Abstract
In addition to its established role in initiating the endocrine arm of the stress response, corticotropin-releasing factor (CRF) can act in the brain to modulate neural pathways that effect coordinated physiological and behavioral adjustments to stress. Although CRF is expressed in a set of interconnected limbic and autonomic cell groups implicated as primary sites of stress-related peptide action, most of these are lacking or impoverished in CRF receptor (CRFR) expression. Understanding the distribution of functional receptor expression has been hindered by the low resolution of ligand binding approaches and the lack of specific antisera, which have supported immunolocalizations at odds with analyses at the mRNA level. We have generated a transgenic mouse that reports expression of the principal, or type 1, CRFR (CRFR1). This mouse expresses GFP in a cellular distribution that largely mimics that of CRFR1 mRNA and is extensively colocalized with it in individual neurons. GFP-labeled cells display indices of activation (Fos induction) in response to central CRF injection. At the cellular level, GFP labeling marks somatic and proximal dendritic morphology with high resolution, and is also localized to axonal projections of at least some labeled cell groups. This includes a presence in synaptic inputs to central autonomic structures such as the central amygdalar nucleus, which is implicated as a stress-related site of CRF action, but lacks cellular CRFR1 expression. These findings validate a new tool for pursuing the role of central CRFR signaling in stress adaptation, and suggest means by which the pervasive ligand-receptor mismatch in this system may be reconciled.
Keywords: Corticotropin Releasing Factor (CRF), Stress, amygdala, HPA axis
Introduction
Corticotropin-releasing factor (CRF) was identified on the basis of its capacity to activate pituitary adrenocorticotropin secretion (Vale et al., 1981), and its localization to hypothalamic neurosecretory neurons defined the central arm of the hypothalamo-pituitary-adrenal (HPA) axis. CRF is broadly distributed in the CNS, including in limbic forebrain and brainstem cell groups that are widely implicated in control of affective behavior and autonomic function, respectively (Merchenthaler et al., 1982; Swanson et al., 1983; Sakanaka et al., 1986). The subsequent demonstration that the peptide could act centrally to elicit stress-like behavioral (e.g., anxiogenic, anorexic) and autonomic (sympathomimetic) responses suggested a broader role for CRF in mediating and/or integrating complementary modes of stress adaptation (Sutton et al., 1982; Brown and Fisher, 1985; for review see: Valentino et al., 1993; Heinrichs and Koob, 2004).
Experiments monitoring expression of immediate-early genes induced in response to central CRF injection or stress, and studies using local peptide/antagonist administration with monitoring of physiological and/or behavioral endpoints have been used to identify stress-related sites of CRF action (Imaki et al., 1993; Heinrichs et al., 1994; Bittencourt and Sawchenko, 2000; Funk et al., 2006). Frequently implicated in this regard is a set of interconnected cell groups, including aspects of the central amygdalar nucleus (CeA), bed nucleus of the stria terminalis (BST), paraventricular hypothalamic (PVH) and lateral parabrachial (PBl) nuclei, locus coereleus (LC), nucleus of the solitary tract (NTS) and ventrolateral medulla (VLM). These are key cell groups in the processing of stress-related sensory information and in effecting adaptive responses to it. As such, they represent core structures in what may be termed the central autonomic system (CAS, Saper, 1995; Sawchenko et al., 2008)
Although CRF is prominently expressed within the CAS network, the evidence regarding the two known CRF receptors (CRFR1, CRFR2) is equivocal. Studies employing hybridization histochemical localization of mRNA identifies only the PBl among the structures listed above as expressing CRFR1, the primary receptor for CRF (Potter et al., 1994; Chalmers et al., 1995; Van Pett et al., 2000). Some immunohistochemical work supports a broader distribution of CRFR1 within the CAS (Chen et al., 2000; Sauvage and Steckler, 2001; Reyes et al., 2008), but the specificity of the available antisera has not been rigorously established. The lack of a clear view of functional CRFR expression represents a major impediment to understanding the role of this signaling system in stress adaptation and the wide range of pathologies associated with it.To address this problem, we have generated a transgenic mouse that reports expression of CRFR1 with green fluorescent protein (GFP). The reporter transgene is based on a bacterial artificial chromosome (BAC) that contains the entire CRFR1 genomic locus (Gong et al., 2002). We find that cellular GFP expression closely matches that of CRFR1 mRNA with few notable exceptions, and that both are poorly expressed in CAS cell groups. However, CRFR1 promoter-driven GFP fills dendritic and axonal projections of labeled cells, including ones that ramify within some CAS cell groups. Although receptor trafficking cannot necessarily be inferred from patterns of transgenic GFP expression, these observations define potential bases for reconciling CRF ligand-receptor misalignment in the CAS.
Materials and Methods
Animals
Transgenic mice (see below) were housed in a temperature-controlled room on a 12:12 hr light:dark cycle with food and water freely available. All experimental procedures were approved by the Institutional Animal Care and Use Committee of the Salk Institute.
Molecular cloning of the CRFR1-GFP reporter transgene
The CRFR1-GFP BAC transgene was generated using methods developed in the laboratory of N. Heintz (Rockefeller University, HHMI, Gong et al., 2002). Briefly, an approximately 500 bp DNA fragment of genomic DNA directly upstream of the ATG start site in CRFR1 was amplified using PCR and subcloned into the Asc I site in the sca-e-b shuttle vector (containing eGFP with a polyadenylation encoding sequence) kindly provided to us by N. Heintz. Similarly, an approximately 500 bp DNA sequence within the first intron of CRFR1 was amplified and subcloned into the Pac I and Fse I sites in sca-e-b. This modified shuttle vector was then transformed into E. coli containing the BAC rp23-4B21, containing the CRFR1 genomic locus, which was obtained from the Children’s Hospital of Oakland Research Institue (bacpac.chori.org). The transformed bacteria were grown in LB containing 30 μg/ml Ampicillin (Amp) and 20 μg/ml Chloramphenicol (Ch), overnight, diluted 1000 fold into LB containing 50 μg/ml Amp and 20 μg/ml Ch for 8 hours and plated onto lb-agar plates containing 50 μg/ml Amp and 20 μg/ml Ch and incubated overnight at 37° C. BAC DNA from single colonies was isolated and PCR was used to confirm that the shuttle vector inserted into the CRFR1 locus by homologous recombination. Four colonies with appropriate insertions were grown in LB containing 20 μg/ml Ch for 1 hr then plated on lb-agar plates containing 20 μg/ml Ch and 5% sucrose, and incubated overnight at 37° C. Resulting colonies were streaked onto lb-agar Amp plates to determine whether the shuttle vector had recombined away from the BAC. DNA was isolated from colonies that did not grow on Amp and used for PCR on both the 5′ and 3′ recombination regions to determine which colonies had recombined appropriately. A single colony was picked from bacteria that showed correct recombination and DNA was prepared using the Nucleobond BAC Maxi Kit (BD Biosciences, PaloAlto, CA). This DNA was diluted to 1 ng/ml into microinjection buffer containing spermine and spermidine, and microinjected into single celled mouse oocytes (CB6F1) by the Salk transgenic core facility using standard procedures. A single mouse was obtained carrying the BAC, and this mouse line was expanded and used in subsequent experiments.
Antisera
Rabbit anti-GFP (1:2000, Molecular Probes, San Diego, CA, cat #A11122 lot 49025A) was generated using Green Fluorescent Protein, purified directly from A. victoria, as the antigen. The antiserum was purified using ion-exchange chromatography to remove non-specific immunoglobulins (product information sheet). Mouse anti-GFP (1:2000, Molecular Probes, San Diego, CA, cat #A11120 lot 73E1-1) is a monoclonal antibody from clone 3E6. The antibody has been shown to specifically recognize GFP in transfected HeLa cells (product information sheet). Goat anti-GFP (1:2000, Rockland, Gilbertsville, PA, cat #600-101-215, lot 17549) was generated using recombinant a. Victoria GFP protein. The immunoaffinity purified antiserum recognizes a single 33Kd protein on a western blot from HeLa cells transfected with GFP that is absent in control lysates. Rabbit anti-CRF (1:1000, rabbit pbl#rC70 pool 397-223) was generated by the Vale lab using a synthetic peptide corresponding to full-length rat/human CRF(1-41) as the antigen. The serum was found to specifically recognize r/h CRF by radioimmunoassay (Vale et al., 1983), and in competition studies with r/h CRF and structurally related peptides (Sawchenko, 1987a and b: Nahon et al., 1989). Rabbit anti-cFos (1:10000, Calbiochem, Temecula, CA, cat #PC38T, lot #D27872) was generated against a synthetic peptide (SGFNADYEASSSRC) corresponding to amino acids 4-17 of human c-Fos. This antiserum recognizes both c-Fos and v-Fos but does not react with the 39 kDa Jun protein (product information sheet). The specificity of the antiserum was confirmed in prior experiments in which preabsorption overnight with 30 μM of the synthetic peptide immunogen eliminated basal and induced staining (Schiltz and Sawchenko, 2007). A mouse-derived monoclonal anti-tyrosine hydroxylase (TH; 1:1000, Boehringer Mannheim, Mannheim, Germany) was produced by immunization with TH purified from a rat pheochromocytoma, and recognized a pattern of neuronal staining in rodent brain stem that corresponds with the consensus distribution of TH by immunohistochemsitry and in situ hybridization (Bittencourt et al., 1991). Rabbit anti-MAP2 (1:1000, Chemicon, Temecula, CA, cat #AB5622, lot 25041049) was generated in rabbit using purified microtubule associated protein from rat brain as the immunogen. The antibody recognizes all isoforms of MAP2 (MAP2a, MAP2b, MAP2c and MAP2d), however, it has the strongest immunoreactivity to MAP2a, b. By Western blot, the antibody recognizes a 280 kD doublet that corresponds to MAP2a and MAP2b as well as a 70kD doublet that corresponds to MAP2c (product information sheet). Mouse anti-sv2 (1:1000, Developmental Studies Hybridoma Bank, Iowa City, IA, cat #sv2-c) is a monoclonal antibody that was generated by injecting mice with purified ommata (electric fish) synaptic vesicles (Buckley and Kelly, 1985). The antibody recognizes a 95kD protein on a Western blot that corresponds to SV2.
