Abstract
Tissue-specific deletion of Fgfr1 results in severe defects in the development of both hair cells and support cells (Pirvola et al., 2002). Despite the importance of Fgfr1 in this early phase of cochlear development, the timing for the requirement for FGF signaling at this stage is not known. Therefore, we investigated the requirement for FGF signaling at early stages of cochlear development using an FGF receptor inhibitor. We find that inhibition of FGF signaling from embryonic day 14 (E14) to E16 has a dramatic effect on the development of the sensory epithelium, causing a severe reduction in hair cells and support cells, similar to that reported for the Fgfr1 deletion. The effects of inhibition of FGF signaling on sensory specification are not explained by increases in cell death or changes in proliferation but lead to a rapid reduction in Pea3 and Erm and a loss of Math1 expression. We also show that a specific FGF, FGF20, is the likely ligand for FGFR1 at this sensory specification phase of cochlear development; Fgf20 is expressed at the right time and place to mediate sensory cell specification, and blocking FGF20 with a specific antibody inhibits hair cell and support cell development in a manner similar to the FGF receptor inhibitor. Our results thus define the period of FGF-dependent sensory cell specification and the ligand that mediates this step in cochlear development.
Keywords: hair cell, support cell, inner ear, sensory development, auditory, hearing
Introduction
The organ of Corti, which mediates auditory function in mammals, develops from the otic placode. The cochlea forms initially from an out-pocketing of the ventromedial otocyst at embryonic day 12 (E12) in the mouse (Morsli et al., 1998). The hair cells and the associated support cells of the organ of Corti are born between E12 and E16, from the apex to the base, with most hair cells birthdated by E14 (Ruben, 1967). A few days later, a temporal gradient of hair cell and support cell differentiation begins in the midbasal region of the cochlea and proceeds along the entire length of the epithelium. The mature complement of hair cells, one row of inner and three rows of outer, is achieved by E17–E18 in the mouse (Ruben, 1967; Sher, 1971; Lim and Anniko, 1985).
Fibroblast growth factors (FGFs) have been shown to be of critical importance for the development of the organ of Corti. FGFs form a large family of proteins that act as signaling molecules in many key developmental processes (Ford-Perriss et al., 2001; Itoh, 2001; Ornitz and Itoh, 2001; Itoh and Ornitz, 2004). There are now >23 different known genes that code for Fgfs in mice and humans and four FGF receptors (Fgfr1–4) (Zhang et al., 2006). In mice, FGF3, FGF10, and FGF8 have been implicated in the early inductive events of the otic vesicle (Mansour et al., 1993; Alvarez et al., 2003; Wright and Mansour, 2003; Ladher et al., 2005; Martin and Groves, 2006; Ohyama et al., 2007). Mice deficient in both Fgf3 and Fgf10 fail to form otic vesicles. Similar defects in the early stages of otocyst development are present in mice with targeted deletion of a specific isoform of Fgfr2 (FGFR2 IIIB) (Pirvola et al., 2000), and it has been proposed that FGF10 and FGF3 act as the ligands for FGFR2 in otic placode formation and patterning (Pauley et al., 2003; Wright et al., 2003).
In the next phase of cochlear development, at the sensory specification phase, FGF signaling is again thought to be required. Tissue-specific deletion of Fgfr1 results in severe defects in the development of both hair cells and support cells, and those sensory cells that develop are found in small clusters (Pirvola et al., 2002). Despite the importance of Fgfr1 in this phase of cochlear development, the ligand for this effect has not been identified. In addition, the precise timing for the requirement for FGF signaling in this process is not known. Therefore, we investigated the requirement of FGF signaling at the sensory specification phase of cochlear development; we find that inhibition of FGF signaling at early stages of development, using an FGF receptor inhibitor, causes a reduction in hair cells and support cells similar to that in the Fgfr1 deletion. We also show that a specific FGF, FGF20, is the likely activator of FGFR1 at this phase of cochlear development. Our results thus define the period of FGF-dependent sensory cell specification and the ligand that mediates this step in cochlear development.
Materials and Methods
Organ cultures of embryonic cochlea.
The explants culture was performed according to Hayashi et al. (2007). In brief, inner ear tissue was isolated from E12.5–E15.0 embryos. The cochlea was treated with 0.1% dispase (Invitrogen), 0.1% collagenase (Invitrogen), and 0.001% DNase (Sigma) for 15 min at 37°C. The cochlear capsule was opened using forceps to expose the cochlear duct. The cochlear ducts were placed on a collagen/Matrigel substrate, along with the mesenchyme surrounding the cochlea. The cochlea duct was opened for the incubation with anti-FGF antibodies to allow the antibodies to reach the surface of the epithelium.
Explants were cultured in modified DMEM/F-12 medium [DMEM/F-12 (Invitrogen), 0.6% glucose, 5 mm HEPES, 0.13% NaHCO3, 800 nm l-glutamine, 100 U/ml penicillin (Sigma), N2 supplement, and 20% fetal bovine serum], 5% CO2, at 37°C, and the medium was replaced each day. To inhibit FGF signaling, 3–30 μm 3-[(3-(2-carboxyethyl)-4-methylpyrrol-2-yl)methylene]-2-indolinone (SU5402; Calbiochem) was added into the culture medium. Anti-FGF8, anti-FGF20, and recombinant FGF20 were obtained from R&D Systems.
Immunofluorescence.
