indicate cap-phyA and underphosphorylated phyA, respectively.
We next explored potential phyA interactions with the nuclear-localized phyA-signaling intermediates FHY3 and FHY1. Anti-FHY3 antibodies detected endogenous FHY3 in the nuclear fraction (Figure 4A). Co-IP with anti-FHY3 antibodies using FRc-grown seedlings predominantly recovered underphosphorylated phyA (Figure 4B, IP1). Similar results were obtained with anti-FHY1 co-IP (Figure 4B, IP2). By contrast, there is an enrichment of cap-phyA in SPA1 co-IP (Figures 4B, IP3 and S4), reminiscent of COP1 co-IP results (Figure 3G). Thus, it appears that underphosphorylated phyA predominantly associates with FHY3 and FHY1, through which phyA signaling may occur. However, the exact molecular mechanisms are currently unknown. On the other hand, cap-phyA seems to preferentially associate with the COP1/SPA complex(es) under FRc.
Under FRc, 97% of phyA is present in the Pr state at the steady-state level (Mancinelli, 1994). It has been proposed that a short-lived signal constantly generated by photo-cycling of phyA between the Pr and Pfr forms mediates FR-HIR (Shinomura et al., 2000). To examine the photoconversion effect on phyA associations with FHY3 and COP1, we exposed protein extracts from FRc-grown seedlings to FR or FR/R pulses before co-IP analysis. Total phyA abundance and the ratio of both phyA forms remained essentially the same during the procedure (Figure 4C). The yield of phyA in FHY3 co-IP was high with FR irradiation but greatly reduced by an R pulse following FR (Figure 4C), suggesting that the endogenous FHY3 associates more stably with Pr-phyA. If FHY3-phyA association supports signal propagation, then their R-induced dissociation is in accordance with the FR/R (but not R/FR) photo-reversibility in the FR-HIR (Shinomura et al., 2000). In contrast, COP1-phyA co-IP was not significantly altered by the R/FR pulse exposure (Figure 4C).
We further examined a potential role of FHY3/FAR1 and FHY1 in the COP1-phyA interaction. We noticed that the ratio of cap-phyA versus underphosphorylated phyA is reduced in total protein extracts from both fhy3 far1 and fhy1 mutants (Figure 4D). The ratio of cap- versus under-phosphorylated phyA in COP1 co-immunoprecipitates largely reflects that in total protein extracts from these mutants (Figure 4D). Nuclear fractionation data suggest that phyA, in particular cap-phyA, is reduced in the nucleus in these mutants (Figure S5). Therefore, FHY3/FAR1 and FHY1 may promote cap-phyA accumulation within the nucleus under FRc, which possibly serves to desensitize light-activated phyA. Strikingly, COP1 associates with high amounts of underphosphorylated phyA in the absence of either FHY3/FAR1 or FHY1. In contrast, there is no discernable alteration in COP1-SPA1 co-IP by these mutations (Figure 4D). These data imply that COP1 may not be intrinsically selective in the recognition of the distinctly phosphorylated phyA forms, but bind phyA pools that are not associated with signaling intermediates. Thus, phyA-signaling intermediates such as FHY3 and FHY1 may sequester and prevent underphosphorylated phyA from associating with the COP1-SPA complex(es) under FRc.
It is unknown how many phosphorylation sites (target amino acid residues) contribute to produce cap-phyA. It could be on one or a combination of multiple sites. On oat phyA, it was reported that light-dependent Ser598 phosphorylation affects phyA interactions with signaling intermediates, and that Ser7 phosphorylation influences phyA protein stability (Kim et al., 2004; Lapko et al., 1999). However, sequence alignment analysis did not reveal obvious corresponding phosphorylation residues between Arabidopsis and oat phyA (Figure S6). We also failed to detect a mobility shift on oat phyA expressed in Arabidopsis seedlings under FRc (Figure S7). Thus it is possible that other as yet unidentified residues are involved in the generation of cap-phyA in Arabidopsis.