Immunohistochemistry
Immunohistochemical localization was carried out on free-floating sections using conventional immunoperoxidase and immunofluorescence techniques (Sawchenko et al., 1990). For avidin-biotin immunoperoxidase localiztion of GFP, sections were pretreated for 10 min with 0.3% hydrogen peroxide to neutralize endogenous peroxidases, followed by 8 minutes exposure to 0.5% sodium borohydride to reduce free aldehydes. They were then incubated in GFP antiserum at 4°C for 48 hours in phosphate-buffered saline (PBS, pH 7.3) containing 0.3% Triton X-100 and 3% blocking serum. The primary antiserum was localized using Vectastain Elite reagents (Vector Laboratories, Burlingame, CA) at the dilutions recommended by the supplier, and the reaction product was developed using a nickel-enhanced glucose oxidase method (Shu et al., 1988). Dual immunoperoxidase labeling for Fos and GFP was carried out by sequentially localizing the antiserum against Fos using a nickel-enhanced diaminobenzidine method (black nuclear reaction product) as above, and the goat antiserum to GFP, without nickel enhancement (brown cytoplasmic product).
For multiple indirect immunofluorescence labeling, sections were incubated in a cocktail of anti-GFP, and one or two additional antisera, each at their optimal dilutions for single staining, for 48 hrs in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 3% blocking serum at 4°C, followed by incubations in species-matched, fluorochrome-conjugated secondary antisera in the following combinations: for dual labeling of goat anti-GFP and rabbit anti CRF, a combination of Cy-2 conjugated donkey anti-goat and Rhodamine red X (RRX) conjugated donkey anti-rabbit secondary antibodies were used, each at a concentration of 1:600 (Jackson Immunoresearch, West Grove, PA). In order to visualize anti-GFP and anti-ACTH together, a combination of Cy2 conjugated donkey anti-goat and Cy3 conjugated donkey anti-mouse secondary antibodies were used, each at a concentration of 1:600 (Jackson Immunoresearch, West Grove, PA). In order to visualize goat anti-GFP, rabbit anti-CRF, and mouse anti-TH together, a combination of Cy2 conjugated donkey anti-goat, RRX conjugated donkey anti-rabbit, and Cy5 conjugated donkey anti-mouse secondary antibodies were used, each at a concentration of 1:600 (Jackson Immunoresearch, West Grove, PA). In supplemental fig. 1, G, H, I, donkey anti-goat conjugated Alexa Flour 488 secondary antibodies were used (1:600, Invitrogen, Eugene, OR).
In situ hybridization
Hybridization histochemical localization was carried out using 35S-labeled cRNA probes that are synthesized, hybridized and applied to tissue as described by Simmons et al. (Simmons, 1989). Sections were mounted onto poly-L-lysine-coated slides, vacuum-dried overnight and post-fixed in 10% formalin for 30 min. Sections are digested with 10 μg/ml proteinase K for 30 min, at 37° C. Probes were labeled to specific activities of ~1–3 × 109 dpm/μg, and applied to tissue at concentrations of ~107 cpm/ml, overnight at 56–58°C in a solution with 50% formamide and 10% dextran sulfate, after which they are treated with 20 μg/ml of ribonuclease A for 30 min, at 37°C, and washed in 15 mM NaCl/1.5 mM sodium citrate, typically at 65–85°C. Sections were dehydrated and exposed to X-ray films (Biomax; Kodak) for 1–2 days. They were defatted, dried, coated with Kodak NTB-2 liquid emulsion, and exposed at 4°C for 3–30 days, depending on the strength of signal on film. Slides were developed with Kodak D-19 for 3.5 min and fixed with Kodak rapid fixer for 2 min, both at 14°C.
Combined immuno- and hybridization histochemistry
Complete series through the brains of 4 CRFR1-GFP mice were prepared for concurrent localization of GFP and CRFR1 mRNA. Combining immunoperoxidase labeling with isotopic in situ hybridization required minor modifications of the constituent methods (Chan et al., 1993). Immunostaining was carried out first, and the protocols modified as follows: (1) normal tissue pretreatments in hydrogen peroxide and sodium borohydride were omitted, (2) blocking sera were replaced in the immunostaining procedure with 2% bovine serum albumin and 2% heparin sulfate, (3) nickel enhancement steps were eliminated, and (4) Nissl counterstaining was omitted.
Intracerebroventricular (icv) injection
Animals (n=6) adapted to handling were stereotaxically implanted with 26 ga guide cannulae (Plastics One) aimed to terminate above a lateral ventricle, and fixed to the skull with dental acrylic. After a minimum of 7 days’ recovery, during which time they were handled and received mock injections, 33 ga injectors pre-fitted to extend just beyond the tip of the guide were inserted and ventricular penetration confirmed by aspirating cerebrospinal fluid. The animals remained undisturbed for 2 hr before receiving icv injections of 500 ng – 1 μg CRF in 5 μl over 60 sec. The animals were sacrificed 2 hr after the injection to detect peak levels of induced Fos (Bittencourt and Sawchenko, 2000).
Immunoelectron microscopy
Unmanipulated CRFR1-GFP mice (n=4) were perfused with saline followed by 2% paraformaldehyde and 2.75% acrolein in 0.1 M phosphate buffer (pH 7.0). 50 μm thick vibratome sections were then prepared for avidin-biotin immunoperoxidase localization of GFP as above, except that tissue was not exposed to detergent during processing. Sections were fixed in 1% osmium tetroxide with 1.5% potassium ferricyanide, dehydrated with ethanol and propylene oxide and infiltrated with Spurr’s resin. The sections were embedded, and blocks were trimmed to isolate the lateral central amygdalar nucleus, thin-sectioned, and counterstained with uranyl acetate and lead citrate. The material was examined in a JEOL 100 CX II transmission electron microscope.
Image acquisition and modification
Photomicrographs were obtained on a fluorescent microscope (Nikon Eclipse E600) using Image-pro plus software or on a confocal microscope (Leica TCS SP2 AOBS) using Leica LCS software. For micrographs covering entire brain sections (Fig. 3, Sup Figs. 2, 3), images were collected with a 10x lens, then digitally stitched together using Autostitch. Images were imported as raw tif files into Adobe Photoshop where all subsequent manipulation was performed. Contrast and brightness of the photomicrographs were adjusted to optimize signal and images were cropped for presentation as figures. Figures were assembled using either Adobe Illustrator or ACD Systems Canvas.
Figure 3.
Comparison of transgenic GFP and CRFR1 mRNA expression. Adjacent sections through two levels of the forebrain in the CRFR1-GFP animal showing immunoperoxidase localization of GFP (A, C) and hybridization histochemical demonstration of CRFR1 mRNA (B, D). The two markers are consistent in revealing major cellular sites of expression in isocortex (Iso), piriform cortex (Pir), hippocampus (Hipp), medial septum (MS), globus palliidus (GP), reticular nucleus of the thalamus (RT, medial n of the amygdala (MeA) and arcuate nucleus of the hypothalamus (ArH). Other abbreviations: CeA, Central n. of the amygdala; CPu, caudoputamen; LA, lateral n. of the amygdala; LS, lateral septum. VMH, ventromedial hypothalamic n.; BLA, basolateral n. of the amygdala; Scale bar: 100 μM.
Results
Transgene design
Using BAC recombination techniques, we replaced the first exon of CRFR1 within BAC RP23-4B21 with sequences encoding GFP (Fig. 1B). This BAC contains approximately 100kb of DNA sequences both upstream (5′) and downstream (3′) of the first exon of CRFR1, and is anticipated to contain most, if not all, promoter and enhancer sequences that regulate the expression of CRFR1 (Fig. 1A). A transgenic mouse line was generated and assayed for expression of GFP. Brain sections were examined by fluorescence microscopy and found to express GFP, which is readily detectable on the basis of its native fluorescence (Supplemental Fig. 1). When we compared the expression pattern of GFP in this transgenic mouse line with a transgenic mouse line generated by the Gensat consortium that used a different, overlapping, BAC construct to target the CRFR1 genomic locus (RP24-239F10), we found a high degree of congruence in regional expression patterns (Supplemental Fig. 1). In order to substantially increase sensitivity and cellular resolution of GFP detection in the CRF1-GFP transgenic mouse, we intensified the signal using immunohistochemistry, allowing visualization of neuronal morphology in fine detail (Fig 2).
Figure 1.
Transgene design. A BAC (rp23-4B21) that contains most, if not all of the genomic locus of CRFR1, including the coding region, promoter and enhancer elements was used to construct the transgene (A). The first exon of CRFR1, which includes the translation start, was replaced with sequences encoding GFP and a translation stop signal such that only GFP would be transcribed from the modified, transgenic, CRFR1 locus (B).
Figure 2.
Morphological detail revealed by CRFR1-driven GFP expression. Immunoperoxidase staining for GFP shows the extent of neuronal labeling provided by the CRFR1-GFP mouse in the lateral hypothalamic area (A) and pontine reticular formation (B). Cell bodies, primary dendrites, dendritic branching and varicose (presumably axonal) processes are all clearly labeled. The degree of cellular and axonal labeling exhibits regional variation. In the dentate gyrus (C), neurons in the hilar region are strongly labeled, while staining of granule cells (gr) is weaker and sporadic. A dense plexus of fine varicosities occupies the outer two-thirds of the molecular layer (mol). This is not seen in the inner third, which receives commissural and associational input from hilar neurons. In cerebellar cortex (D) labeling of small cells in the granule cell layer (gr) and punctate (axonal) elements in the molecular layer (mol) contrast with a distinct lack of staining of Purkinje cells (pcl). The latter have been reported as sites of CRFR1 expression in some histochemical studies. Other abbreviations: fx, fornix, hf, hippocampal fissure. Scale bars: 25 μm.
GFP expression in the CRFR1-GFP transgenic mouse brain conforms to that of CRFR1 mRNA
To evaluate the fidelity with which the CRFR1-GFP mouse reports CRFR1 expression, we first carried out immunoperoxidase localization of GFP and hybridization histochemical localization of CRFR1 on adjacent series of sections through brain and pituitary (n=4). Semi-quantitative assessments of regional GFP and CRFR1 mRNA expression were generated and compared with results of prior in situ hybridization studies (Van Pett et al., 2000). In general, acknowledged major sites of CRFR1 mRNA expression, including in isocortex, cerebellum and some of its major targets and afferent cell groups, and certain sensory systems, were closely recapitulated in patterns of transgenic GFP expression (Table 1, Fig. 3, Sup. Figs. 2, 3). The sensitivity with which the two methods revealed sites of receptor expression seemed generally comparable, though cellular resolution and detail was far superior in the transgenic mouse. In addition, axonal projections that often could be inferred as arising from sites of cellular receptor expression were filled in the CRFR1-GFP mouse, which, as elaborated below, may offer insights into potential sites of CRFR1 localization. A more comprehensive comparison of cellular expression patterns revealed by the two localization methods is provided in Table 1.
