Abstract
TMPRSS2/ERG gene fusions are found in the majority of prostate cancers; however, there is significant heterogeneity in the 5′ region of the alternatively spliced fusion gene transcripts. We have found that there is also significant heterogeneity within the coding exons as well. There is variable inclusion of a 72-bp exon and other novel alternatively spliced isoforms. To assess the biological significance of these alternatively spliced transcripts, we expressed various transcripts in primary prostatic epithelial cells and in an immortalized prostatic epithelial cell line, PNT1a. The fusion gene transcripts promoted proliferation, invasion and motility with variable activities that depended on the structure of the 5′ region encoding the TMPRSS2/ERG fusion and the presence of the 72-bp exon. Cotransfection of different isoforms further enhanced biological activity, mimicking the situation in vivo, in which multiple isoforms are expressed. Finally, knockdown of the fusion gene in VCaP cells resulted in inhibition of proliferation in vitro and tumor progression in an in vivo orthotopic mice model. Our results indicate that TMPRSS2/ERG fusion isoforms have variable biological activities promoting tumor initiation and progression and are consistent with our previous clinical observations indicating that certain TMPRSS2/ERG fusion isoforms are significantly correlated with more aggressive disease.
Keywords: prostate cancer, invasion, ERG, proliferation, fusion gene
INTRODUCTION
Chromosomal rearrangements resulting in gene fusions and expression of functional proteins are common in non-epithelial malignancies (1). For certain malignancies, such as chronic myelogenous leukemia, the presence of the fusion gene (BCR-ABL) is critical for diagnosis and the fusion gene protein product is a key therapeutic target. The discovery of recurrent fusion of the androgen-regulated TMPRSS2 gene to the ETS transcription factors, particularly the ERG gene, in the majority of prostate cancer (PCa) lesions, has led to a paradigm shift in the study of PCa (2). The TMPRSS2/ERG fusion gene occurs in 15-80% of PCa lesions, depending on the clinical stage (3-14). A smaller percentage of cases contain fusions with genes for other ETS transcription factors (2, 4, 7, 9, 14-16), often with promoter fusion partners other than TMPRSS2 (17).
The TMPRSS2/ERG gene fusion arises by fusion of the promoter and 5′ portions of the TMPRSS2 gene (21q22.3) with the coding sequence of the ERG gene (21q22.2). Fusion of these two genes occurs by both intrachromosomal deletion and translocation (2, 6, 14, 18). The TMPRSS2 promoter, which contains androgen receptor (AR)-responsive promoter elements (18), can mediate the overexpression of ETS family members in PCa in response to androgens (2). The ubiquitous activity of AR in PCa cells would then result in the constitutive expression of ERG fusion transcripts in the neoplastic prostatic epithelium bearing this fusion gene.
There is significant heterogeneity in the structure of the 5′ end of the mRNA transcripts of the fusion gene (3, 5, 15, 19). Some prostate cancers express a single mRNA isoform, while others express multiple isoforms of the fusion gene that arise via alternative splicing of the initial fusion transcript. We have characterized 8 fusion types in PCa (3), which have been confirmed by others (4, 12), and other isoforms have been identified, as well. In all cases, the fusion mRNA includes the TMPRSS2 exon 1 and often exon 2, as well (5, 13). The most common transcript contains the TMPRSS2 exon 1 fused to ERG exon 4, such that translation would have to arise from an internal ATG codon and give rise to a slightly truncated protein which we have designated as the Type III isoform. Of particular interest is an isoform in which TMPRSS2 exon 2 is fused with ERG exon 4 (designated Type VI). This variant was present in 26% of our cases with fusion gene expression (3). For this isoform, translation can be initiated from the TMPRSS2 translation initiation codon and results in a true fusion protein containing the first five amino acids of the TMPRSS2 gene fused to a slightly truncated ERG protein. We found that expression of this isoform is associated with aggressive disease.
The TMPRSS2/ERG fusion can be detected in high grade prostatic intraepithelial neoplasia (4, 20) and in 40-60% of surgically treated prostate carcinomas. These findings argue that the fusion gene plays a critical role in prostate carcinogenesis. ETS transcription factors are generally mitogenic (21) and should promote tumor progression. Most, but not all, studies have shown an association between the presence of the TMPRSS2/ERG fusion and aggressive disease (3, 8, 9, 11-14, 22) although in some studies RT-PCR, and not fluorescence in-situ hybridization was used to assess fusion status, and it is possible that in some cases only a fraction of genes in the tumor may have fusion gene but generate enough transcript to give an RT-PCR product. We have shown that among cases with the TMPRSS2/ERG fusion, those expressing the Type VI isoform were more aggressive than those expressing Type III alone (3). Furthermore, some cases expressing only the Type III isoform had high levels of fusion gene expression that was also associated with aggressive disease (3).
Recent studies have characterized some of the biological activities of the most common isoform (Type III) in benign and transformed prostatic epithelial cells (23), (24). In order to better understand the biological activities of all of the alternatively spliced TMPRSS2/ERG fusion gene isoforms in PCa, we cloned the variant isoforms described above into expression vectors. During this process we noted a significant heterogeneity of the coding exons of the fusion gene. Systematic investigation of the biological activities of the various isoforms indicate that they have variable biological activities that can promote tumor initiation and progression, consistent with our previous clinical observations that certain TMPRSS2/ERG fusion isoforms are associated with more aggressive disease.
MATERIALS and METHODS
Prostate tissue samples
The clinical and pathological data for radical prostatectomy cancer samples has been described previously (3). RNAs were prepared as described previously (3).