Our findings support a model in which FHY3/FAR1 and FHY1 contribute to keep underphosphorylated phyA bound in a plausible signaling complex(es), limiting its vulnerability to COP1/SPA complex(es)-mediated proteolysis under FRc. Selective recruitment of underphosphorylated phyA into associations with signaling intermediates, such as FHY3 and FHY1 (Figure 4B), may lead to signal propagation. As a result, COP1 access to underphosphorylated phyA is blocked. In response to prolonged FR, a pool of cap-phyA accumulates within the nucleus and dissociates from the presumed signaling complex(es). This phyA pool is available for recognition by COP1/SPA complex(es). Thus, together with phyA-induced SPA1 accumulation (Figure 2A), phyA phosphorylation represents a feedback control to desensitize phyA signaling under FRc (Figure S8). It remains to be elucidated whether phyA-COP1 interaction plays another role beyond phyA degradation. It is also possible that cap-phyA rather acts as a decoy to trap the COP1/SPA complex(es) with this phyA form, thereby protecting underphosphorylated phyA. Accordingly, loss of FHY3/FAR1 or FHY1 may no longer be able to maintain the proposed phyA-signaling complex(es) in the nucleus, and thus allows access of COP1/SPA complex(es) to underphosphorylated phyA. This model is consistent with the enrichment of cap-phyA in co-IP with COP1 and SPA1 under FRc (Figures 3G and 4B).
The lack of a detectable change in total phyA abundance in the cop1 and spa triple mutants under FRc (Figure 3C) suggests that phyA recruitment into the COP1/SPA complex(es) is not sufficient to activate phyA ubiquitination and degradation. We suggest that the Pfr state of phyA may stimulate phyA ubiquitination/degradation in vivo. Under R or white light, the majority (~88%) of phyA photo-converts to the Pfr state, while under FRc only 3% of phyA is in the Pfr state (Mancinelli, 1994). This drastic difference (about 30 fold) in the Pfr-phyA abundance under distinct light conditions might explain the vastly different rates of phyA proteolysis. Upon R-induced phyA photoconversion from the Pr to Pfr form, its interaction with COP1/SPA complex may lead to rapid ubiquitination/degradation of phyA. Furthermore, Pfr-phyA dissociation from FHY3 (Figure 4C) and light-induced FHY1 destabilization (Shen et al., 2005) should facilitate targeting of underphosphorylated phyA for degradation. Alternatively, but not mutually exclusively, Pfr-phyA may transiently act as an active signaling form prior to its degradation, as proposed with a role in PIF3 phosphorylation (Al-Sady et al., 2006). In this scenario, phyA associations with FHY3 and FHY1 may support the supply of this active phyA form. In both models, phyA dissociation from its downstream signaling components presumably accelerates phyA recruitment, regardless of its phosphorylation state, to both COP1-dependent and independent degradation pathways (Figure S8).
Experimental Procedures
Plant material
Seed sterilization and plant growth were performed as previously described (Saijo et al., 2003). Detailed description of plant materials is provided in the Supplemental Material.
Protein biochemical experiments
Immunoblot, IP, and gel filtration analyses were performed essentially as previously described (Saijo et al., 2003). Further detailed procedures are described in the Supplemental Material. All IP experiments in Figure 4 were performed under dim green safe light unless otherwise stated. Quantification of immunoblots was carried out with ImageJ (http://rsb.info.nih.gov/ij/). Band intensities of total lysates and IP samples were respectively normalized with the value of background regions and that of corresponding regions for mock IP control. Relative band intensities were then calculated for each immunoblot panel. All IP and immunoblot experiments were repeated at least 3 times essentially with the same conclusions and representative results are shown.
Purification of TSC
We introduced a functional TAPSPA1 construct (Saijo et al., 2003) into a COP1 overexpression background (McNellis et al., 1994). Five-day-old dark- or continuous white light-grown seedlings (approximately 500 g fresh weight for mass spectrometry and 20 g for an E3 assay sample) were used for the TAP procedures as described previously (Saijo et al., 2003) with the modifications described in the Supplemental Material.