Table 1.
Expression of GFP in the CRFR1-GFP transgenic mouse Expression of the GFP transgene examined throughout the brain and compared with previous reports of CRFR1 expression detected by in situ hybridization. Each area was scored using the following system of + symbols: + − very few, scattered cells are positive; ++ − a substantial number of cells within the nucleus are positive; +++ − the majority of cells in the nucleus are positive; ++++ − all of the cells within the nucleus are positive. The area was scored (−) if no cells were present in the nucleus expressing GFP.
| CELL GROUP | CRFR1-GFP | CRFR1 mRNA (Van Pett et al.) |
|---|---|---|
| I. Forebrain | ||
| A. Isocortex | ||
| I | − | − |
| II-III | ++ | ++ |
| IV | ++++ | +++ |
| V | ++ | + |
| VI | + | + |
| Claustrum | + | + |
| B. Olfactory regions | ||
| 1. Main bulb | ||
| Periglomerular layer | +++ | ++ |
| Outer plexiform layer | + | + |
| Mitral layer | ++++ | +++ |
| Inner plexiform layer | + | − |
| Granular layer | +++ | +++ |
| 2. Anterior olfactory n. | ++ | ++ |
| 3. Olfactory tubercle | ++ | |
| I | − | |
| II | ++ | |
| III | ++ | |
| 4. Piriform cortex | ++ | |
| I | − | |
| II | +++ | |
| III | ++ | |
| Endopiriform nucleus | + | − |
| 5. Taenia tecta | +++ | +++ |
| C. Hippocampal formation (cortex) | ||
| 2. Subiculum (dorsal) | + | ++ |
| Subiculum (ventral) | + | - |
| 3. CA1 | +++ | ++ |
| 4. CA3 | + | ++ |
| 5. Dentate gyrus | ||
| Granular layer | − | − |
| Polymorph layer | +++ | ++ |
| 6. Induseum griseum/fasciola cinerea | − | − |
| D. Amygdala | ||
| 1. Medial nucleus | ||
| Anterior part | ++ | ++ |
| Posterodorsal part | + | + |
| 3. Cortical nucleus | ||
| Anterior part | ++ | ++ |
| Posterior part | + | ++ |
| 4. N. lat. olfactory tract | ? | ++ |
| 5. Anterior amygdaloid area | ++ | ++ |
| 6. Central nucleus(lat/med) | +/++ | −/+ |
| 7. Lateral nucleus | ++ | − |
| 8. Basolateral nucleus | +/− | + |
| 9. Basomedial nucleus | ++ | + |
| 10. Intercalated nuclei | ++ | − |
| E. Septum | ||
| 1. Lateral nucleus | ||
| Dorsal part | + | − |
| Intermediate part | ++ | + |
| Ventral part | − | − |
| 2. Medial n./n. diagonal band | +++ | ++ |
| 3. Bed n. Stria terminalis | ||
| Rostromedial region | + | + |
| Rostrolateral region | ++ | + |
| Posterodorsal region | ++ | ++ |
| Posteroventral region | ++ | + |
| 4. Bed n. anterior commissure | + | − |
| 5. Septofimbrial nucleus | +++ | − |
| 6. Triangular nucleus | + | − |
| 7. Subfornical organ | − | − |
| F. Basal ganglia | ||
| 1. Caudoputamen | ||
| Posteroventral part | ++ | − |
| Nucleus accumbens | ++ | + |
| Fundus of striatum | ++ | − |
| 2. Globus pallidus | ++++ | +++ |
| 3. Subthalamic nucleus | +++ | ++ |
| 4. Substantia nigra | ||
| Compact part | +++ | ++ |
| Reticular, latertal parts | ++ | + |
| Ventral tegmental area | ++ | + |
| G. Thalamus | ||
| 1. Medial habenula | − | − |
| 2. Lateral habenula | ++ | − |
| 3. Anterior group | - | - |
| 4. Mediodorsal nucleus | + | - |
| 5. Lateral group | ||
| Lateral dorsal n. | − | − |
| Lateral posterior n. | − | − |
| 6. Midline group | ||
| Paraventricular n. | − | + |
| Parastaenial n. | − | ++ |
| N. reunions | +/− | ? |
| Rhomboid n. | − | − |
| N. gelatinosa | − | − |
| 7. Ventral group | ||
| Ventral anterior/v. lat. | − | + |
| Ventral medial | − | − |
| Ventral posterior | − | + |
| Gustatory nucleus | − | − |
| 8. Posterior complex | − | − |
| 9. Medial geniculate n. | ||
| Dorsal part | ++ | − |
| Ventral part | + | − |
| Medial part | ++ | ++ |
| 10. Lateral geniculate n. | ||
| Dorsal part | − | − |
| Intergeniculate leaflet | ++ | ++ |
| Ventral part | +/− | ++ |
| 11. Intralaminar nuclei | ||
| Central medial n. | − | − |
| Paracentral n. | − | − |
| Central lateral n. | − | − |
| Parafascicular n. | ++ | − |
| 12. Reticular nucleus | ++++ | +++ |
| 13. Zona incerta | ||
| Rostral | ++ | ++ |
| Caudal | + | + |
| 14. N. fields of Forel | − | − |
| H. Hypothalamus | ||
| 1. Periventricular zone | ||
| Median preoptic n. | + | − |
| Anteroventral periventricular n. | + | − |
| Preoptic periventricular nucleus | + | + |
| Suprachiasmatic n. | ++ | + |
| Supraoptic nucleus | − | − |
| Paraventricular n. | ++ | + |
| Anterior periventricular nucleus | ++ | − |
| Arcuate nucleus | +++ | ++ |
| Posterior periventricular n. | ++ | − |
| 2. Medial zone | ||
| Medial preoptic area | + | − |
| Medial preoptic n. | ||
| Lateral part | ++ | − |
| Medial part | + | + |
| Central part | + | − |
| Anterior hypo. n. | ||
| Anterior part | + | − |
| Central, posterior parts | ++ | + |
| Retrochiasmatic area | − | + |
| Ventromedial n. | − | − |
| Dorsomedial n. | ++ | + |
| Tuberomammillary n. | − | − |
| Premammillary n. | ||
| Dorsal part | + | − |
| Ventral part | ++ | + |
| Supramammillary n. | ||
| Lateral part | + | ++ |
| Medial part | ++ | ++ |
| Lateral mammillary n. | − | + |
| Medial mammillary n. Median part | − | − |
| Medial part | − | − |
| Lateral part | − | − |
| Posterior part | − | − |
| 3. Lateral zone | ||
| Lateral preoptic area | + | + |
| Lateral hypothalamic a. | ++ | + |
| Posterior hypo. area | ++ | ++ |
| I. Pituitary Gland | ||
| 1. Anterior lobe | ++ | ++ |
| 2. Intermediate lobe | ++ | ++ |
| 3. Posterior lobe | − | − |
| II. Brainstem | ||
| A. Sensory | ||
| 1. Visual | ||
| Superior colliculus | ||
| Superficial Gray | ++ | ++ |
| Intermediate Gray | +++ | + |
| Deep Gray | + | ? |
| Parabigeminal n. | − | ++ |
| Pretectal region Olivary n. | + | − |
| N. optic tract | +++ | − |
| Anterior n. | ++ | − |
| Medial pretectal a. | ++ | − |
| N. posterior commissure | − | − |
| 2. Somatosensory | ||
| Mesencephalic n. V | − | − |
| Principal sensory n. V | ++++ | ++++ |
| Spinal n. V Oral part | ++ | +++ |
| Interpolar part | +++ | +++ |
| Caudal part | ++ | ++++ |
| Dorsal column n. | ++ | ++++ |
| External cuneate n. | +++ | ++++ |
| 3. Auditory | ||
| Cochlear nuclei | ||
| Dorsal | +/− | ++ |
| Ventral | ++ | + |
| N. trapezoid body | − | + |
| Superior olive | + | + |
| N. lateral lemniscus | ||
| Ventral | ++ | ++ |
| Dorsal | ++ | + |
| Inferior colliculus | ||
| External | +/− | + |
| Dorsal | + | + |
| Central | ++ | ++ |
| N. brachium inf. coll. | +/− | − |
| N. saguluum | ++ | − |
| 4. Vestibular | ||
| Medial n. | ++ | ++ |
| Lateral n. | +/− | ++ |
| Superior n. | + | ++ |
| Spinal n. | − | + |
| 5. Gustatory | ||
| N. solitary tract, ant. | +/− | − |
| 6. Visceral | ||
| N. solitary tract | ||
| Medial part | + | + |
| Commissural part | + | − |
| Lateral part | − | + |
| Area postrema | − | − |
| Parabrachial n. | ||
| Lateral | ++ | +++ |
| Medial | ++ | ++ |
| Kölliker-Fuse n. | + | + |
| B. Motor | ||
| 1. Eye | ||
| Oculomotor (III) | − | − |
| Trochlear (IV) | − | − |
| Abducens (VI) | − | + |
| 2. Jaw | ||
| Motor n. V | + | − |
| 3. Face | ||
| Facial n. (VII) | + | +/++ |
| 4. Pharynx/larynx | ||
| N. ambiguus | − | ++ |
| 5. Tongue | ||
| Hypoglossal n. (XII) | − | − |
| 6. Viscera | ||
| Dorsal motor n. X | − | ++ |
| C. Reticular Core (including central, gray and raphé) | ||
| 1. Periaqueductal gray - assoc. w/PAG | + | + |
| Interstitial nucleus of Cajal | + | + |
| N. Darkschewitsch | ++ | ++ |
| Dorsal tegmental n. | − | − |
| Ventral tegmental n. | + | ++ |
| N. incertus | ++ | + |
| Laterodorsal teg. n. | +++ | ++ |
| Barrington’s n. | ++ | − |
| Locus coeruleus | +/− | − |
| 2. Raphé | ||
| Interfascicular n. | − | − |
| Rostral linear n. | + | + |
| Dorsal raphé | ++ | − |
| Median raphé | ++ | − |
| N. raphé magnus | + | + |
| N. raphé obscurus | +/− | + |
| N. raphé pallidus | − | − |
| 3. Interpeduncular n. | + | |
| Rostral subnucleus | + | |
| Apical subnucleus | − | |
| Dorsomedial subnucleus | − | |
| Lateral subnucleus | − | |
| Intermediate subnucleus | ++ | |
| Central subnucleus | − | |
| 4. Reticular formation | ||
| Central teg. field Retrorubral part | − | + |
| Peripeduncular n. | − | − |
| Pedunculopontine n. | +++ | ++ |
| Cuneiform n. | + | + |
| Pontine reticular | + | + |
| Linear n. medulla | +++ | ++ |
| Parvicellular ret. field | + | ++ |
| Gigantocellular ret. field | − | + |
| Lateral paragigantocellular | +/− | + |
| D. Pre- and Postcerebellar | ||
| 1. Pontine gray | ++++ | ++++ |
| 2. Tegmental reticular n. | ++ | + |
| 3. Inferior olive | +/− | − |
| 4. Lateral reticular n. | +++ | +++ |
| 5. Red nucleus | +++ | ++ |
| 6. N. Roller | − | − |
| 7. N. Prepositus | +++ | +++ |
| III. Cerebellum | ||
| A. Deep nuclei | ++++ | +++ |
| B. Cortex | ||
| Molecular layer | ++ | − |
| Purkinje layer | − | + |
| Granular layer | +++ | ++ |
Isocortex
The strong and widespread expression throughout the isocortical mantle, which is perhaps the most salient feature of the CRFR1 mRNA distribution in rodent brain (Van Pett et al. 2000), was mirrored in transgenic GFP expression, albeit with slightly different laminar emphases (Fig 3A, C). Both displayed a dominant expression in layer IV, with scattered cellular labeling in deeper layers, but the secondary (to layer IV) focus of labeling throughout layer II/III at the mRNA level was less expansive and robust when visualized by immunolocalization of GFP (Fig 4C, D) Labeling of apical dendrites of GFP-stained pyramidal neurons was commonly observed, with some extending as far as layer I; basilar dendrites were less extensively filled. First, and occasionally higher, order dendritic branching was regularly observable in GFP labeled material.