Cloning and Expression of TMPRSS2/ERG Isoforms
The fusion transcript isoforms were amplified and subcloned into pcDNA 3.1/V5-His-Topo vector (Invitrogen, Carlsbad, CA) using patient cDNA samples (3). Primers used are listed in Table S1. Primary prostatic epithelial cells (PrEC) were purchased from the Lonza (Walkersville, MD) and maintained in media from the same supplier. Fusion isoforms were expressed in PrEC cells using the retrovirus pBMN-IRES- EGFP, which has a 5′LTR driving expression of cDNAs, followed by an IRES then GFP (25). High infection efficiency (greater than 80%) was shown in PrEC cells based on GFP fluorescence so cells were analyzed without selection. Primers used for the retrovirus construct were TMPERG Vir F XhoI: 5′- CCG CTCGAGCGCCTAAGCAGGAG-3′ and TMPERG Vir R NotI: 5′-CCCAGAATGCGGCCGCT TAGTAGTAAGTGCCC-3′. Amplified fragments were digested by XhoI and NotI before being ligated into pBMN-I-GFP vector. Retroviruses carrying these two isoforms were generated in Phoenix A packaging cells. PNT1a cells, were maintained in RPMI 1640 with 10% fetal bovine serum (FBS), and were stably transfected with fusion isoforms with a V5-tag, either individually or in combination as described previously (26) followed by G418 selection (100 ug/ml). Pooled cells were used for all experiments. The transcriptional level of the fusion isoforms in both types of cell lines were evaluated by real-time PCR normalized to β–actin as described previously (26).
Proliferation Assay
Cells (2.5 × 104) of each cell line were plated in 35 mm dishes in complete medium. Cells were trypsinized and counted using a Coulter counter at different time points in triplicate. The experiment was repeated 3 times.
Matrigel® Invasion Assay
The Matrigel invasion assays were performed in triplicate as described previously (27). The experiment was repeated 3 times.
Migration Assay
Motility was assessed using a scratch wound method as described previously (27). This experiment was repeated 4 times.
Soft Agar Colony Formation Assay
35mm dishes with 0.5% base agar layer mixed with 1X culture media plus 10% FBS were prepared before the seeding of cells. PNT1a cells (2.5 × 104) expressing the TMPRSS2/ERG fusion genes or vector controls were plated in 0.35% top agar layer each agar dishes. Plates were stained with crystal violet and cell colonies were counted after incubation at 37°C in humidified incubator for three weeks. This experiment was repeated twice.
Western Blot and Immunoprecipitation
Western blot was performed as described previously (27). Primary antibodies were anti-V5 monoclonal antibody (1:5000 dilution, Invitrogen), anti-Flag M2 monoclonal antibody (1:2000, Stratagene) or anti-β-actin (1:5000, Sigma, St Louis, MO). For immunoprecipitation, 1 mg of protein lysate from each sample was incubated with anti-V5 antibody (1:500) for 2 hours at 4°C. Then, 20 ul of protein A/G Agarose beads (Santa Cruz, SC-2003) were applied to each sample and incubated at 4°C overnight. Pellets were collected by centrifugation at 3000 rpm for 30 seconds. Supernatant was carefully aspirated and discarded. Pellets were washed 3 to 4 times with RIPA buffer. After the final wash, pellets were resuspensed in 25 ul 2x Laemmli sample buffer (Bio-Rad, Hercules, CA) analyzed by standard Western blot protocol (24).
Generation of shRNA against Type III TMPRSS2/ERG fusion mRNA
We designed single-stranded primer oligos targeting sequence around the fusion junction site by using Invitrogen's website BLOCK-iT™ RNAi Designer2 . The primers are shIIITop 5′- CACCGCGGCAGGAAGCCTTATCAGTTCGAA AACTGATAAGGCTTCCTGCCGC-3′ shIIIBot 5′- AAAAGCGGCAGGAAGCCTTATCAGTTTTCG AACTGATAAGGCTTCCTGCCGC-3′. The negative control shRNA target sequence has been described previously (28). Double stranded oligos were cloned into pENTR/U6 vector containing the U6 promoter and Pol III terminator. pENTR/U6 vector was transferred to plenti6/BLOCKit-DEST vector during LR recombination following the manufacturer's protocol (Invitrogen). Lentivirus was generated in 293FT cells by cotransfection with the packaging mix from Invitrogen and the final plenti6/BLOCKit-DEST expression construct. VCaP cells were infected with lentiviruses and stably selected in 2ug/ml Blasticidin media.
VCaP-Luc orthotopic mice model
pEF1-Luc-IRES-Neo luciferase expression plasmid has been described previously (29). The luciferase-encoding cassette was released by NheI/NotI digestion and inserted into a lentiviral vector pCDH-MCS1-EF1-Puro (SBI, Mountain View, CA) using the same digestion sites to create pCDH-Luc-EF1-Puro. By cotransfection of pCDH-Luc-EF1-Puro with necessary packaging plasmids in 293 cells, lentiviruses were generated. VCaP-sh-III cells and VCaP-sh-con cells were both infected with lentivirus carrying luciferase and stably selected in puromycin. Eight to ten-week old SCID mice were used for orthotopic injection. 1×106 VCaP-Luc-sh-III or VCaP-Luc-sh-con cells were used for each mouse. Total volume of 20 ul was orthotopically injected into the mouse prostate. In each group 20 mice were used. Tumor growth was assessed at 2, 3 and 4 weeks after injection using the IVIS imaging system (Xenogen, Alameda, CA). Mice were anesthetized and imaged ten minutes after D-Luciferin (Molecular Probes, Eugene, OR) injection (25 mg/kg i.p.). Mice were sacrificed at 4 weeks after imaging and tumors collected. Primary tumor weight was recorded and a full necropsy performed to identify metastasis. Differences in mean tumor size and image signal were examined by t-test.
Characterization of ERG isoforms in prostate
Based on NCBI published sequences and exon analysis through the Ensembl website1 we aligned 17 ERG exons to the genomic DNA sequence located on Chr.21 25732292-25364248. Primers to amplify ERG isoforms were designed and are listed in Table S1. PCR was performed based on a standard protocol and subcloning was done into Topo2.1 vector from Invitrogen. Vectors with insert were sequenced.