Whole-mount staining of cultured cochleas was performed according to Hayashi et al. (2007). The primary antibodies used in this study were as follows: rabbit anti-Prox1 (Millipore Bioscience Research Reagents) used at 1:1000 dilution; rabbit anti-Myo6 used at 1:2000 dilution; goat anti-Sox2 (Santa Cruz Biotechnology) used at 1:1000 dilution; rabbit anti-p75 (Millipore Bioscience Research Reagents) used at 1:2000 dilution. The secondary antibodies used were goat anti-mouse Alexa 594, chicken anti-rabbit Alexa 594, donkey anti-goat Alexa 594, and donkey anti- rabbit Alexa 488, all from Invitrogen and used at 1:750. Images were captured on a Zeiss Pascal confocal or on an Olympus FluoView system and processed using ImageJ and Photoshop.
After in situ hybridization, the slides were fixed with 4% paraformaldehyde (PFA) for 1 h and washed in PBS. The slides were then incubated with 10% fetal bovine serum and 2% nonfat dry milk in PBS/0.1% Triton X-100 (PBST) for 30 min. After an overnight incubation with the primary antibody (rabbit anti-Myo6 or mouse anti-p27kip1; BD Transduction Laboratories) at 1:300 dilution at 4°C, the sections were rinsed with PBST, incubated for 90 min with a fluorescent-conjugated secondary antibody, rinsed with PBST, and coverslipped in Fluoromount G (Southern Biotechnology). Images were captured on a Zeiss Axioplan microscope using a SPOT CCD camera and processed using Adobe Photoshop.
For terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) staining, the cochlear explants were fixed in 4% PBS for 2 h and washed in PBST. The explants were incubated with 3 μ/ml terminal deoxynucleotidyl transferase (Promega) and 5 mm Alexa 546-conjugated dUTP in PBST for 2 h at 37°C.
In situ hybridization. The cochlear explants were fixed with 4% PFA for 2 h and dehydrated in ethanol/xylene series. The samples were rehydrated in PFA. For the section in situ, whole heads (E13.5, E15.5, and E18.5) were fixed in a modified Carnoy's solution for 6 h at room temperature. The samples were washed and dehydrated in 100% ethanol overnight at 4°C and embedded in paraffin, and 6 μm sections were collected. At least three animals were examined at each time point. Mouse ERM and Pea3 cDNAs were obtained from Open Biosystems, and cDNA coding for mouse Fgfr1 and Fgfr2 was a gift from Dr. P. Soriano (Fred Hutchinson Cancer Research Center, Seattle, WA). cDNA for mouse Sox2 was a gift from Dr. H. Kondoh (Osaka University, Osaka, Japan). cDNA for mouse Fgf20 was a gift from Dr. S. L. Mansour (University of Utah, Salt Lake City, UT), and cDNA for Fgfr4 was a gift from Dr. A. McMahon (Harvard University, Boston, MA). Digoxigenin (DIG)-labeled probes were prepared according to the manufacturer's manual for DIG-11-UTP (Roche), and the hybridization was performed according to Hayashi et al. (2007). The in situ product was visualized using anti-DIG alkaline phosphatase-conjugated secondary antibody (Roche) and NBT (nitroblue tetrazolium)/BCIP (5-bromo-4-chloro-3-indolyl phosphate).
Quantitative PCR analysis of cochlea RNA.
Total RNA was extracted from pools of six to eight cochleas (E13.0) cultured with/without 30 μm SU5402 using TRIzol (Invitrogen). Three independent RNA pools were prepared for each experiment. cDNA was synthesized by Superscript III reverse transcriptase (Invitrogen) using random primers and 2.5 μg of total RNA as the template, and quantitative PCRs (Q-PCRs) were performed using the cDNAs and the following primer sets (Primer Bank identification): ERM (24528550a1) and Pea3 (6679271a2). Q-PCR was performed by using an Opticon monitor (Genetic Technologies), and the cycle in which log phase was attained was recorded. A SYBR Green-based mastermix (Applied Biosystems) was used for the PCRs. All samples were normalized to expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Results
In vitro analysis of FGF signaling
Much of what we know about the role of FGFs in cochlear development in mice comes from analysis of knock-out mice. Although these experiments have been invaluable for implicating a specific FGF or receptor in the process of cochlear development, it has been difficult to define the temporal requirements for FGF signaling, because the gene is deleted throughout cochlear development. However, several groups have used an in vitro approach to define key periods in cochlear development during which inductive signaling events are required. To define the period of cochlear development when FGF signaling is required for the formation of the sensory epithelium, we treated explant cultures with a small molecule inhibitor of FGF receptors (SU5402). SU5402 is a highly effective inhibitor of FGF receptors (Mohammadi et al., 1997) and has been used previously at later stages of cochlear development to examine the requirement for FGFR3 in support cell development (Mueller et al., 2002; Jacques et al., 2007). We verified that SU5402 treatment phenocopies the effects of FGF receptor deletion at later stages of cochlear development, by treating E18 cultures with 30 μm SU5420 for 48 h; consistent with the previous studies, the effect on cochlear development is similar to the Fgfr3 knock-out: pillar cells fail to develop their characteristic morphology (there is no clear space between the inner and outer hair cell rows indicative of pillar heads) and do not express p75 (supplemental Fig. 1A, available at www.jneurosci.org as supplemental material).