Figure 4.
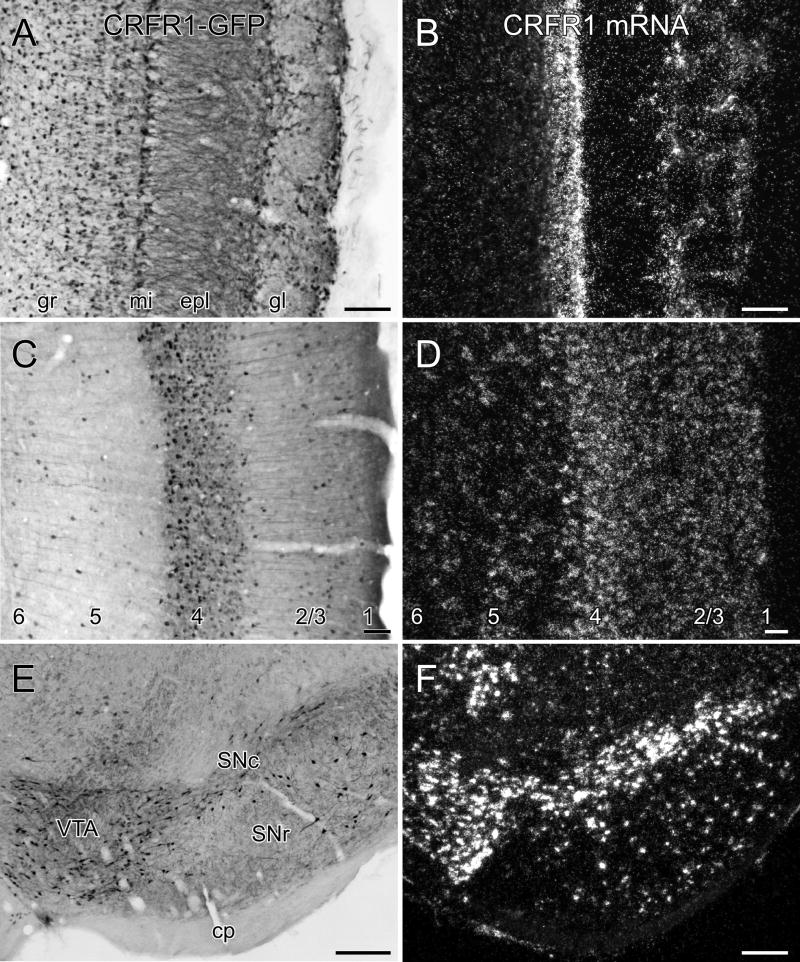
Finer grained comparison of GFP and receptor mRNA distributions. Adjoining sections through the olfactory bulb (A, B), temporal cortex (C, D) and ventral midbrain (E, F) prepared for immunoperoxidase localization of GFP (left column) and CRFR1 mRNA (right). The distribution and relative strength of labeling for the two markers in similar in most layers of the olfactory bulb, isocortex, the substantia nigra (SNc, SNr) and ventral tegmental area (VTA). Transgenic GFP expression labels granule cells in the olfactory bulb much more decisively than in situ hybridization. In contrast, relatively strong secondary focus of CRFR1 mRNA expression in layer 2/3 of isocortex (B) is not matched by comparable GFP labeling (A). Other abbreviations: cp, cerebral penuncle; epl, external plexiform layer, gl, glomerular layer, gr, granule cell layer; mi, mitral cell layer; SNc, substantia nigra, compact part; SNr, substantia nigra, reticular part. Scale bars: 100 μm,
Hippocampus
Major cellular sites of transgenic GFP expression in hippocampus included CA1 pyramidal neurons, and polymorph neurons in the hilar region of the dentate gyrus (Fig 2C). Expression in the CA3 field of Ammon’s Horn and in the granule cell layer of the dentate gyrus was consistently weaker and more variable. This overall pattern was similar to that seen in hybridization histochemical preparations, and described previously (Van Pett et al., 2000). The most salient feature of labeling in hippocampus was that of an extremely dense meshwork of punctate elements in the outer two-thirds of the molecular layer of the dentate gyrus and in stratum lacunosum-moleculare of Ammon’s Horn, ostensibly corresponding to axonal projections arising from the entorhinal cortex (Amaral and Witter, 1995, Fig. 2C). This was consistently the most prominent labeling of axonal elements observed in our material. In well-stained preparations, mossy fiber projections of dentate granule neurons were also weakly to moderately labeled.
Limbic forebrain
The medial septal nucleus and nucleus of the diagonal band were found to contain a moderate concentration of GFP labeled neurons, while fewer were seen in the lateral septal nuclei (Fig. 3A). Most subregions of the bed nucleus of the stria terminalis (BST) displayed at least moderate densities of GFP positive cells throughout their rostrocaudal extents, with one notable exception. The oval nucleus of the BST (BSTov), a central autonomic cell group in which CRF is richly expressed in cells and fibers (Swanson et al., 1983; Sakanaka et al., 1986), exhibited little or no cellular GFP labeling, but was invested with a moderately dense GFP-stained innervation (see below).
The amygdala was also a seat of substantial immunolabeling in the CRFR1-GFP mouse, which again displayed substantial regional variation. The greatest densities of cellular expression were seen in the medial and basomedial nuclei (Fig. 3C). Very few cells within the basolateral nucleus were labeled for GFP, an observation at odds with present and past findings based on CRFR1 mRNA expression (Van Pett et al., 2000; Sup. Fig 3), whereas the lateral and cortical nuclei contained moderate densities of GFP positive cell bodies. In the CeA, another central autonomic cell group and the best established site of CRF action in effecting a range of stress responses (Funk et al., 2006; Pawlyk et al., 2006; Ji and Neugebauer, 2007), only the medial part of the nucleus contained a small-moderate number of CRFR1-GFP positive neurons. The lateral CeA, which is the principal site of cellular CRF expression and which contains a dense CRF innervation (Swanson et al., 1983; Sakanaka et al., 1986), contained few, if any, GFP-labeled cell bodies, but was invested with an apparent GFP-ir innervation. Because of the disparity between the expression of endogenous CRF and CRFR1-driven GFP in the lateral CeA, and the importance of this cell group in CRF-dependent stress responses, additional characterization of GFP elements in the region was carried out (see below).
Basal ganglia
A low-moderate density of cells labeled positively for GFP was seen in both the caudoputamen and nucleus accumbens (Fig 3A). Both contained a moderately dense GFP positive innervation (Fig 3A), which in the caudoputamen was focused dorsomedially. Accordingly, prominent accumulations of GFP-labeled neurons were labeled in the substantia nigra (primarily the pars compacta) and ventral tegmental area, major sources of inputs to the basal ganglia (Fig 4E). Virtually all neurons of the globus pallidus were strongly labeled, and prominent GFP staining of cell bodies and axons/terminals was seen in the subthalamic nucleus (not shown). Cellular GFP expression in this system was again in good accord with patterns and relative strengths of mRNA expression (Van Pett et al., 2000).
Hypothalamus
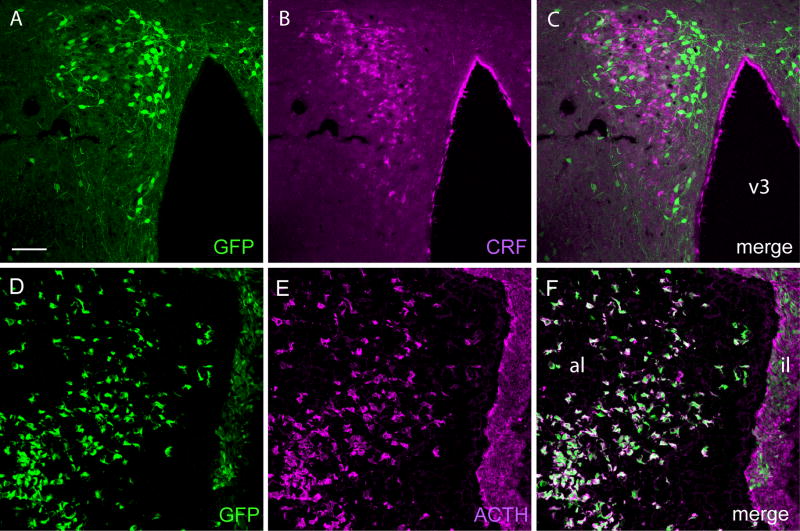
Most major cell groups of the hypothalamus and preoptic area contained low-moderate densities of GFP-stained neurons, a notable exception being the ventromedial nucleus, which was devoid of cellular labeling (Fig 3C). Particularly prominent accumulations of labeled neurons were seen in the arcuate nucleus, whose ventromedial aspect contained an abundance of intensely stained cells (Fig 3C). The perifornical aspect of the lateral hypothalamic area harbors a group of large neurons whose primary dendrites extended up to 1 mm from the soma (Fig 2A, B). In the PVH, the seat of parvocellular neurosecretory neurons that govern HPA output, prior analyses at the mRNA level have suggested that basal levels of CRFR1 expression are low-equivocal but may be induced in response to a range of stressors (Rivest et al., 1995; Van Pett et al., 2000; Sup. Fig. 4). The PVH of the unperturbed CRFR1-GFP mouse displays a moderate complement of heavily stained neurons (Fig. 5A). Intriguingly, dual-labeling experiments revealed that only a small minority of this CRFR1 population expresses CRF (Fig 5B, C).