RESULTS
Alternative splicing of the coding exons of the TMPRSS2/ERG fusion gene
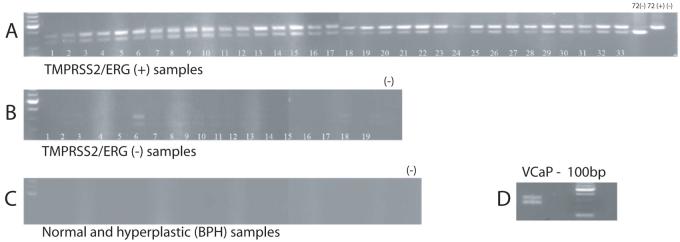
We have previously identified alternative splicing of the 5′ portion of the TMPRSS2/ERG fusion gene in PCa that was correlated with clinical aggressiveness. In order to examine the biological functions of the Type III and VI TMPRSS2/ERG isoforms, we cloned the coding portion of these variants into expression vector constructs. During this process we noted the variable presence of a 72-bp fragment by sequence analysis. We have denoted the variants with this fragment, which corresponds to exon 11 of the ERG genomic sequence, Type III+72 and VI+72. To assess the extent of the expression of the 72-bp exon in PCa, we designed an additional primer pair (ERG RT F and ERG RT R, Table S1), which spans this region. As shown in Figure 1A, all TMPRSS2/ERG fusion-positive tumor RNAs revealed two bands, the higher band corresponding to the isoform with the 72-bp exon. Fusion negative cancer RNAs showed rare very weak bands (Fig 1B) while benign tissue RNAs were completely negative (Fig 1C). This primer pair detects both the fusion gene and native ERG transcripts and the vast majority of ERG coding transcripts arise from the fusion gene, based on comparison of the fusion gene positive and fusion gene negative cancers (Fig 1A versus Fig 1B). However, to confirm that expression of the +72-bp exon was present in the fusion transcript in we used another primer pair with forward primer located in the first exon of the TMPRSS2 gene (TMPERG RT-f) and reverse primer within the 72-bp exon (ERG 72R; Table S1). All samples were positive (Supplemental data, Fig S1A). A third primer pair, one located in the TMPRSS2 exon 2 (TMPEx2 F) and the second located in genomic exon 14 (ERG69R2), which will not amplify the Type III and III+72 isoforms but will amplify the Type VI or VI+72 isoforms, was also used. For the previously identified (3) Type VI positive samples tested, all samples demonstrated double bands with stronger expression of the upper band containing the 72-bp exon (Fig S1B). The VCaP cell line expresses both Type III and III+72 fusion isoforms (Fig 1D). No other 5′ fusion isoforms such as the Type VI isoform were identified in VCaP cells.
Figure 1. Expression of a 72-bp exon in prostate cancer tissues expressing the TMPRSS2/ERG fusion gene.
RT-PCR amplification of ERG alternatively spliced isoforms with or without 72-bp exon in using primers spanning this exon. A. Fusion gene expressing cancer tissues; B. Fusion negative cancer tissues; C. Benign tissues from the peripheral zone (14 tissues) or hyperplastic transition zone (6 tissues) which were free of cancer on pathological examination. D. VCaP cells.
Increased cell proliferation in primary and immortalized prostate epithelial cells expressing TMPRSS2/ERG fusion genes
A critical step for cancer cell development is the dysregulation of proliferation. Therefore, we initially evaluated the effects of the alternatively spliced fusion gene isoforms on cell proliferation. Using retroviral constructs we stably expressed TMPRSS2/ERG fusion proteins in primary prostate epithelial cells (PrEC). Three groups of PrEC cells were generated: vector negative control, Type III+72 and Type (III+72) +(VI+72) groups. The fusion gene transcript levels are shown in Figure 2A. PrEC cells expressing the Type III+72 or Type (III+72) + (VI+72) fusion mRNA had significantly increased proliferation rates (Fig 2B), when compared to vector controls (P<0.01, t-test, day 9).
Figure 2. TMPRSS2/ERG fusion isoforms increase primary prostatic epithelial cell (PrEC) proliferation.
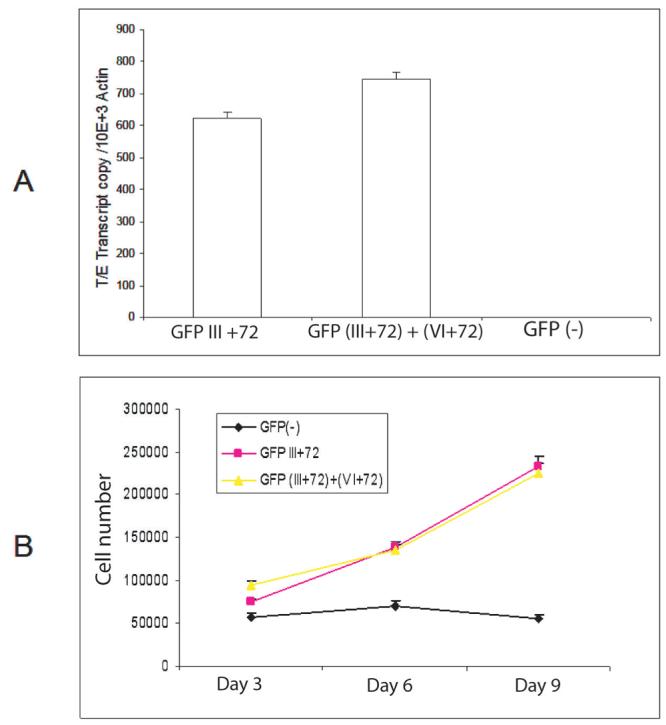
A. Expression level of fusion gene in PrEC cells by real-time PCR, normalized to β-actin. B. Proliferation of the three groups of PrEC cells expressing TMPRSS2/ERG fusion Type III+72, (III+72) + (VI+72) or empty vector were measured using a Coulter counter. Cells (2.5 × 104) of each cell group were plated in 35mm dishes in complete medium. Cells were trypsinized and counted at day 3, 6 and 9 in triplicate. Mean +/− standard deviation is shown
To confirm this finding, immortalized normal prostate epithelial cells (PNT1a) cells were transfected with plasmids encoding V5 epitope-tagged Type III+72, Type VI+72, or Type (III+72) + (VI+72) fusion genes or the empty vector, and G418-resistant lines were established. As shown in Figure 3A, equivalent expression of the fusion isoforms at the transcriptional level was confirmed by real-time PCR. PNT1a cells expressing the Type III+72 isoform alone showed no increase in cell growth while cells expressing the Type VI+72 or (III+72) + (VI+72) had significant increases in growth rate compared to negative control cells (Fig 3A). Interestingly, the PNT1a line expressing both fusion isoforms displayed a three-fold increase in cell number at day 7 relative to the vector control group (P<0.01, t-test) and almost double the cell number versus the group expressing Type VI+72 only (P <0.01, t-test). To further investigate the potential cooperativity among different isoforms, we overexpressed the four fusion isoforms individually or in combination in PNT1a cells, and proliferation rates were determined in the nine stable PNT1a cell lines (Fig 3B). Equivalent levels of the two fusion isoforms were seen on RT-PCR in all double transfectants, indicating that there is no interference of expression or stability of fusion mRNAs when they are co-expressed (Supplemental data, Fig S2). Interestingly, Type VI alone appears to even decrease proliferation, perhaps by sequestering factors needed for proliferation. It should be noted that the Type VI isoform is never expressed without other isoforms in vivo (3). Among four lines expressing individual fusion isoforms, only the cells expressing Type VI+72 isoform showed an increased growth rate relative to the negative control group Three groups of cotransfected cells, Type (III+72)+VI; Type III+(VI+72) and Type (III+72)+(VI+72) demonstrated significantly higher cell growth rates than the negative control group, which demonstrates synergism between these isoforms in promoting cell proliferation, especially between the Type III+72 and VI+72 isoforms.