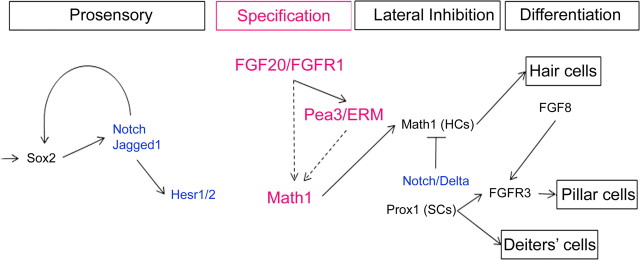
To determine the time during the prosensory period when FGF signaling is required, we made explant cultures from E12.5, E14, or E15 and treated them for 40 h; the SU5402 was then removed, and the explant was cultured for varying lengths of time as shown in Figure 1A. Explants were fixed and labeled with the combination of Sox2 and Myosin6 (Myo6) or of Sox2 and Prox1 to determine the overall extent of the sensory epithelium (defined as the Sox2+ region) and the number of hair cells and support cells within this region (Myo6+ or Prox1+, respectively). We found that treatment of cultures with 30 μm SU5402 in each of the three periods caused a reduction in the number of hair cells (Fig. 1B), but the most dramatic reduction was found when the explants were treated from E14 to E16 (Fig. 1C,D). The number of hair cells was reduced from 1422 (±263.3) in the control cultures to 52 (±20.2) in the 30 μm SU5402-treated cultures of E14–E16 (Fig. 1B). The reduction of hair cells during this most sensitive period (E14–E16) was dose dependent (control, 1422 ± 263.3; 3 μm, 1204 ± 343; 15 μm, 778 ± 192; 30 μm, 52 ± 20). The treatment groups of 15 μm (p < 0.002) and 30 μm (p < 0.0001) showed significant differences compared with the control. Those hair cells that form are organized in small clusters (Fig. 1D), similar to what has been seen in the conditional Fgfr1 deletion (Pirvola et al., 2002). All of the explants (12 of 12) treated with SU5402 from E14 to E16 show clusters of hair cells, and these clusters are seen throughout the explant, from base to apex. When explants were treated at E12, 6 of 10 explants showed reduced truncated sensory epithelia without clusters, and 4 of 10 explants showed clusters of hair cells in addition to a truncated epithelium.
Figure 1.
Effects of FGF receptor inhibition on hair cell development. A, Experimental design. Three different treatment regimens were used to define the sensitive period for FGF receptor inhibition on hair cell development. The beginning of the line indicates the time at which the cochlea was explanted and treatment was started; the yellow part of the line shows the period when SU5402 was added to the culture. All explants were cultured until the equivalent of E18.5. B, Graph of hair cell number in control (blue bars) and treated (yellow bars) explants for the different treatment groups. Inhibiting FGF receptor from E14 to E16 had the greatest effect on hair cell number, although all treatment regimens showed significant effects (*p < 0.0005). Each bar in the graphs represents the mean of six explants. Error bars indicate SD. C, D, Control (C) and treated (D) cultures. Explant cultures were immunolabeled with antibodies to Sox2 (red) and Myo6 (green). C′, D′, Higher-magnification images of the regions boxed in panels C and D. E, F, Control (E) and treated (F) explants from Math1–GFP mice and immunolabeled with p75 to show pillar cells.
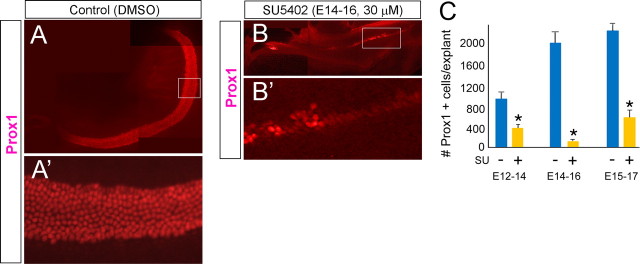
Reduction in hair cell numbers was confirmed using explants from a Math1–GFP strain (Fig. 1E,F). Interestingly, the support cells that develop in the SU5402-treated cultures are primarily pillar cells, as shown by p75 immunoreactivity (Fig. 1E,F). We also found that SU5402 treatment causes a severe reduction in the number of support cells that develop in the cultures. We labeled treated and control cultures with an antibody against Prox1 (Fig. 2), a support cell marker (Bermingham-McDonogh et al., 2006), and found a significant reduction in the number of Prox1+ cells (Fig. 2C). Like the hair cells in the treated cultures, the remaining support cells were also, at times, clustered in small rosettes (Fig. 2B,B′,C).
Figure 2.
Effects of FGF receptor inhibition on support cell development. Support cells in explant culture are identified by Prox1 immunolabeling (red). A, B, Examples of control (A) and treated (A) explant cultures. A′, B′, Higher-magnification images of the boxed areas in A and B. C, Graph of Prox1-positive support cell number in control (blue bars) and treated (yellow bars) explants for the different treatment groups. Each bar in the graphs represents the mean of six explants. Error bars indicate SD (*p < 0.0001).
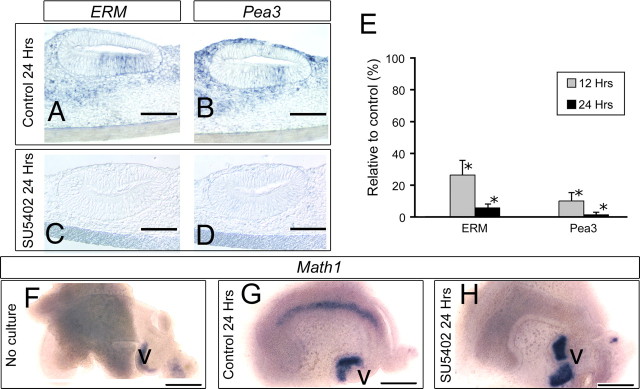
The effects of FGF receptor inhibition on sensory cell development are rapid. We assayed the expression of two transcription factors known to be downstream of FGF signaling in other systems, Pea3 and ERM. After the SU5402 treatment, expression levels of both Pea3 and ERM were downregulated in the epithelium and the mesenchyme (Fig. 3A–D). By Q-PCR analysis, we confirmed that both were significantly reduced within 12 h of treatment with SU5402 (Fig. 3E). In addition, we assayed Math1 expression using in situ hybridization after SU5402 treatment (Fig. 3F–H). Math1 is not expressed in the cochlea at the time of dissection (Fig. 3F), but in control cultures, it is expressed within 24 h (Fig. 3G); however, in SU5402-treated cultures, Math1 is not expressed in the cochlea (Fig. 3H). Interestingly, the expression of Math1 in the vestibular epithelia is not affected at this time (Fig. 3F–H, V).