Figure 5.
Characterization of CRFR1-GFP expression in the paraventricular nucleus (PVH) and pituitary. A-C: Confocal microscopic images through the PVH of the CRFR1-GFP mouse showing immunofluorescence staining for GFP, CRF and merged channels. The substantial population of GFP-stained cells does not overlap appreciably with the CRF-expressing contingent. D-F: Sections though the pituitary co- stained for GFP (A) and ACTH (B) reveal substantial congruence in the anterior lobe (al; white cells in merged image, C), but that only a subset of melanotropes in intermediate lobe (il) are GFP-positive. Scale bar: 50 μM.
Sensory systems
Transgenic GFP expression was consistent with present and past analyses in revealing abundant CRFR1 expression at multiple levels of select sensory systems (Van Pett et al., 2000). For example, throughout the main and accessory olfactory bulbs, mitral cells, periglomerular cells and a substantial subset of granule cells, were prominently stained (Fig 4A). Axonal labeling was reliably detected in the lateral olfactory tract and its projections to piriform cortex, which itself exhibited strong cellular labeling in superficial portions of layer 2 and scattered layer 3 cells (not shown). Cellular labeling was also extensive in the anterior olfactory nucleus and the olfactory tubercle (not shown).
Cell groups involved in the processing of somatic sensory information, including the dorsal column and external cuneate nuclei, along with major trigeminal sensory structures, all comprised sites of prominent cellular GFP expression. This was also seen at multiple levels of the primary auditory path (ventral cochlear, lateral superior olivary, and lateral lemniscal nuclei), with lesser densities of cells scattered in the central part of the inferior colliculus and medial division of the medial geniculate. GFP expression in the visual system was more limited, with the superior colliculus (particulary its intermediate gray) and pretectum (anterior nucleus, nucleus of the optic tract) comprising the main sites of expression. Cellular GFP expression in the vestibular system was most prominent in the medial vestibular nucleus, and was generally weaker and less widespread than CRFR1 mRNA signal. Interestingly, the vestibular nuclei are quite richly invested with GFP positive fibers.
Among central autonomic cell groups involved in processing visceral sensory information, expression in the NTS is limited to a discrete column of cells (central subnucleus) related to esophageal function (Cunningham and Sawchenko, 1990). This paucity of expression in the NTS contrasts with high levels of expression in the PB, which receives massive input from the NTS (Herbert et al., 1990). GFP staining in the external lateral subnucleus of PB was particularly prominent.
Brainstem
The LC is a noradrenergic cell group at which cellular effects of CRF are best established (Valentino et al., 1992; Jedema and Grace, 2004). Here, however, we find no evidence for either CRFR1 mRNA or transgenic GFP expression (Fig 6A; Van Pett et al., 2000). This was confirmed in material stained concurrently for GFP, CRF and tyrosine hydoxylase (TH, a marker for catecholaminergic neurons), in which we fail to detect neurons co-labeled for GFP and TH (Fig. 6). These preparations do, however, reveal a field of CRFR1-GFP fibers intermingled with CRF positive ones amid TH-positive cell bodes and dendrites in LC (Fig. 6). Processes (presumably dendritic) of TH neurons do extend into adjoining cell groups (e.g., laterodorsal tegmental nucleus) in which cellular GFP expression is seen, though the extent to which this may comprise a substrate for interaction is unclear.
Figure 6.
GFP expression in and around the locus coeruleus (LC). Confocal images of a section through the LC showing concurrent immunoflourescence labeling for GFP (A), CRF (B), tyrosine hydroxylase (TH; C) and merged channels (D). GFP-stained cell bodies are numerous in adjoining regions, but none co-stain for TH, a marker of LC neurons. Punctate, presumably axonal, elements are labeled for both GFP and CRF within the LC, defining potential substrates for interactions between peptidergic terminals and presynaptic CRFR1 receptors in this cell group. Scale Bar: 100 μM.
Among other brainstem regions of interest, the midbrain raphe nuclei harbor relatively small numbers of GFP-stained neurons, an observation somewhat at odds with more widespread, albeit weak and diffuse, CRF mRNA signal over these cell groups. This contrasts with strong labeling for both receptor message and transgenic GFP expression of nearby cholinergic neurons of the laterodorsal tegmental and pedunculopontine nuclei.
Cerebellum
CRFR1-GFP transgene expression in cerebellum included an enormous number of densely packed granule cells (Fig 2D), and essentially all cells of the deep cerebellar nuclei (not shown) were intensely labeled. We did not observe GFP staining of Purkinje cells (Fig 2D), which is at odds with the appearance of our hybridization histochemical material and prior studies at this level of analysis (Van Pett et al., 2000; Sup. Table 1).
Expression of GFP in the CRFR1-GFP animal is also particularly high in a number of pre- and post-cerebellar nuclei. This includes dense accumulations of strongly labeled cells in the red, lateral reticular, and perihypoglossal nuclei, as well as a great majority of cells in the pontine gray. This is again consistent with reports of CRFR1 mRNA localization in these nuclei (Van Pett et al., 2000).
Pituitary gland
The anterior pituitary of the CRFR1-GFP transgenic mouse contains a substantial population of GFP-positive cells of stellate morphology, which were identified in dual staining preparations as conforming to corticotropes, based on their coexpression of ACTH (Fig. 5). The intermediate lobe also displays moderate levels of GFP expression, consistent with in situ hybridization results for CRFR1 mRNA (Potter et al., 1994; Van Pett et al., 2000). Not all ACTH positive cells in the intermediate lobe are positive for GFP, suggesting that CRFR1 expression may differ in subpopulations of melanotropes. In line with previous reports, the posterior lobe was seen to be lacking in CRFR1-driven GFP expression (Van Pett et al., 2000).
Coincidence of CRFR1-GFP and CRFR1 mRNA expression
Transgenic GFP and CRFR1 mRNA expression were compared directly in combined immuno- and hybridization histochemical preparations (n=4). In most areas, GFP-labeled cells were overlain by accumulations of reduced silver grains indicative of the presence of CRFR1 transcripts. The degree of overlap was greatest, and nearly complete, in cell groups which both markers were robustly expressed in singly stained material. These included mitral cells of the olfactory bulb, CA1 pyramidal neurons in hippocampus, neurons of the deep cerebellar nuclei and a number a sensory and pre-cerebellar nuclei of the brainstem (Fig. 7A). A few areas that displayed strong labeling for each marker independently (globus pallidus, reticular nucleus of the thalamus) displayed less complete correspondence (50–75%) when examined concurrently, with the relative strengths of CRFR1 and GFP labeling of individual neurons bearing no consistent relationship to one another (Fig. 7B). In other areas in which strong hybridization signals contrasted with weak GFP expression (e.g., CA3 layer of hippocampus, layer II/III of isocortex) or vice versa (e.g., pretectal nuclei) when examined separately, the expected decrement in sensitivity of localization in doubly labeled preparations resulted in a minority of cells being doubly labeled. Among central autonomic cell groups highlighted above, dual labeling was observed only in the parabrachial nucleus. Overall, the results of single and dual labeling experiments support transgenic GFP expression as an accurate reporter of the CRFR1 distribution, although some differences in the relative strength of transgenic GFP and endogenous CRFR1 mRNA expression were noted (see discussion).
Figure 7.
Overlay of transgenic GFP with molecular and functional indices of CRFR1 expression. Left column: Concurrent dual labeling for GFP (brown) and CRFR1 mRNA (black grains) showing extensive overlap between the two markers in the dentate nucleus of the cerebellum (DN; panel A), the globus pallidus (GP; B) and gracile nucleus (Gr; C). Arrowheads mark examples of doubly-labeled neurons. Note the lack of expression of either marker in the nucleus of the solitary tract (NTS; C), a central autonomic cell group identified as a site of CRF action. Right column: Dual immunoperoxidase staining for GFP (brown cytoplasm) and the inducible activation marker, Fos (black nuclei), in sections from CRFR1-GFP mice that received an icv injection of CRF (1μg), 2 hr before perfusion. Nearly all Fos-positive cells in the red nucleus (RN; D) and reticular part of the substania nigra (SNr; E) are GFP positive. The central nucleus of the amygdala (CeA; F), another central autonomic cell group, shows a strong activational response, but little or no GFP expression. Some cells immediately adjoining the CeA (arrowhead) are double labeled. Scale bar: 25 μm.
CRF activates GFP-positive and -negative neurons in the CRFR1-GFP mouse
To probe the utility of the CRFR1-GFP mouse in an experimental setting, we assessed their capacity to facilitate phenotyping neurons activated in response to icv CRF administration. A prior study in rats indicates that central CRF evokes a pattern of cellular activation (Fos expression, Morgan and Curran, 1991) in brain that conforms to the CRFR1 distribution, with a few notable exceptions (Imaki et al., 1993; Bittencourt and Sawchenko, 2000). Here, again using Fos induction as a generic marker of neuronal activation, we see a correlation between activated cells and cells expressing CRFR1-GFP, whose strength varied with peptide dose. At lower doses of CRF (500 ng; n=3), prominent sites of Fos induction included GFP-stained neurons of cell groups immediately adjoining the cerebral ventricles (e.g., hippocampus) or meninges (e.g., pontine gray). Engagement of other GFP-positive cells tended to occur as a function of proximity to the ventricular or meningeal surfaces, with aspects of structures such as the reticular nucleus of the thalamus or globus pallidus showing few, if any, Fos-positive neurons. Higher doses of CRF (1 μg; n=3) gave rise to widespread Fos induction, including in many cells that were positive for GFP at sites distant to the ventricles (Fig 7D, E).
The principal exceptions to the general correspondence between CRF-induced Fos and GFP expression were central autonomic cell groups. These structures all displayed robust Fos induction in response to both doses of peptide, but only a very limited capacity for CRFR1-driven GFP expression (Fig. 7F). In the near absence of GFP expression, doubly labeled cells were never observed in the BSTov, CeAl, locus coeruleus or NTS. Even in the PVH, substantial complements of both activated and GFP-stained neurons were present yet comprised essentially separate populations. The only CAS cell group in which GFP neurons were reliably engaged following icv peptide administration was the parabrachial nucleus, where doubly labeled neurons were numerous in its lateral division.