Figure 3. TMPRSS2/ERG fusion isoforms affect PNT1a cell proliferation, invasion and motility.
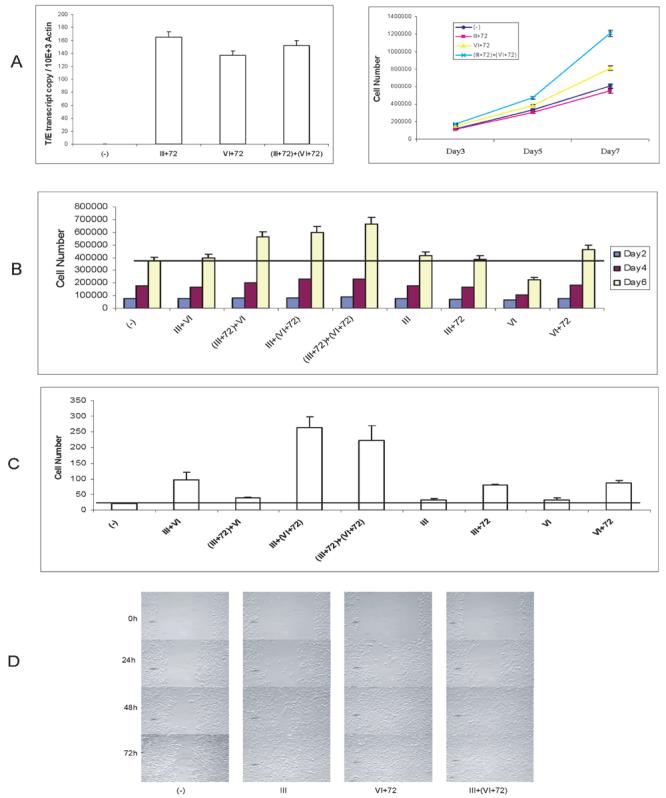
A. Expression level of fusion gene in PNT1a cells was evaluated by real-time PCR, normalized to β-actin. The growth PNT1a cells expressing TMPRSS2/ERG fusion Type III+72, VI+72, (III+72) + (VI+72) or empty vector was measured by using a Coulter counter. Cells (2.5 × 104) of each cell group were plated in 35mm dishes in complete medium. Cells were trypsinized and counted at day 3, 5 and 7 in triplicate. Mean +/− standard deviation is shown.
B. Proliferation of 9 groups of PNT1a cells with overexpression of TMPRSS2/ERG isoforms individually or in combination and control cells. Cell numbers were counted at day 2, 4 and 6.
C. Matrigel invasion of 9 groups of PNT1a cells with overexpression of TMPRSS2/ERG isoforms individually or in combination and control cells. Cells (2.5 × 104) of each group were plated into each well at day 0, and after 48h, the non-invading cells were removed from the upper surface of the membrane and the invading cells were stained and counted under the microscope. Assays were performed in triplicate. Mean +/− standard deviation is shown.
D. PNT1a cells transfected with empty vector or TMPRSS2/ERG fusion types III, VI+72 and III+(VI+72) were seeded at 2.5×106 in 60-mm diameter culture dishes in complete medium. Cells were gently scraped with a plastic tip. The medium was removed, and cells were washed twice with PBS. Complete medium was added and cells were allowed to scatter/migrate into the area of clearing for a total of 72 hr and photomicrographs taken at 0h, 24h, 48h, and 72h time points. Scratch assays were performed four times and representative results are shown.
Invasion and motility of PNT1a cell lines expressing TMPRSS2/ERG fusion genes
Another hallmark of neoplastic transformation is the ability to invade the extracellular matrix. We therefore evaluated the invasiveness of the PNT1a cell lines using a Matrigel invasion assay (26). A typical result of one such experiment is shown in Figure 3C. Cell expressing any fusion isoform showed significantly higher invasiveness through Matrigel when compared with vector control PNT1a cells (Type III, P<0.02; Type VI, P<0.04; all others P< 0.01, t-test). Of the cells expressing a single isoform, Type VI+72 was most invasive. Of the cells expressing two isoforms, Type III + (VI +72) or Type (III +72) + (VI+72) fusion isoforms were most invasive. Total cell numbers at 48 hours after plating were within 20% of each other (Fig 3B), so that even accounting for these minor differences in cell numbers, the invasive ability of all groups were still significantly higher than the vector control cells.
To evaluate cell motility, we performed a wounding assay (26) using Type III, Type VI+72, Type III + (VI+72) and negative control cell lines. In this assay, the ability of cells to migrate and fill a defect in the epithelial monolayer is assessed. As shown in Figure 3D, PNT1a cells expressing the Type VI+72 isoform covered the defect within 48h after the scratch. Cells expressing Type III or Type III + (VI+72) required 72h for full closure. The latter result is in contrast to the Matrigel invasion assays, in which the coexpression of the VI+72 isoform with the Type III isoform significantly enhanced invasion, and indicates that while motility and invasion in vitro have some common pathways there are almost certainly unique effectors for these two phenotypes as well. All three groups expressing the TMPRSS2/ERG fusion consistently closed the wound quicker than the vector control group, which required greater than 72 hrs to completely close the defect.