Figure 3.
SU5402 treatment rapidly downregulates ERM and Pea3 and suppresses expression of Math1. A–D, Expression patterns of ERM and Pea3 by in situ hybridization in E14.0 cochlea explants cultured without (A, B) or with (C, D) 30 μm SU5402 for 24 h. Scale bars, 100 μm. E, Q-PCR shows a decrease in the expression of ERM and Pea3 as early as 12 h after treatment with SU5402 (*p < 0.02). F–H, In situ hybridization for Math1 in cochlear explants. F, Explant just before culture showing that, at this stage (E14), Math1 is not yet expressed. G, Explant cultured under control conditions shows onset of Math1 after 24 h of culture. H, SU5402-treated culture shows no expression of Math1 after 24 h of culture, although expression in vestibular epithelium (V) is unaffected by the treatment. Scale bars, 500 μm.
To determine whether the reduction of hair cells and support cells in the SU5402-treated cultures was attributable to cell death, we quantified the number of apoptotic cells in the Sox2+ domain in treated and control E14 explant cultures using TUNEL staining. We did not find a significant difference between the treated and control cultures in the number of apoptotic cells within the Sox2+ domain after 40 h of SU5402 treatment (control: 96 ± 20, n = 5; SU5402 treated: 139 ± 20, n = 5; p = 0.124, Student's t test). It is also unlikely that the effects of SU5402 treatment on hair and support cell development are attributable to an inhibition in the proliferation of the progenitors to these cells, because we see effects of SU5402 treatment at a time (E14) when, in most of the cochlea, the precursors of hair cells and support cells have already withdrawn from the cell cycle (Ruben, 1967; Lee et al., 2006). We cultured E14.0 Math1–GFP cochlea with bromodeoxyuridine (BrdU) for 40 h with BrdU added to the medium the entire period. We found a few BrdU+ (<20) cells in the sensory epithelium, as defined by the Math1–GFP-expressing hair cells, but these were confined to the base and were not in the hair cells themselves (supplemental Fig. 2C, available at www.jneurosci.org as supplemental material). At early stages of cochlear development (E12–E14), there are still mitotic cells within the sensory epithelium, and so some of the effects of blocking FGF receptors may be attributable to a reduction in proliferation of hair cell progenitors (supplemental Fig. 2A,B, available at www.jneurosci.org as supplemental material). However, at later stages of cochlear development (>E14), the vast majority of hair cell progenitors have exited the mitotic cycle, and so the effects we observe with SU5402 treatment are not attributable to inhibition of proliferation.
Expression patterns of Fgf20 and its signaling components in the developing sensory epithelium
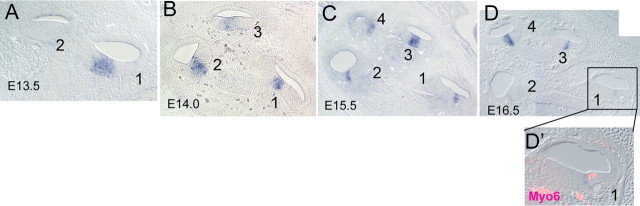
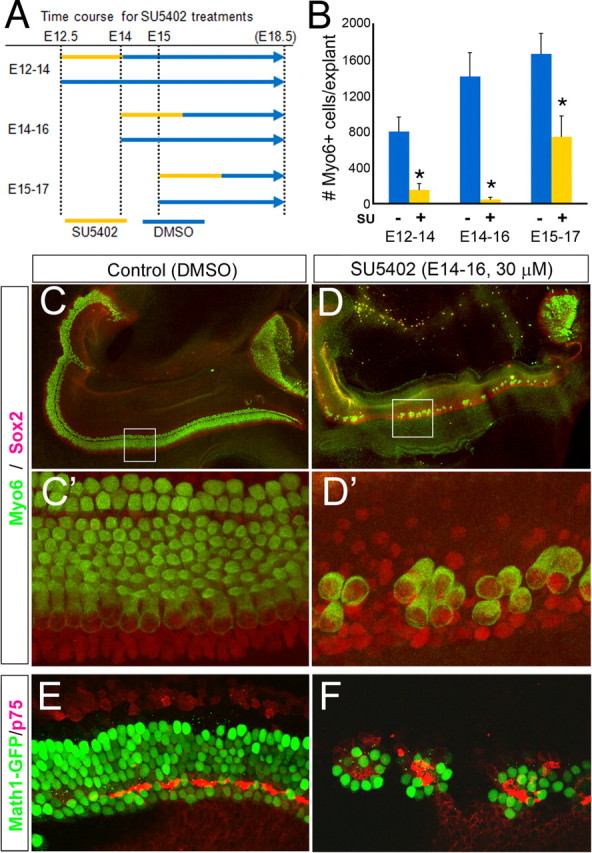
In an in situ screen of known FGFs that would make good candidates for regulating cochlear development, we discovered that Fgf20 is highly expressed for a distinct period of cochlear development, from E13.5 to E16.5 in the presumptive sensory epithelial domain (Fig. 4). FGF20 is a member of the FGF9 subfamily of FGFs. The FGF9 subfamily activates all FGFRc isoforms (Zhang et al., 2006). At E13.5, the expression of Fgf20 is present at the base (Fig. 4A, turn 1) but not the apex (Fig. 4A, turn 2). The expression of Fgf20 persists through all turns of the cochlea at E14.0–E15.5 (Fig. 4B,C). At E16.5, Fgf20 is still expressed in the apical turns (Fig. 4D, turns 3 and 4) but shows only a very low level of expression in the base (Fig. 4D, turns 1 and 2, D′). Thus, Fgf20 is expressed as a wave that spreads from base to apex and then declines in a similar pattern. To identify the temporal and spatial relationship among Fgf20, Fgfr1, and Sox2, a prosensory marker, we compared the expression patterns of these genes in adjacent sections. Fgf 20 and Sox2 are essentially overlapping in their expression patterns at E14 and E15 (Fig. 5A–D,G). Fgfr1 shows a more diffuse expression pattern (Fig. 5E); however, it is upregulated in the region of the developing sensory epithelium at E15.0 (Fig. 5F), and this upregulation sweeps from base to apex. Fgf20/Sox2 expression overlaps with Fgfr1 expression; however, the stronger expression of Fgfr1 in the developing sensory epithelium extends further laterally than the Fgf20 domain (Fig. 5H,I).