GFP is found in axonal and dendritic profiles in CAS cell groups
The paucity of evidence supporting cellular expression of either CRFR1 mRNA or CRFR1 promoter-driven GFP in central autonomic cell groups implicated as stress-related sites of CRF action prompted us to further characterize GFP-stained fibers in the CeAl and BSTov as potential substrates for reconciling the apparent ligand-receptor mismatch. In series of sections co-stained for GFP and CRF, we detected no more than an occasional isolated GFP-stained neuron within the confines of the CeAl and BSTov, though labeled cells were numerous in immediately adjoining areas (Fig. 8 A–C, D–F). Instead, we observed a network of GFP-positive fibers intermingled within a denser field of fibers labeled for CRF, suggesting the presence of CRFR1-expressing axons and/or dendrites (Fig 8). Co-staining for GFP and the dendritic marker, MAP2, revealed that most of the fibers in the CeA are not dendrites (Fig. 8. G–I), suggesting that the axons of CRFR1 positive cells invade the lateral CeA. Alternate series of sections were co-stained for GFP, CRF and SV2, a synaptic vesicle protein enriched in axon terminals, to determine if synapses containing CRF were adjacent to CRFR1-GFP fibers in the CeAl. We observed numerous instances in which synapses were opposed to GFP positive fibers, nearby to CRF puncta (Fig. 8J–L). This organization supports the possibility that CRFR1 is present in axon terminals in a position to interact with CRF that is locally released in the lateral CeA and BSTov.
Figure 8.
Some central autonomic cell groups harbor axonal and dendritic, but not somatic, expression of CRFR1-GFP. Top two rows: Sections through the oval nucleus of the BTS (BSTov; top) and CeA (second row) co-stained for transgenic GFP (A, D) and endogenous CRF (B, E) expression; merged images are shown at the right (C, F). Both cell groups are surrounded by GFP-labeled perikarya, but contain very few within their borders. Both are invested with GFP-positive processes, which intermingle with a denser CRF-immunoreactive innervation. G-I: Dual staining for GFP and Map2, a dendritic marker, reveals that some of the coarser elements in the CeA are identifiable as CRFR1-expressing dendrites (arrow) while many GFP fibers in the CeA are Map2 negative, indicating they are axonal (arrowheads). J-L: Higher power confocal micrography reveals a fine GFP positive fiber (J) containing puncta positive for SV2 (K, arrow, blue), a marker of presynaptic terminals, adjacent to puncta containing CRF (K, magenta, arrowhead). The arrow in the merged micrograph (I) points to an example of a presynaptic terminal from a CRFR1 expressing neuron that might respond to locally released CRF (arrowhead). Scale Bars: A-F, 50 μM; G-I, 2 μM; J-L, 1 μm.
To further evaluate this hypothesis, series of vibratome sections through the lateral CeA of CRFR1-GFP mice (n=4) were prepared for pre-embedding immunoperoxidase localization of GFP at the electron microscopic level. In this material, we regularly saw GFP positive axon terminals, in which reaction product was distributed diffusely amid small, clear vesicles (Fig 9C–D). These were most frequently apposed to one or more fine, unlabeled dendritic profiles, with some contacts displaying synaptic specializations, indicating that these GFP positive terminals are pre-synaptic. We also observed GFP-stained dendritic profiles of varying caliber within the confines of CeAl (Fig. 9A-B) These were commonly surrounded by rosettes of unlabeled axon terminals containing mainly small (~ 30 nm diameter), round and electron-lucent vesicles, some of which formed (mainly asymmetric) synaptic contacts with the labeled dendrite. Together with the confocal microscopic analyses, these findings suggest the possibility that CRFR1 expression in the lateral CeA may occur on axonal (presynaptic) or dendritic (postsynaptic) elements, both of which derive from neurons located outside the CeAl, proper.
Figure 9.
Fine structure of GFP labeling in the central nucleus of the amygdala. Electron micrographs showing preembedding immunoperoxidase localization of transgenic GFP expression in dendrites and axon terminals in the central nucleus. A, B: We frequently observed smaller caliber dendritic processes (D*) that were commonly encircled by unlabeled axon terminals (open arrows), some of which formed (mainly asymmetric) synaptic contacts with the GFP+ dendrite. In addition, GFP-stained axon terminals (AT*) were also found apposed (curved arrows) to one or more unlabeled dendrites, with some contacts displaying clear synaptic specializations (D). Scale bars: 1 μm in A and C; 0.5μm in B and D.
Discussion
We have generated a transgenic mouse line that reports the expression of CRFR1 with GFP. The pattern of cellular GFP localization conforms closely to that of CRFR1 mRNA with a few notable exceptions, and of Fos induction evoked by central administration of CRF. The availability of an independent method for establishing the CRFR1 expression pattern highlights misalignments between ligand and receptor in established sites of stress-related peptide action. The enhanced resolution provided by transgenic GFP expression in revealing axonal and dendritic projections of CRFR1-expressing neurons suggests ways in which such mismatches may be reconciled.
Methodological considerations
To generate a CRFR1-reporter mouse line, we opted to use a BAC-based approach as this allows for the inclusion of most, if not all, promoter and enhancer sequences that regulate endogenous receptor expression. BAC transgenic expression of GFP has been used extensively to map gene expression in the CNS (Gong et al., 2003). When the accuracy of transgenic reporting has been compared using shorter promoter sequences versus BAC-based approaches, the shorter promoter sequences tend to direct expression in subsets of neurons that express the gene of interest, whereas BAC transgenics more completely recapitulate the endogenous expression pattern (Lee et al., 2001; Liu et al., 2003). Indeed, the general agreement observed here between transgenic GFP and endogenous CRFR1 mRNA expression is consistent with this view.
This correspondence was incomplete, however. Our analysis indicated that 70% (141/199) of all brain regions listed in Table 1 displayed scores for relative strength of transgenic GFP and endogenous CRFR1 mRNA expression that were within one unit of one another on a five-point rating scale. Disparities almost invariably occurred in the lower half of the scale, as the percentage increases to 87% (173/199) when the distinction between “−“ (no labeling) and “+” (widely scattered cells) is eliminated. Moreover, no differences exceeding one scale point were noted for any of the strongest (+++ - ++++) sites of expression. It may also be noted that the degree of concordance would increase substantially if more detailed subregional expression patterns (e.g., in individual isocortical fields) are considered. Under any circumstances, however, areas of disagreement are evident (see Table 1, Sup Table 1). In view of numerous cases where BAC based transgenic expression only partially matches the endogenous pattern (gensat.org), possible bases for them warrant consideration.
Discrepancies could reflect problems inherent in mouse genetic techniques, such as the failure to include all promoter and enhancer sequences that are required to express GFP in a manner that faithfully mimics the CRFR1 mRNA distribution, and/or position effects that may arise due to the insertion site of the transgene. Alternatively, they could derive in part from regional differences in the processing and/or stability of mRNAs versus protein. Finally, technical factors could be at play, such as the facts that (1) the sensitivity of immuno- and hybridization histochemical methods are each diminished when the two are applied concurrently, and (2) fixation protocols that provide for optimal sensitivity of each may be quite different. Lacking a basis to sort among these alternatives, we suggest that before using transgenic GFP expression as a surrogate marker for an endogenous protein that the degree of overlap be examined explicitly in cell groups of interest.
The availability of a method for localizing CRFR1 expression independent of in situ hybridization should help fill a lingering void in understanding, particularly in view of uncertainties associated with attempts to achieve antiserum-based detection. A substantial number of studies have been published using immunohistochemical approaches (e.g. Chen et al., 2000; Sauvage and Steckler, 2001; Reyes et al., 2008), and while many of the localizations reported are consistent with the hybridization histochemical literature, some are not, and these prominently include central autonomic cell groups such as the central nucleus of the amygdala and locus coeruleus. This may reflect limitations of currently available CRFR1 antisera, none of whose specificity has been decisively established, e.g., in knockout mice. CRFR1 exhibits substantial structural relatedness to CRFR2 and other members of the secretin clan of G protein-coupled receptors (Fredriksson et al., 2003), several of whose regional expression profiles overlap in part with that of CRFR1, including in the cell groups of contention highlighted above (Merchenthaler et al., 1999; Joo et al., 2004).
In labeling receptor-expressing neurons more completely and with far greater resolution than hybridization histochemical methods, the CRFR1-GFP transgenic should facilitate such applications as phenotyping and/or morphological characterization in electrophysiological, tract tracing and colocalization studies. It must be emphasized, however, that the transgenic reports receptor expression, and not subcellular localization. While we use such findings as the filling of axonal projections of GFP neurons to identify sites where presynaptic receptor mechanisms may be in play, it remains an open question as to whether receptors are trafficked and localized in a similar manner.
An amended view of CRFR1 expression in the adult brain
The resolution provided by the CRFR1-GFP BAC transgenic refines current understanding of the central CRFR1 distribution in several ways. As noted above, while the relative strength of GFP and receptor mRNA were often in accord, there were instances in which one or the other predominated. For example, while cell groups in the visual pretectum yielded evidence of weak-moderate CRFR1 mRNA expression, transgenic GFP labeling was more robust in terms of both cell number and signal strength. Conversely, where comparably strong hybridization signals were seen over CA1 and CA3 pyramidal neurons in hippocampus, robust GFP labeling was seen only over the CA1 field. Other instances in which the relative strengths of mRNA signals substantially exceeeded those of the GFP reporter were encountered in superficial layers of isocortex and the basolateral nucleus of the amygdala. It is of interest to note that in the case of the hippocampus, the expression pattern suggested by the transgenic is more compatible with the standard ligand binding study in rodent (rat) brain (De Souza et al., 1984) than are the receptor mRNA data, while the reverse is the case in the basolateral nucleus of the amygdala. Such observations highlight the limitations associated with inferring receptor distributions on the basis of any single, indirect marker.
Second, GFP labeling in the transgenic calls into question at least one major site of CRFR1 expression consistently indicated in immuno- and hybridization histochemical studies, namely in cerebellar Purkinje cells (Bishop et al., 2000; Chen et al., 2000; Van Pett et al., 2000). Large neurons that are densely packed and/or organized in a laminar manner are common sources of false positive artifact in isotopic hybridization histochemistry, and the absence of labeling of this cell type in the CRFR1-GFP mouse is consistent with recent findings using non-isotopic in situ hybridization to detect receptor mRNA (Lein et al., 2007, brainmap.org).