Colony formation in soft agar of PNT1a cell lines expressing TMPRSS2/ERG fusion genes
The PNT1a cell line is immortalized but not fully transformed and will not form colonies in soft agar (30). To examine the transforming activity of TMPRSS2/ERG fusion in PNT1a cells, we assessed colony formation in soft agar. No foci were formed in all 9 stable cell lines groups. As a positive control, we used PNT1a cells expressing Huntington interacting protein-1 (31), which did form colonies in soft agar (data not shown).
Interactions between TMPRSS2/ERG isoforms
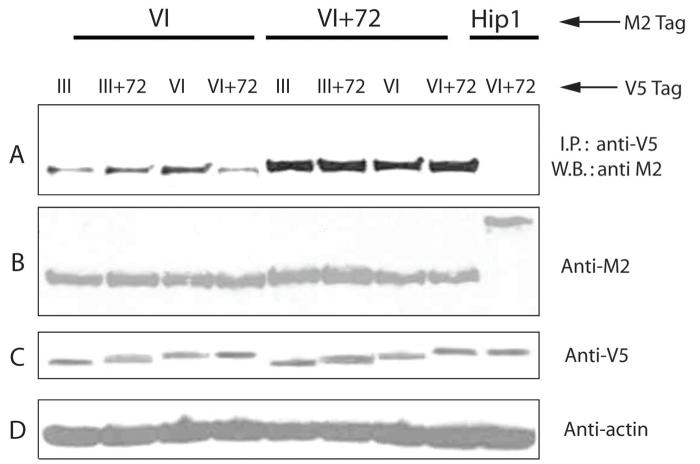
ETS family transcriptional factors are known to be able to form homo- or heterodimers. To evaluate whether the alternative fusion gene isoforms showed differences in protein-protein interactions we transiently co-transfected M2 (Flag) and V5-tagged fusion gene constructs in 293 cells. Immunoprecipitation was then performed on cell lysates with anti-V5 antibody followed by Western blotting with anti-M2 antibody. As shown in Figure 4A, all isoforms are able to bind to each other, to form homo- or heterodimers. However, significantly more protein was bound to the Type VI+72 than Type VI isoform, despite equal protein expression levels (Fig 4B-D), indicating this 72-bp fragment has potentially important function regulating protein-protein interactions, which may account for its more potent biological activities. No binding to M2-tagged Hip1, which was used as a negative control, was detected.
Figure 4. Homo- and heterodimerization of TMPRSS2/ERG fusion isoforms.
A. M2 (Flag) tagged Type VI and VI+72 fusion genes or Hip1 protein were co-transfected with four fusion isoforms with a V5 tag in 293 cells, immunoprecipitated with anti-V5 antibody, and Western blot performed using anti-M2 antibody. Hip1 tagged with M2, cotransfected with Type VI+72 with a V5 tag was the negative control. B. Expression level of type VI, VI+72 and Hip1 proteins detected by anti-M2 antibody; C. Expression level of Type III, III+72, VI and VI+72 proteins detected by anti-V5 antibody; D. β-actin input control for each sample.
Function of the TMPRSS2/ERG fusion in VCaP cells
VCaP is the only commonly used PCa cell line which expresses the TMPRSS2/ERG fusion gene, specifically the Type III and III+72 isoforms. To understand the biological role of the fusion gene in vivo in fully transformed cells, we designed a specific oligonucleotide sequence against the Type III fusion junction. As shown in Figure 5A, lentiviruses expressing this shRNA can efficiently knock down more than 60% of Type III fusion protein expression when tested in transiently transfected 293 cells by Western blot using anti-V5 antibody. Transcriptional levels of fusion were also assessed in VCaP cells by quantitative real-time PCR using the primers specific for the fusion gene mRNA (3). Similarly, ∼60% knock down efficiency was found in VCaP cells infected with the shRNA lentivirus compared to control VCaP infected with scrambled shRNA and native VCaP cells (Fig 5A). To determine if the fusion gene enhances proliferation in VCaP cells, proliferation was assessed in VCaP expressing shRNA or scrambled control. Decreased cell growth was found in VCaP expressing fusion gene shRNA compared to the control group (P<0.01, day 8), indicating these fusion isoforms can affect cell proliferation (Fig 5B). Experiments were repeated three times with the same result.
Figure 5. Knockdown of the TMPRSS2/ERG fusion in VCaP cells decreases cell proliferation in vitro and tumor progression in vivo.
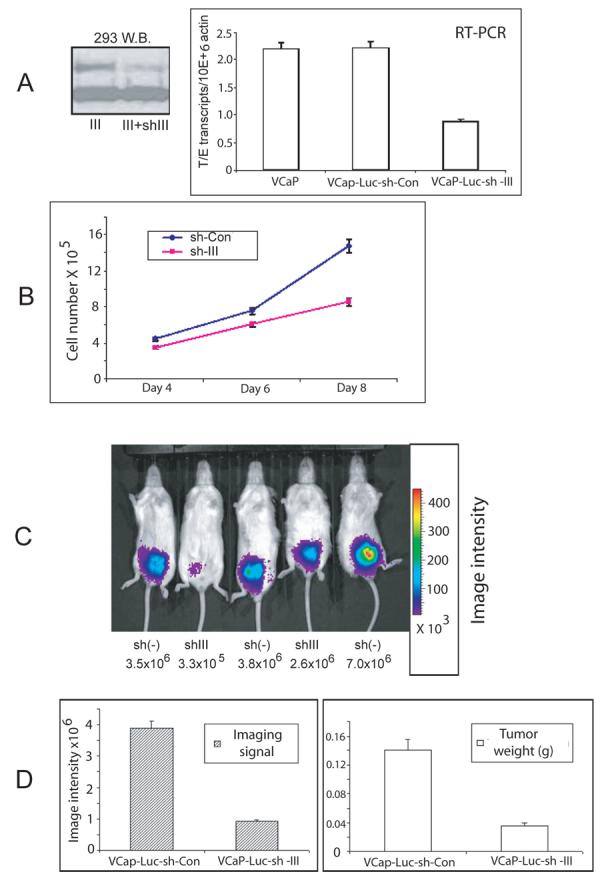
A. 293 cells were transiently transfected with V5-tagged Type III fusion gene expression construct and infected with lentivirus carrying shRNA against Type III fusion and protein expression evaluated using Western blotting with anti-V5 antibody. β-actin loading control is shown. Expression level of fusion gene mRNA in VCaP shRNA negative control or VCaP cells sh-III cells by real-time PCR normalized to β-actin. Mean +/− standard deviation. B. Cell growth curve comparing VCaP-Luc-sh-con and Vcap-Luc-sh-III cell numbers. Mean +/− standard deviation. C. Luciferase imaging of tumor growth in live mice from one cage by IVIS imaging system at 4 week time point. Three control (sh(−)) and two shRNA expressing (shIII) tumors are shown along with the corresponding image intensity. D. Luciferase imaging signal at 4 weeks after orthotopic injection and tumor weight at same time point.