Figure 4.
Developmental expression pattern of Fgf20. The cochlear turns are numbered from the base (1) to apex (2–4). A, Expression of Fgf20 at E13.5 in the base of the developing cochlea. B, C, Expression of Fgf20 at E14.0 (B) and E15.5 (C) extends to the apex. D, Expression of Fgf20 at E16.5 is reduced in the apex and almost extinguished in the base, where the hair cells are differentiating as indicated by Myo6 staining of this section (D′). In all panels, the apex is at top left.
Figure 5.
Fgf20, Sox2, and Fgfr1 are expressed in a similar domain in the developing cochlea. A, C, E, Adjacent sections of E14.0 cochlea. B, D, F, Adjacent sections of E15.0 cochlea. The cochlear turns are numbered from the base (1) to the apex (3). G–I, To allow direct comparison of the expression domains, the photomicrographs B′, D′, and F′ are pseudocolored and overlaid using Adobe Photoshop. G, Fgf20 (B′) versus Sox2 (D′). H, Fgf20 (B′) versus Fgfr1 (F′). I, Sox2 (D′) versus Fgfr1 (F′). Scale bars, 100 μm.
Expression patterns of FGF receptors and their downstream targets in the developing sensory epithelium
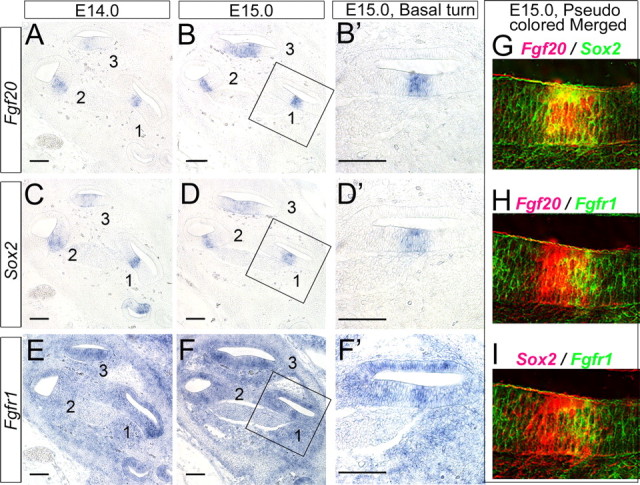
We examined the expression patterns of all four FGF receptors in an effort to see which FGF receptor(s) could potentially act as the receptor for FGF20. Fgfr1 and Fgfr2 are both expressed at early embryonic stages in cochlear development (Fig. 6A,B). Fgfr1 has a somewhat diffuse pattern of expression through the cochlear epithelium and at E13.5 (Fig. 6A and supplemental Fig. 3, available at www.jneurosci.org as supplemental material) but is more highly expressed in the ventral region of the cochlea epithelium (Fig. 6A, arrowheads). This higher ventral expression becomes more pronounced at E 15.5 (Fig. 6F, arrow). Fgfr1 is also expressed in the mesenchymal cells (Fig. 6A). In contrast, Fgfr2 is expressed in a dorsolateral domain of the cochlear duct, in the presumptive stria vascularis and developing spiral prominence, at all stages we examined (Fig. 6B,G,M). Fgfr3 is expressed in otic capsule cells but not in the cochlear duct at E13.5–E15.0 (Fig. 6C, arrowheads, and supplemental Fig. 3, available at www.jneurosci.org as supplemental material), but at later stages of development (E15.5 and E18.5), Fgfr3 expression delineates the sensory domain (Fig. 6H,N and supplemental Fig. 3, available at www.jneurosci.org as supplemental material). These expression patterns suggest that Fgfr1 is the most likely receptor for FGF20, because it is expressed at the right time and place. Fgfr4 is expressed in the mesenchymal cells but is never expressed in the developing sensory epithelium (supplemental Fig. 3, available at www.jneurosci.org as supplemental material).
Figure 6.