Third, the transgenic sheds light on uncertainties regarding CRFR1 status in a pivotal central autonomic cell group, the PVH. Prior studies report that receptor mRNA expression in PVH is weak or undetectable under basal conditions, but may be induced over a period of hours following exposure to any of a range of acute stressors (Van Pett et al., 2000; Rivest et al., 1995; Sup. Fig. 4). We observe a sizable population of GFP-stained cells in the PVH of unmanipulated mice. Intriguingly, this population does not co-express CRF, raising questions as to whether CRFR1 signaling at this locus is in position to directly modulate stress hormone secretion under resting conditions. Ongoing experiments indicate that these neurons may nevertheless alter their expression of GFP in response to stress and/or perturbations in circulating adrenal steroid hormone levels (Justice et al., unpublished).
Finally, the labeling of axonal projections of GFP neurons in the transgenic demands consideration of presynaptic receptor mechanisms. In the dentate gyrus, for example, the massive labeling of punctate elements in the outer two-thirds of the molecular layer strongly indicates CRFR1 expression by the neurons in entorhinal cortex whose projections distribute in this manner (Amaral and Witter, 1995). This contrasts sharply with the lack of axonal/terminal labeling in the inner third of the molecular layer occupied by projections of cells in the hilar region of the dentate gyrus, despite the fact that hilar neurons comprised the most intensely GFP-stained cell bodies observed the hippocampus of transgenic mice. Such observations underscore the need to develop and validate antisera with which to probe for authentic presynaptic CRFR1 disposition in immunohistochemical studies.
The mismatch problem in central autonomic cell groups
Core cell groups of the CAS (i.e., NTS, PB, PVH, CeAl and BSTov) are extensively interconnected (Saper, 1995; Sawchenko et al., 2008), in part by way of CRF-containing projections. They are heavily involved in processing visceral sensory information, and quite directly in effecting adaptations to it (Sawchenko et al., 1993; Saper, 1995). All are responsive to a wide range of stressors (Sawchenko et al., 1993) and all have been identified in microinjection studies as sites at which CRF can act to elicit stress-related endocrine, autonomic and/or behavioral responses (Lewis et al., 2002; Heinrichs and Koob, 2004; Pawlyk et al., 2006). Though less elaborately interwoven into this system, the locus coeruleus warrants consideration here, as it is interconnected with several core CAS cell groups, including by virtue of receiving a CRF innervation (Valentino et al., 1992; Van Bockstaele et al., 1996). It, too, is activated by a variety of (mainly emotional) stressors, and its widespread noradrenergic output is involved in executing relevant arousal-related functions (Valentino et al., 1993).
Each of the cell groups mentioned above was found in the present and previous studies to be engaged by central CRF administration (Imaki et al., 1993; Bittencourt and Sawchenko, 2000). Interpreting theses responses is severely complicated by the evidence from hybridization histochemistry and transgenic GFP expression that cellular expression of CRFR1 within this system is extremely limited, with only the lateral parabrachial nucleus representing a consensus site of receptor expression (Potter et al., 1994; Chalmers et al., 1995; Van Pett et al., 2000). While it is possible that the global activation of the CAS seen in response to icv peptide administration could be secondary to a direct effect on receptor-bearing parabrachial neurons, such a mechanism cannot readily explain the sensitivity of these cell groups to local peptide/antagonist administration (Lewis et al., 2002; Jedema and Grace, 2004; Liu et al., 2004). Additionally, it is unlikely that local activation of other CRF receptors can account for this sensitivity as the CeA, BSTov and LC are lacking in CRFR2 cellular expression (Van Pett et al., 2000).
Two potential means of reconciling the apparent ligand-receptor mismatch were proffered by the CRFR1-GFP transgenic mouse. In the central nucleus of the amygdala, in particular, we observed at the light and EM levels dendritic processes of GFP-labeled neurons in adjoining cell groups that invade the central nucleus, some of which were apposed by CRF-stained axon terminals. This has some precedent in Golgi studies of amygdalar neurons whose distal dendrites may fail to respect nuclear boundaries (McDonald, 1984), and may indicate interaction of some CRF-containing terminals in CeA with receptor-expressing neurons in other subregions of the amygdala (Jochman et al., 2005). Secondly, three CAS components, the CeAl, BSTov, and locus coeruleus, that were lacking or limited in cellular CRFR1 expression, were found to be invested with GFP-stained axons and terminals, again suggesting possible presynaptic CRFR1 localization at these sites. A presynaptic signaling mechanism has been suggested for the GABA-B-1b receptor, a class C GPCR, which shares a common sushi domain structure with the N-terminus of CRFR1 that has been suggested to be involved in the presynaptic targeting of this receptor isoform (Perez-Garci et al., 2006; Perrin et al., 2006; Grace et al., 2007). Additionally, there exists electrophysiological evidence supporting presynaptic CRFR mechanisms in modulating glutamatergic and/or GABAergic transmission in several CNS cell groups, including the central nucleus (Liu et al., 2004; Orozco-Cabal et al., 2006). Likely sources of receptor-bearing projections to the central nucleus include the lateral parabrachial nucleus, which contains strongly GFP positive neurons in the CRFR1-GFP mouse that can be retrogradely labeled after flourogold injections in the CeA (NJJ JFZ, unpublished observations, Sarhan et al., 2005). The CRFR1-GFP transgenic will be valuable in ascertaining the origin(s) and functional importance of receptor-bearing projections in CAS responses to central peptide administration and stress.
Supplementary Material
Acknowledgments
The authors thank C. Peto and C. Arias for excellent technical assistance, K. Trulock for assistance with preparation of the figures, and M. Huising and R. Berdeaux for critical reading of the manuscript. We thank N. Heintz for developing the BAC recombination techniques we used, and the Developmental Studies Hybridoma Bank for the SV2 antibody. N. Justice is supported by the Hewitt Foundation for Medical Research and the Adler Foundation. This work was supported in part by NIDDK P01 Program Project Grant DK26741 and in part by the Clayton Medical Research Foundation, Inc. W.W.V. and P.E.S. are CMRF, Inc. Senior Investigators.
Footnotes
CRFR1 promoter-based GFP expression in mouse
Conflict of Interest Statement: W.W.V. is a cofounder, consultant, equity holder, and member of the Board of Directors of Neurocrine Biosciences and Acceleron Pharma, Inc. The following have been licensed by The Salk Institute for Biological Studies and/or The Clayton Medical Research Foundation: CRF to Ferring Pharmaceuticals, CRF1 receptor and Ucn 2 to Neurocrine Biosciences, and Ucn 3 to Johnson & Johnson.
Acknowledgement of Support: N. Justice is supported by the Hewitt Foundation for Medical Research and the Adler Foundation. This work was supported in part by NIDDK P01 Program Project Grant DK26741 and in part by the Clayton Medical Research Foundation, Inc. W.W.V. and P.E.S. are CMRF, Inc. Senior Investigators.
Literature Cited
- Amaral DG, Witter MP. Hippocampal Formation. In: Paxinos G, editor. The Rat Nervous System. San Diego: Academic Press; 1995. pp. 443–493. [Google Scholar]
- Bishop GA, Seelandt CM, King JS. Cellular localization of corticotropin releasing factor receptors in the adult mouse cerebellum. Neuroscience. 2000;101:1083–1092. doi: 10.1016/s0306-4522(00)00413-9. [DOI] [PubMed] [Google Scholar]
- Bittencourt JC, Benoit R, Sawchenko PE. Distribution and origins of substance P-immunoreactive projections to the paraventricular and supraoptic nuclei: partial overlap with ascending catecholaminergic projections. J Chem Neuroanat. 1991;4:63–78. doi: 10.1016/0891-0618(91)90032-8. [DOI] [PubMed] [Google Scholar]
- Bittencourt JC, Sawchenko PE. Do centrally administered neuropeptides access cognate receptors?: an analysis in the central corticotropin-releasing factor system. J Neurosci. 2000;20:1142–1156. doi: 10.1523/JNEUROSCI.20-03-01142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Fisher LA. Corticotropin-releasing factor: effects on the autonomic nervous system and visceral systems. Fed Proc. 1985;44:243–248. [PubMed] [Google Scholar]
- Chalmers DT, Lovenberg TW, De Souza EB. Localization of novel corticotropin-releasing factor receptor (CRF2) mRNA expression to specific subcortical nuclei in rat brain: comparison with CRF1 receptor mRNA expression. J Neurosci. 1995;15:6340–6350. doi: 10.1523/JNEUROSCI.15-10-06340.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RK, Brown ER, Ericsson A, Kovacs KJ, Sawchenko PE. A comparison of two immediate-early genes, c-fos and NGFI-B, as markers for functional activation in stress-related neuroendocrine circuitry. J Neurosci. 1993;13:5126–5138. doi: 10.1523/JNEUROSCI.13-12-05126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Brunson KL, Muller MB, Cariaga W, Baram TZ. Immunocytochemical distribution of corticotropin-releasing hormone receptor type-1 (CRF(1))-like immunoreactivity in the mouse brain: light microscopy analysis using an antibody directed against the C-terminus. J Comp Neurol. 2000;420:305–323. doi: 10.1002/(sici)1096-9861(20000508)420:3<305::aid-cne3>3.0.co;2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ET, Jr, Sawchenko PE. Central neural control of esophageal motility: a review. Dysphagia. 1990;5:35–51. doi: 10.1007/BF02407391. [DOI] [PubMed] [Google Scholar]