To determine if knock down of the fusion gene expression decreased growth in vivo, we employed an orthotopic injection model. Orthotopic injection models, in which cells are injected directly into the mouse prostate, have the advantage of developing tumors in the native prostatic microenvironment. We have successfully generated two stable cell lines, VCaP-Luc-sh-III and VCaP-Luc-sh-Con. By using an IVIS imaging system, we were able monitor the tumor growth in real time (Fig 5C). Luciferase signal was easily detectable at 2 weeks after injection for the majority of mice in the control group. After 4 weeks, the average imaging signal was 4-fold higher in the mice injected with VCaP-Luc-sh-Con cells compared to VCaP-Luc-sh-III cells. Tumor weight was also 4-fold higher in the control group when compared to shRNA expressing group, consistent with the imaging data (Fig 5D). We examined expression of four genes (CACNA1D, KCNS3, PLA1A and LAMC2) that Tomlins et al (23) have shown to be downregulated by 35-70% in vitro in their si-ERG treated VCaP cells by quantitative RT-PCR using mRNAs from 7 control and 5 shRNA expressing tumors. Three of the four genes identified by Tomlins et al (23) as downregulated in vitro (CACNA1D, KCNS3, PLA1A) were also downregulated in vivo by ∼50% (data not shown). LAMC2 was only slightly downregulated (∼20%) in vivo but was actually downregulated 60% in vitro (data not shown), implying that in some cases alternative pathways for activation of genes may be selected for or activated in tumors in which the fusion gene is downregulated. The significantly reduced cell growth both in vitro and in vivo indicates that the TMPRSS2/ERG fusion gene has an important role in regulating primary tumor progression in vivo.
Heterogeneity of coding exons in ERG transcripts in PCa
To further explore the extent of variability in alternative splicing in the coding exons of the ERG gene in PCa, we cloned and sequenced ERG transcripts from human PCa tissues and VCaP cells, almost all of which arise from the fusion gene, using several primer sets designed to detect known ERG isoforms. Nine alternatively spliced ERG isoforms have been reported to date. We carried out an analysis of the NCBI published cDNA sequences and exon analysis through the Ensembl website1 and aligned the ERG exons on the genomic sequence located on Chr.21 from 25732292 to 25364248. Figure 6A shows all reported ERG isoforms and their relation to the genomic sequence. Prior reports regarding TMPRSS2/ERG gene fusion transcripts have all used the ERG2 sequence for reporting the structure of 5′ alternatively spliced TMPRSS2/ERG transcripts and the exon numbers of this isoform are shown. ERG 6 and 9 were excluded from the study because of their nonfunctional transcripts. Primer pairs were used to amplify different ERG isoforms (Table S1) which were cloned and sequenced. Results are summarized in Figure 6B. We found expression of ERG3, with and without the 72-bp exon 11. ERG4 was also expressed. We also detected a novel transcript which contains a previously unreported 61-bp exon. Of note, we were unable to amplify ERG3 or ERG4 transcripts using the exon 17 primer that we used for ERG2. Whether this indicates truncation of these transcripts or a technical problem with the primer pair is currently unclear. We also detected ERG7 with and without the 72-bp exon, as well as a novel isoform missing both genomic exons 10 and 11. ERG8 was also detected. We did not detect ERG1. Thus, there is significant variability in the coding sequence of the ERG transcripts in PCa, as well in the region of the 5′ fusion.
Figure 6. Summary of reported ERG transcripts and identified ERG isoforms in prostate.
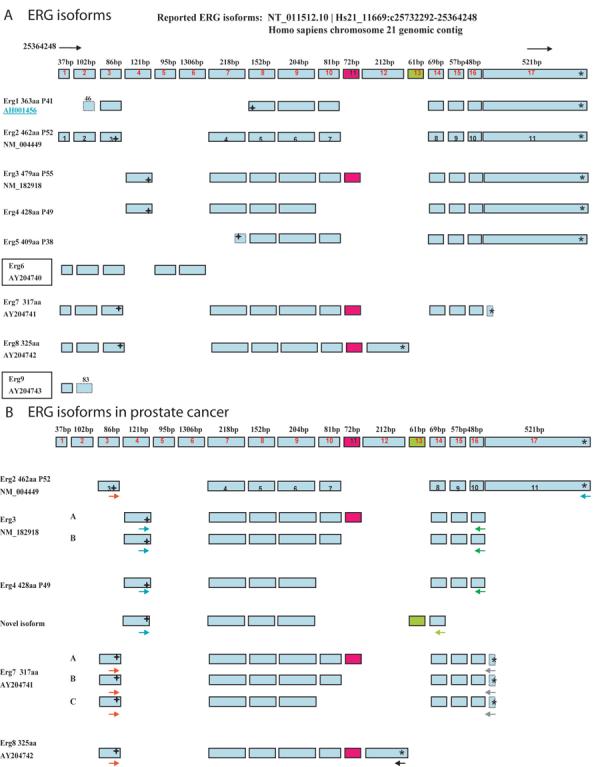
A. The nine reported ERG isoforms are listed in left column with NCBI accession number and predicted protein weight; 17 reported exons are aligned on Chr.21 genomic sequence from 25364248 to 257322921, the size of each exon listed on the top of each exon. The 72-bp exon is highlighted by red color, the novel 61bp exon is shown in green. The 11 exons of ERG 2 are indicated. In-frame start codons are indicated by (+) and stop codons by (*). B. ERG isoforms in prostate cancer: arrows with different colors stand for different primers used to amplify the ERG transcripts. Sub-variants are indicated by letters.