Developmental expression patterns of Fgf receptors and downstream targets. All panels show the basal turn of the cochlear ducts. A, Fgfr1 is expressed broadly in the developing cochlea at E13.5 as well as in the surrounding mesenchyme. F, L, The expression becomes more concentrated to the ventral cochlear duct and, in particular, in the area of presumptive sensory epithelium at E15.5 (F); the expression is then downregulated as the hair cells differentiate at E18.5 (L). B, G, M, Fgfr2 is expressed only in the lateral domain of the developing cochlear duct at E13.5 (B), at E15.5 (G), and at E18.5 (M). This is the region that will give rise to the stria vascularis. C, H, N, Fgfr3 is not expressed at E13.5 (C) but is expressed specifically in the sensory region at E15.5 (H), and this expression is confined to pillar cells and Deiters' at E18.5 (N). D, E, I, J, O, P, ERM (D, I, O) and Pea3 (E, J, P), downstream targets of FGF signaling, are also expressed from E13.5 (D, E) and concentrated to the presumptive sensory domain (delineated by p27 staining in K) at E15.5 (I, J) and continue to be expressed in developing pillar and Hensen cells at E18.5 (O, P). Q, The developing hair cells are labeled with Myo6, which is the same section shown in P.
We also examined the expression of potential downstream targets/effectors of FGF signaling at the time of Fgf20 expression. Pea3 and Erm are known to be transcriptionally regulated by FGF signaling. Both Erm and Pea3 show widespread expression in a ventral domain of the cochlear duct at E13.5 (Fig. 6D,E). This expression becomes restricted over the next few days to the region of the duct that will give rise to the sensory cells (Fig. 6I–K). At E18.5, Erm and Pea3 are only expressed by developing pillar cells and in the Claudius/Hensen cell region (Fig. 6O–Q).
Fgf20 is required for sensory epithelial development
The expression pattern of Fgf20 suggested to us that this is a likely candidate for the ligand for FGFR1 during sensory specification in the developing cochlea. To test this hypothesis, we cultured explants of cochlea starting at E12.5–E13.0, as described above. We then added a blocking antibody against FGF20 to some of the cultures at varying concentrations, whereas sister cultures received an equivalent concentration of an IgG control antibody (Fig. 7). The IgG-alone (60 μg/ml) treated cultures had similar numbers of hair cells as untreated explants; when explanted at E13, the number of hair cells that develop in culture is between the values for E12 and E14 (compare Fig. 1B). However, we found a dose-dependent inhibition of hair cells in the cultures treated with the anti-FGF20 antibody (control IgG, 1011 ± 200 vs 60 μg/ml anti-FGF20, 88 ± 42). Moreover, those hair cells that formed were organized in small clusters (Fig. 7B,B′), similar to what we observed in the SU5402-treated cultures. The inhibition of the hair cell development in the cochleas cultured in the presence of anti-FGF20 was rescued by the addition of recombinant FGF20 protein. A 1:1 antibody to FGF20 ratio resulted in 423 ± 64 hair cells versus 88 ± 42 in antibody alone (Fig. 7C,D). We were unable to use higher concentrations of FGF20 (typical of antibody rescue experiments) because of the robust effects on proliferation of mesenchymal cells, in the explant culture, leading to their rapid overgrowth. Together, our data support the hypothesis that FGF20 is required for normal hair cell development.
Figure 7.
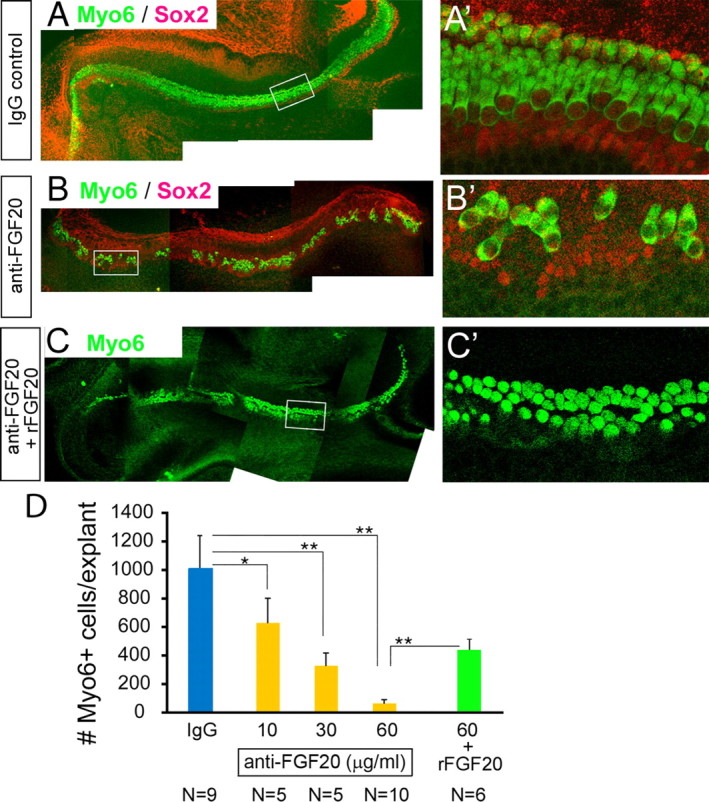
Anti-FGF20 antibody inhibits formation of hair cells in explant cochlear cultures. A–C′, Explants were treated with control IgG (A, A′), 30 μg/ml of a blocking antibody to FGF20 (B, B′), or 60 μg/ml of the blocking antibody with 9 μg/ml recombinant FGF20 (C, C′) from E13 for 5 d A′, B′, and C′ are higher-magnification images of the regions boxed in A–C. There is a high background in the red channel because the Sox2 and the FGF20 antibodies are both produced in goat. Each bar represents the mean of five explants. The hair cells were immunolabeled with Myo6 (green) and Sox2 (red) antibodies. D, Graph of hair cell number with increasing concentrations of anti-FGF20 (yellow bars), 60 μg/ml IgG control antibody (blue bar), 60 μg/ml anti-FGF20 antibody plus 9 μg/ml recombinant FGF20 (rFGF20; green bar). N, Number of explants. Error bars indicate SDs. *p < 0.02; **p < 0.00002.