- De Souza EB, Perrin MH, Insel TR, Rivier J, Vale WW, Kuhar MJ. Corticotropinreleasing factor receptors in rat forebrain: autoradiographic identification. Science. 1984;224:1449–1451. doi: 10.1126/science.6328656. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Funk CK, O’Dell LE, Crawford EF, Koob GF. Corticotropin-releasing factor within the central nucleus of the amygdala mediates enhanced ethanol self-administration in withdrawn, ethanol-dependent rats. J Neurosci. 2006;26:11324–11332. doi: 10.1523/JNEUROSCI.3096-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome Res. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Grace CR, Perrin MH, Gulyas J, Digruccio MR, Cantle JP, Rivier JE, Vale WW, Riek R. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci U S A. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs SC, Koob GF. Corticotropin-releasing factor in brain: a role in activation, arousal, and affect regulation. J Pharmacol Exp Ther. 2004;311:427–440. doi: 10.1124/jpet.103.052092. [DOI] [PubMed] [Google Scholar]
- Heinrichs SC, Menzaghi F, Pich EM, Baldwin HA, Rassnick S, Britton KT, Koob GF. Anti-stress action of a corticotropin-releasing factor antagonist on behavioral reactivity to stressors of varying type and intensity. Neuropsychopharmacology. 1994;11:179–186. doi: 10.1038/sj.npp.1380104. [DOI] [PubMed] [Google Scholar]
- Herbert H, Moga MM, Saper CB. Connections of the parabrachial nucleus with the nucleus of the solitary tract and the medullary reticular formation in the rat. J Comp Neurol. 1990;293:540–580. doi: 10.1002/cne.902930404. [DOI] [PubMed] [Google Scholar]
- Imaki T, Shibasaki T, Hotta M, Demura H. Intracerebroventricular administration of corticotropin-releasing factor induces c-fos mRNA expression in brain regions related to stress responses: comparison with pattern of c-fos mRNA induction after stress. Brain Res. 1993;616:114–125. doi: 10.1016/0006-8993(93)90199-w. [DOI] [PubMed] [Google Scholar]
- Jedema HP, Grace AA. Corticotropin-releasing hormone directly activates noradrenergic neurons of the locus ceruleus recorded in vitro. J Neurosci. 2004;24:9703–9713. doi: 10.1523/JNEUROSCI.2830-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G, Neugebauer V. Differential effects of CRF1 and CRF2 receptor antagonists on pain-related sensitization of neurons in the central nucleus of the amygdala. J Neurophysiol. 2007;97:3893–3904. doi: 10.1152/jn.00135.2007. [DOI] [PubMed] [Google Scholar]
- Jochman KA, Newman SM, Kalin NH, Bakshi VP. Corticotropin-releasing factor-1 receptors in the basolateral amygdala mediate stress-induced anorexia. Behav Neurosci. 2005;119:1448–1458. doi: 10.1037/0735-7044.119.6.1448. [DOI] [PubMed] [Google Scholar]
- Joo KM, Chung YH, Kim MK, Nam RH, Lee BL, Lee KH, Cha CI. Distribution of vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide receptors (VPAC1, VPAC2, and PAC1 receptor) in the rat brain. J Comp Neurol. 2004;476:388–413. doi: 10.1002/cne.20231. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lewis MW, Hermann GE, Rogers RC, Travagli RA. In vitro and in vivo analysis of the effects of corticotropin releasing factor on rat dorsal vagal complex. J Physiol. 2002;543:135–146. doi: 10.1113/jphysiol.2002.019281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kishi T, Roseberry AG, Cai X, Lee CE, Montez JM, Friedman JM, Elmquist JK. Transgenic mice expressing green fluorescent protein under the control of the melanocortin-4 receptor promoter. J Neurosci. 2003;23:7143–7154. doi: 10.1523/JNEUROSCI.23-18-07143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yu B, Neugebauer V, Grigoriadis DE, Rivier J, Vale WW, Shinnick-Gallagher P, Gallagher JP. Corticotropin-releasing factor and Urocortin I modulate excitatory glutamatergic synaptic transmission. J Neurosci. 2004;24:4020–4029. doi: 10.1523/JNEUROSCI.5531-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AJ. Neuronal organization of the lateral and basolateral amygdaloid nuclei in the rat. J Comp Neurol. 1984;222:589–606. doi: 10.1002/cne.902220410. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I, Lane M, Shughrue P. Distribution of pre-pro-glucagon and glucagon-like peptide-1 receptor messenger RNAs in the rat central nervous system. J Comp Neurol. 1999;403:261–280. doi: 10.1002/(sici)1096-9861(19990111)403:2<261::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I, Vigh S, Petrusz P, Schally AV. Immunocytochemical localization of corticotropin-releasing factor (CRF) in the rat brain. Am J Anat. 1982;165:385–396. doi: 10.1002/aja.1001650404. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Orozco-Cabal L, Pollandt S, Liu J, Shinnick-Gallagher P, Gallagher JP. Regulation of synaptic transmission by CRF receptors. Rev Neurosci. 2006;17:279–307. doi: 10.1515/revneuro.2006.17.3.279. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Sanford LD, Brennan FX, Morrison AR, Ross RJ. Corticotropin-releasing factor microinjection into the central nucleus of the amygdala alters REM sleep. Pharmacol Rep. 2006;58:125–130. [PubMed] [Google Scholar]
- Perez-Garci E, Gassmann M, Bettler B, Larkum ME. The GABAB1b isoform mediates long-lasting inhibition of dendritic Ca2+ spikes in layer 5 somatosensory pyramidal neurons. Neuron. 2006;50:603–616. doi: 10.1016/j.neuron.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Perrin MH, Grace CR, Riek R, Vale WW. The three-dimensional structure of the N-terminal domain of corticotropin-releasing factor receptors: sushi domains and the B1 family of G protein-coupled receptors. Ann N Y Acad Sci. 2006;1070:105–119. doi: 10.1196/annals.1317.065. [DOI] [PubMed] [Google Scholar]
- Potter E, Sutton S, Donaldson C, Chen R, Perrin M, Lewis K, Sawchenko PE, Vale W. Distribution of corticotropin-releasing factor receptor mRNA expression in the rat brain and pituitary. Proc Natl Acad Sci U S A. 1994;91:8777–8781. doi: 10.1073/pnas.91.19.8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes BA, Valentino RJ, Van Bockstaele EJ. Stress-induced intracellular trafficking of corticotropin-releasing factor receptors in rat locus coeruleus neurons. Endocrinology. 2008;149:122–130. doi: 10.1210/en.2007-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivest S, Laflamme N, Nappi RE. Immune challenge and immobilization stress induce transcription of the gene encoding the CRF receptor in selective nuclei of the rat hypothalamus. J Neurosci. 1995;15:2680–2695. doi: 10.1523/JNEUROSCI.15-04-02680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakanaka M, Shibasaki T, Lederis K. Distribution and efferent projections of corticotropin-releasing factor-like immunoreactivity in the rat amygdaloid complex. Brain Res. 1986;382:213–238. doi: 10.1016/0006-8993(86)91332-6. [DOI] [PubMed] [Google Scholar]
- Saper CB. Central autonomic system. In: Paxinos G, editor. The Rat Nervous System. 2. San Diego: Academic Press; 1995. pp. 107–128. [Google Scholar]
- Sarhan M, Freund-Mercier MJ, Veinante P. Branching patterns of parabrachial neurons projecting to the central extended amgydala: single axonal reconstructions. J Comp Neurol. 2005;491:418–442. doi: 10.1002/cne.20697. [DOI] [PubMed] [Google Scholar]
- Sauvage M, Steckler T. Detection of corticotropin-releasing hormone receptor 1 immunoreactivity in cholinergic, dopaminergic and noradrenergic neurons of the murine basal forebrain and brainstem nuclei--potential implication for arousal and attention. Neuroscience. 2001;104:643–652. doi: 10.1016/s0306-4522(01)00137-3. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Arias C, Bittencourt JC. Inhibin beta, somatostatin, and enkephalin immunoreactivities coexist in caudal medullary neurons that project to the paraventricular nucleus of the hypothalamus. J Comp Neurol. 1990;291:269–280. doi: 10.1002/cne.902910209. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Imaki T, Potter E, Kovacs K, Imaki J, Vale W. The functional neuroanatomy of corticotropin-releasing factor. Ciba Found Symp. 1993;172:5–21. doi: 10.1002/9780470514368.ch2. discussion 21–29. [DOI] [PubMed] [Google Scholar]
- Sawchenko PE, Yuan ZF, Laplante FRAR, Bittencourt JC. Corticotropin-releasing factor and the integration of neuroendocrine, autonomic and behavioral responses to stress. In: Squire L, editor. New Encyclopedia of Neuroscience. London: Elsevier; 2008. [Google Scholar]
- Schiltz JC, Sawchenko PE. Specificity and generality of the involvement of catecholaminergic afferents in hypothalamic responses to immune insults. J Comp Neurol. 2007;502:455–467. doi: 10.1002/cne.21329. [DOI] [PubMed] [Google Scholar]
- Simmons D, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radiolabeled single-stranded RNA probes. Journal of Histotechnology. 1989;12:169–181. [Google Scholar]
- Sutton RE, GFK, ML, Rivier J, Vale W. Corticotropin Releasing Factor (CRF) produces behavioral activation in rats. Nature. 1982;297:331–333. doi: 10.1038/297331a0. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Sawchenko PE, Rivier J, Vale WW. Organization of ovine corticotropin-releasing factor immunoreactive cells and fibers in the rat brain: an immunohistochemical study. Neuroendocrinology. 1983;36:165–186. doi: 10.1159/000123454. [DOI] [PubMed] [Google Scholar]
- Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981;213:1394–1397. doi: 10.1126/science.6267699. [DOI] [PubMed] [Google Scholar]
- Vale W, Vaughan J, Yamamoto G, Bruhn T, Douglas C, Dalton D, Rivier C, Rivier J. Assay of corticotropin releasing factor. Methods Enzymol. 1983;103:565–577. doi: 10.1016/s0076-6879(83)03040-2. [DOI] [PubMed] [Google Scholar]
- Valentino RJ, Foote SL, Page ME. The locus coeruleus as a site for integrating corticotropin-releasing factor and noradrenergic mediation of stress responses. Ann N Y Acad Sci. 1993;697:173–188. doi: 10.1111/j.1749-6632.1993.tb49931.x. [DOI] [PubMed] [Google Scholar]
- Valentino RJ, Page M, Van Bockstaele E, Aston-Jones G. Corticotropin-releasing factor innervation of the locus coeruleus region: distribution of fibers and sources of input. Neuroscience. 1992;48:689–705. doi: 10.1016/0306-4522(92)90412-u. [DOI] [PubMed] [Google Scholar]
- Van Bockstaele EJ, Colago EE, Valentino RJ. Corticotropin-releasing factor-containing axon terminals synapse onto catecholamine dendrites and may presynaptically modulate other afferents in the rostral pole of the nucleus locus coeruleus in the rat brain. J Comp Neurol. 1996;364:523–534. doi: 10.1002/(SICI)1096-9861(19960115)364:3<523::AID-CNE10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Van Pett K, Viau V, Bittencourt JC, Chan RK, Li HY, Arias C, Prins GS, Perrin M, Vale W, Sawchenko PE. Distribution of mRNAs encoding CRF receptors in brain and pituitary of rat and mouse. J Comp Neurol. 2000;428:191–212. doi: 10.1002/1096-9861(20001211)428:2<191::aid-cne1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.