DISCUSSION
The discovery that TMPRSS2/ERG gene fusion occurs in 40-60% of clinically localized PCas makes this one of the most common genetic lesion in PCa and elucidating the clinical and biological consequences of this gene fusion is critical for understanding the pathogenesis of PCa and the development of targeted therapies. We have now shown that TMPRSS2/ERG fusion transcripts have an important biological function in promoting cell proliferation and/or invasion and motility of primary prostatic epithelial cells and immortalized prostatic epithelial cells. These biological activities are consistent with oncogenic activity of the TMPRSS2/ERG fusion gene in prostatic epithelial cells and indicate the TMPRSS2/ERG gene fusion is driving neoplastic progression via expression of ERG proteins. This is confirmed by our finding that knockdown of expression of the TMPRSS2/ERG fusion gene decreases primary tumor growth after orthotopic injection of VCaP cells in SCID mice.
Studies by our group and others have shown significant heterogeneity of the alternatively spliced isoforms at the 5′ portion of the TMPRSS2/ERG transcript. We have now shown a similar heterogeneity of the coding sequence. In particular, alternative splicing leading to inclusion or exclusion of a 72-bp exon (genomic exon 11) is common. A recent report (32) using exon arrays to analysis ERG expression in PCa has shown variable expression of this exon in an independent sample set, indicating that alternative splicing at this exon is common enough to lead to detectable alterations in overall expression of this exon. A similar variability in expression was noted for genomic exon 10, and we have noted variable inclusion of this exon in our studies as well. We have also detected other novel isoforms of the TMPRSS2/ERG fusion transcript. The quantitative extent of expression of these alternative isoforms is difficult to assess at present, and will require larger scale sequencing.
While detection of alternatively spliced transcripts is of interest, a critical question is whether there are differences in the biological activities of these transcripts within benign or transformed prostatic epithelial cells. We have now shown that the Type VI+72 enhances proliferation of PNT1a cells compared to cells expressing Type III+72 at similar transcript levels. This is consistent with our finding that Type VI isoform is associated with more aggressive disease. Similarly, inclusion of the 72-bp exon in the Type VI significantly enhances proliferation in PNT1a cells relative to Type VI without this exon. It should be noted that while the Type III isoform without the 72-bp exon has no activity in promoting proliferation, it can enhance both invasion and motility, although less effectively than the Type VI+72, indicating pleiotropic activities of each specific isoform. Finally, it should be noted that the biological activity of each isoform is related to cellular context. For example the Type III+72 isoform enhances proliferation in primary prostatic epithelial cells but not in immortalized normal prostatic epithelial cells (PNT1a). PNT1a cells are immortalized with SV-40 T-antigen and as such have alterations of the p53 and retinoblastoma gene pathways that may impact on the biological activities of individual fusion gene isoforms. Thus, alternatively spliced isoforms, both at the 5′ portion of the gene and in the coding exons, have variable, pleiotropic activities that can enhance various aspects of the transformed phenotype depending on the cellular context.
During the analysis of the TMPRSS2/ERG isoforms in PCa tissues we noted that expression of the Type VI isoform was always accompanied by expression of the Type III isoform. To determine whether this co-expression had any biological significance or simply was a reflection of the common expression of the Type III isoform, we cotransfected PNT1a cells with various combinations of Type VI +/−72-bp and Type III +/−72-bp and compared them to cells transfected with individual isoforms. At equivalent transcript levels, this resulted in both increased proliferation and invasion. Also, we showed that these fusion isoforms can form homo- or heterodimers in vitro with different binding strength, which might be partial reason for this variable ability of promoting cell growth and invasion.
The ERG protein is a member of ETS family, and the common feature of ETS family proteins is their DNA-binding ETS domain. The ETS domain can bind to the purine rich GGA (A/T) core sequence, and these proteins function as transcription factors(21). The ETS domain, the amino pointed (PNT) domain, and the ERG protein central domain, have all been reported to be involved in dimerization (33). The 72-bp exon is located in CAE (Central Alternative Exon) region of ERG gene, which is directly 5′ of the central domain of the ERG gene. The ERG central domain has been shown to act as an inhibitory domain for protein-protein interaction (34). Therefore, the 72-bp exon may be able to change the protein folding structure and affect interaction with other proteins. ERG proteins and other ETS family members can form heterodimeric or homodimeric complexes to regulate their transcriptional activity (21), which is modulated by the competition for homodimerization versus heterodimerization, depending on the relative intracellular concentrations of ETS proteins. Therefore, we posit that heterodimers between these fusion protein isoforms may have more potent transcriptional activity toward critical target genes promoting proliferation and invasion through extracellular matrix compared to either Type VI, VI+72, Type III, or III+72 homodimers. Obviously, this hypothesis requires direct testing and validation.
Recent studies in vitro have indicated that expression of ERG and other ETS transcription factors are associated with increased invasion (17, 23, 24) in both primary prostatic epithelial cells (23) and immortalized prostatic epithelial cells (24,25) that may, in part, be linked to increased expression of matrix metalloproteases and activation of the plasminogen activator pathway (23, 24). This is consistent with our finding that all isoforms tested increase invasion. Studies of proliferation have yielded variable results in other studies. Expression of the Type III isoform, which probably include the 72-bp exon, resulted in increased proliferation in BPH1 (24), but not RWPE cells (23), both immortalized normal prostatic epithelial cell lines. Of note, RWPE overexpressing the fusion gene did not form colonies in soft agar, similar to our results in PNT1a cells, or tumors following orthotopic injection (23). Tomlins et al (15) did not see changes in proliferation in primary prostatic epithelial cells; however, they used transient expression with an adenovirus, while we used stable expression with a retrovirus, and only observed significant differences after 6 days of growth. Similarly, Tomlins et al (15) did not see decreased proliferation in VCaP after knockdown with siRNA for 3 days, however by using stable shRNA expressing cell lines we were able to examine a longer time course and observed a significant decrease after 8 days. Of note, recent studies by Sun et al (35) using transfection of an siRNA targeting all ERG mRNAs in VCaP cells showed both decreased proliferation in vitro and decreased tumorigenicity in vivo in a subcutaneous xenograft model, consistent with our results. Overall the data indicates that the Type III+72 isoform can enhance proliferation in some contexts and that the Type VI+72 is even more potent in this regard.