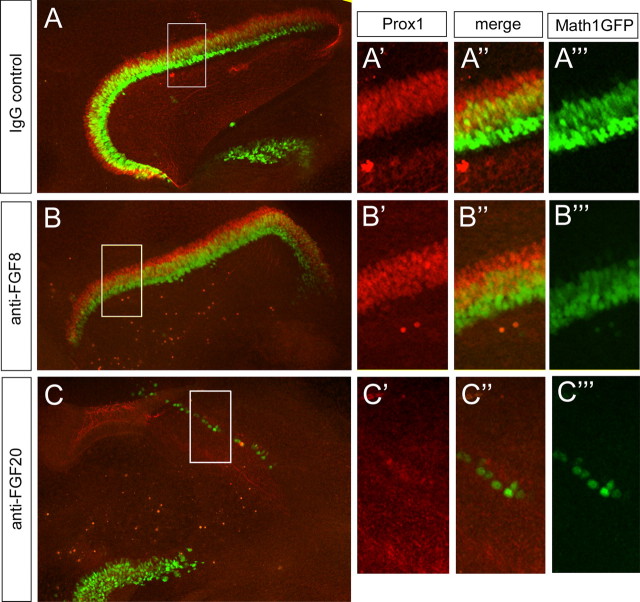
To determine whether inhibiting FGF20 blocked support cell development, as well as hair cells, we used explants of cochlea from Math1–GFP reporter mice and colabeled them with a marker for support cells, Prox1. We found that treatment of cultures with anti-FGF20 led to a profound inhibition in Math1 expression (Fig. 8C–C″) compared with the IgG control. In addition to the effects on hair cell development, we found that inhibition of FGF20 also blocked support cell development; the number of Prox1+ support cells was greatly reduced in the cultures treated with anti-FGF20 compared with the IgG control cultures (Fig. 8C–C″). We also treated cultures with an antibody against FGF8 (30 μg/ml); anti-FGF8 had no effect on the development of either hair cells or support cells at this stage of treatment (Fig. 8B′–B‴), whereas anti-FGF8 prevented the pillar cell development in the later stage as reported previously (supplemental Fig. 1B, available at www.jneurosci.org as supplemental material).
Figure 8.
Blocking FGF20 inhibits support cells development in explant culture. Explants were treated with anti-FGF20 antibody (C–C‴), anti-FGF8 antibody (B–B‴), or control goat IgG (A–A‴) from E13 for 5 d. Explants were from a Math1–GFP mouse strain to show hair cells (green) and were labeled with Prox1 antibody (red) to reveal support cells. High-power images are taken from boxed regions in adjacent low-magnification images.
Discussion
We investigated the requirement for FGF signaling at early stages of cochlear development. We found that there is a particularly sensitive period in development (E14–E16) when inhibition of FGF signaling has a dramatic effect on the development of the sensory cells. The effects of inhibition of FGF signaling on sensory specification are not attributable to increases in cell death or changes in proliferation, but rather result from a rapid reduction in Pea3 and Erm and an inhibition of Math1 onset. We also show that a specific FGF, FGF20, is expressed at the right time and place to mediate sensory cell specification; in addition, blocking FGF20, with a specific antibody, inhibits hair cell and supports cell development in a manner similar to the FGF receptor inhibitor. Our results thus define the period of FGF-dependent sensory cell specification and the ligand that mediates this step in cochlear development.
Previous experiments by Pirvola et al. (2002) have shown that tissue-specific deletion of Fgfr1 results in severe defects in the development of both hair cells and support cells. Although these experiments defined a role for FGF signaling in sensory cell specification, they were unable to define the precise period when this receptor was required. Our in situ hybridization results indicate that Fgfr1 is expressed in the presumptive sensory domain from E13 until E16, and our experiments with SU5402 define the most sensitive period from E14 to E16. Whereas Pirvola et al. (2002) proposed that FGF10 might be the ligand for FGFR1 in the early phase of cochlear development, more recent studies have shown that mice deficient in Fgf10 have no hair cell defects in cochlear development (Pauley et al., 2003). Another FGF that is also expressed in the sensory epithelium during embryonic development is FGF8. We found that treatment of the cultures with anti-FGF8 had no effect on hair cell development, although we did find an inhibition of pillar cell development that was reported in a previous publication. Instead, our results indicate that FGF20 is the likely ligand for FGFR1 in the sensory cell specification phase of cochlear development for several reasons: (1) Fgf20 is highly expressed for a very brief period, from E13.5 to E16, in the presumptive sensory epithelial domain, which (2) coincides with Fgfr1 expression and the expression of two potential downstream targets of FGF signaling, ERM and Pea3; and (3) inhibition of FGF20 with a specific antibody blocks sensory cell development. FGF20 has been shown to have activity on all c-forms of the FGF receptors, with the preferred receptor FGFR3c (Zhang et al., 2006). Fgfr3 is also expressed in the developing cochlea, close to the time when inhibition of FGF signaling has an effect on sensory cell development; however, we do not think it is involved in this process for three reasons. (1) Fgfr3 is not expressed in the cochlea until E15.5, and then it is only expressed in the base (supplemental Fig. 3, available at www.jneurosci.org as supplemental material), so Fgfr3 could only be important at the end of sensory specification. (2) More importantly, Fgfr3 knock-out mice do not show a reduction in the number of hair cells and support cells (Hayashi et al., 2007); rather, there is an increase in hair cell number in these mutants. This is the opposite result one would expect if the effects we observe with SU5402 or anti-FGF20 treatments were mediated through Fgfr3. (3) Although FGF20 is more active on FGFR3 than FGFR1, there is only a twofold difference, and we found that Fgf20 is highly expressed and thus likely to activate FGFR1c in the developing sensory epithelium.