In vitro E3 assays
The N-terminal GST-tagged fusion protein with Arabidopsis phyA was expressed in E. Coli and purified using a glutathione matrix (Amersham). In vitro E3 assays were performed essentially as described previously (Saijo et al., 2003; Yang et al., 2005). Further details are provided in the Supplemental Materials.
Supplementary Material
Acknowledgments
We thank P. H. Quail, P. S. Song, and K. Sakaguchi for providing anti-phyA antibody, Arabidopsis lines expressing oat phyA, and E2, respectively, J. A. Sullivan, W. Gong and O.-S. Lau for assisting with in vitro E3 assays, and V. J. Karplus for proof reading this manuscript. Our work was supported by a grant from the National Institutes of Health (GM47850) to XWD. Y.S. was a Japanese Society for Promotion of Science Postdoctoral Fellow, and V.R. was a Human Frontier Science Program Organization Postdoctoral Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Sady B, Ni WM, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasorne-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KCS, Adam E, Fejes E, Schafer E, Nagy F. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. FHY1: a phytochrome A-specific signal transducer. Genes Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff K, Laubinger S, Nixdorf M, Fackendahl P, Baumgardt RL, Batschauer A, Hoecker U. Functional and expression analysis of Arabidopsis SPA genes during seedling photomorphogenesis and adult growth. Plant J. 2006;47:577–590. doi: 10.1111/j.1365-313X.2006.02812.x. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schafer E. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hoecker U. Regulated proteolysis in light signaling. Current Opinion in Plant Biology. 2005;8:469–476. doi: 10.1016/j.pbi.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Holm M, Hardtke CS, Gaudet R, Deng XW. Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 2001;20:118–127. doi: 10.1093/emboj/20.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabben M, Shanklin J, Vierstra RD. Ubiquitin-phytochrome conjugates - pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989;264:4998–5005. [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Shen Y, Han YJ, Park JE, Kirchenbauer D, Soh MS, Nagy F, Schafer E, Song PS. Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell. 2004;16:2629–2640. doi: 10.1105/tpc.104.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapko VN, Jiang XY, Smith DL, Song PS. Mass spectrometric characterization of oat phytochrome A: Isoforms and posttranslational modifications. Protein Sci. 1999;8:1032–1044. doi: 10.1110/ps.8.5.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell. 2004;16:2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancinelli AL. In: The physiology of phytochrome action. in photomorphogenesis in plants. 2. Kendrick RE, Kronenberg GHM, editors. Kluwer Academic Publishers; The Netherlands: 1994. pp. 211–269. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Deng XW. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development - evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell. 1994;6:1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Nagy F, Schafer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu Rev Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Ryu JS, Kim JI, Kunkel T, Kim BC, Cho DS, Hong SH, Kim SH, Fernandez AP, Kim Y, Alonso JM, et al. Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell. 2005;120:395–406. doi: 10.1016/j.cell.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang HY, Yang JP, Shen YP, Rubio V, Ma LG, Hoecker U, Deng XW. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- Shen YP, Feng SH, Ma LG, Lin RC, Qu LJ, Chen ZL, Wang HY, Deng XW. Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 2005;139:1234–1243. doi: 10.1104/pp.105.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome a for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Deng XW. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002;21:1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Ma LG, Habashi J, Li JM, Zhao HY, Deng XW. Analysis of far-red light-regulated genome expression profiles of phytochrome A pathway mutants in Arabidopsis. Plant J. 2002;32:723–733. doi: 10.1046/j.1365-313x.2002.01462.x. [DOI] [PubMed] [Google Scholar]
- Yang JP, Lin RC, James S, Hoecker U, Liu BL, Xu L, Deng XW, Wang HY. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh KC, Lagarias JC. Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Nat Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.