In summary, we have demonstrated complex alternative splicing of the TMPRSS2/ERG fusion gene in PCa, both at the 5′ fusion junction and in the coding exons that can significantly impact the biological activities of the encoded proteins. Depending on the isoform and system examined, the TMPRSS2/ERG fusion gene can enhance proliferation, invasion and motility. Finally, knockdown of the fusion gene in a cancer cell line inhibits primary tumor growth, indicating that the TMPRSS2/ERG fusion gene is a potential therapeutic target, which is present in the majority of prostate cancers.
ACKNOWLEDGEMENTS
We gratefully acknowledge the assistance of Isaiah Schauer and Dr David Rowley with retroviral infections of primary prostatic epithelial cells.
Grant Support: This work was supported by grants from the Department of Defense Prostate Cancer Research program (DAMD W81XWH-08-1-0055), the National Cancer Institute to the Baylor Prostate Cancer SPORE (P50CA058204) and the Dept of Veterans Affairs Merit Review program (MI) and by the use of the facilities of the Michael E. DeBakey VAMC.
Footnotes
Ensembl website: http://www.ensembl.org
Invitrogen RNAi Designer website: http://rnaidesigner.invitrogen.com/rnaiexpress
REFERENCES
- 1.Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res. 2000;462:247–53. doi: 10.1016/s1383-5742(00)00006-5. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Cai Y, Ren C, Ittmann M. Expression of variant TMPRSS2/ERG fusion messenger RNAs is associated with aggressive prostate cancer. Cancer Res. 2006;66:8347–51. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 4.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–32. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark J, Merson S, Jhavar S, et al. Diversity of TMPRSS2-ERG fusion transcripts in the human prostate. Oncogene. 2007;26:2667–73. doi: 10.1038/sj.onc.1210070. [DOI] [PubMed] [Google Scholar]
- 6.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimoto M, Joshua AM, Chilton-Macneill S, et al. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia. 2006;8:465–9. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–43. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–44. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 10.Nam RK, Sugar L, Wang Z, et al. Expression of TMPRSS2:ERG gene fusion in prostate cancer cells is an important prognostic factor for cancer progression. Cancer Biol Ther. 2007;6:40–5. doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- 11.Lapointe J, Kim YH, Miller MA, et al. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007;20:467–73. doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- 12.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 13.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–9. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 14.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 16.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 18.Lin B, Ferguson C, White JT, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4. [PubMed] [Google Scholar]
- 19.Liu W, Ewing CM, Chang BL, et al. Multiple genomic alterations on 21q22 predict various TMPRSS2/ERG fusion transcripts in human prostate cancers. Genes Chromosomes Cancer. 2007;46:972–80. doi: 10.1002/gcc.20482. [DOI] [PubMed] [Google Scholar]
- 20.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 21.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Lapointe J, Li C, Giacomini CP, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–10. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- 23.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klezovitch O, Risk M, Coleman I, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer research. 2005;65:8887–95. doi: 10.1158/0008-5472.CAN-05-1702. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10:6169–78. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Cai Y, Penland R, Chauhan S, Miesfeld RL, Ittmann M. Increased expression of the metastasis-associated gene Ehm2 in prostate cancer. Prostate. 2006;66:1641–52. doi: 10.1002/pros.20474. [DOI] [PubMed] [Google Scholar]
- 28.Mitra SK, Lim ST, Chi A, Schlaepfer DD. Intrinsic focal adhesion kinase activity controls orthotopic breast carcinoma metastasis via the regulation of urokinase plasminogen activator expression in a syngeneic tumor model. Oncogene. 2006;25:4429–40. doi: 10.1038/sj.onc.1209482. [DOI] [PubMed] [Google Scholar]
- 29.Winter SF, Acevedo VD, Gangula RD, Freeman KW, Spencer DM, Greenberg NM. Conditional activation of FGFR1 in the prostate epithelium induces angiogenesis with concomitant differential regulation of Ang-1 and Ang-2. Oncogene. 2007;26:4897–907. doi: 10.1038/sj.onc.1210288. [DOI] [PubMed] [Google Scholar]
- 30.Ropiquet F, Berthon P, Villette JM, et al. Constitutive expression of FGF2/bFGF in non-tumorigenic human prostatic epithelial cells results in the acquisition of a partial neoplastic phenotype. International journal of cancer. 1997;72:543–7. doi: 10.1002/(sici)1097-0215(19970729)72:3<543::aid-ijc26>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Rao DS, Bradley SV, Kumar PD, et al. Altered receptor trafficking in Huntingtin Interacting Protein 1-transformed cells. Cancer cell. 2003;3:471–82. doi: 10.1016/s1535-6108(03)00107-7. [DOI] [PubMed] [Google Scholar]
- 32.Jhavar S, Reid A, Clark J, et al. Detection of TMPRSS2-ERG translocations in human prostate cancer by expression profiling using GeneChip Human Exon 1.0 ST arrays. J Mol Diagn. 2008;10:50–7. doi: 10.2353/jmoldx.2008.070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer research. 2006;66:10658–63. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 34.Iwamoto M, Higuchi Y, Koyama E, et al. Transcription factor ERG variants and functional diversification of chondrocytes during limb long bone development. The Journal of cell biology. 2000;150:27–40. doi: 10.1083/jcb.150.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun C, Dobi A, Mohamed A, et al. TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene. 2008 doi: 10.1038/onc.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]