How does FGF20–FGFR1 regulate sensory cell specification? Pirvola et al. (2002) concluded that the loss of hair cells and associated support cells in the conditional Fgfr1 deletion was attributable to a reduction in proliferation of their progenitors. Whereas at early stages of development it is likely that Fgfr1 mediates cell proliferation, we do not think that a reduction in the proliferation of the hair cell progenitors can explain our results, because SU5402 treatment inhibits hair cell and support cell formation most dramatically at a time in cochlear development, E14 to E16, when nearly all of the precursors of hair cells and support cells have withdrawn from the cell cycle (Ruben, 1967; Lee et al., 2006) (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). It is also unlikely that inhibition of FGF20 or FGF receptors causes the reduction in hair and support cells by increasing cell death in the cultures, because we did not find a significant difference in the number of apoptotic cells between the control and treated cultures. We propose instead that FGF signaling is responsible either directly or indirectly for the up-regulation in Math1 expression that normally occurs during this phase of development in the cochlea. It is possible that the activation of FGFR1 by FGF20 acts via the upregulation of E26 transformation-specific (Ets)-domain transcription factor expression. In the developing Drosophila chordotonal organ, a low level of atonal is expressed in the proneural clusters. The upregulation of atonal (and consequent commitment to the chordotonal precursor fate) is triggered by the activation of the receptor tyrosine kinase EGFR, which in turn upregulates Pnt (the fly Pea3/ERM homolog) expression via the MAPK (mitogen-activated protein kinase) pathway. The combination of low-level atonal and high Pnt form a complex and act together on a neighboring E-box and Ets-binding site in the atonal promoter to drive the high-level atonal necessary for formation of chordotonal precursors (zur Lage et al., 2004). A similar process may occur at the onset of hair cell differentiation in the mammalian cochlea. Consistent with this model, we found that inhibition of FGF signaling in mammalian cochlea causes a rapid reduction in the Pnt homologs Pea3 and ERM and blocks the onset of Math1 expression.
Our model for how FGF20 fits into the current data on cochlear development is shown in Figure 9. One of the earliest factors known to be essential for the development of the sensory epithelium in the cochlea is Sox2 (Kiernan et al., 2005a). This transcription factor is reduced in its expression in the Jagged1 knock-out mouse, which itself is critical for the prosensory phase of development (Morrison et al., 1999; Brooker et al., 2006; Kiernan et al., 2006). Hesr1 and Hesr2 are regulated by Notch at this stage of development and may mediate the Jagged1 signal (Hayashi et al., 2008). Shortly after the expression of Hesr1/2, we found that Fgf20 is expressed by cells within the presumptive sensory epithelial domain, as a subset of the Sox2+-expressing cells. At this stage in cochlear development, our data show that FGFR1 is the only FGF receptor expressed in this domain, and thus it appears likely that FGFR1 is the receptor for FGF20. The signal of FGF20 to FGFR1 activates downstream members of the Ets-domain transcription factor family, Pea3 and Erm, which, we propose, go on to activate Math1 expression, either directly or through a still unidentified factor. Math1 expression in the sensory epithelium leads to hair cell development (Bermingham et al., 1999; Chen et al., 2002), and these cells signal to the surrounding cells via Jagged2 and Dll1 (Lanford et al., 1999; Kiernan et al., 2005b) to direct support cell (Prox1+) fate (Bermingham-McDonogh et al., 2006). Support cells are further specialized through a later FGF signal, FGF8 from the inner hair cells to FGFR3 in the pillar and Deiters' cells (Colvin et al., 1996; Mueller et al., 2002; Shim et al., 2005; Hayashi et al., 2007). Although this model is still speculative, it is clear from our data that there is an FGF20-dependent step in sensory epithelial development just before the onset of Math1; additional investigations will be necessary to determine the specifics of the model.
Figure 9.
Model for the role of FGF signaling in the sensory specification stage of cochlear development.
Footnotes
This work was supported by National Institutes of Health Grants DC005953, P30DC00466, and P30HD002274 and the Hearing Regeneration Initiative. We thank Dr. Jane Johnson for the Math1-GFP mouse strain; Drs. H. Kondoh, P. Soriano, A. McMahon, and S. Mansour for DNA probes; Dr. Thomas Reh for comments on this manuscript; and members of the Reh laboratory for helpful discussions.
References
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, Hayashi T. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496:172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin Exp Pharmacol Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Dev Dyn. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Kokubo H, Hartman BH, Ray CA, Reh TA, Bermingham-McDonogh O. Hesr1 and Hesr2 may act as early effectors of Notch signaling in the developing cochlea. Dev Biol. 2008;316:87–99. doi: 10.1016/j.ydbio.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N. FGFs as multifunctional signaling molecules: diversity of structure and function. Seikagaku. 2001;73:525–535. [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Jacques BE, Montcouquiol ME, Layman EM, Lewandoski M, Kelley MW. Fgf8 induces pillar cell fate and regulates cellular patterning in the mammalian cochlea. Development. 2007;134:3021–3029. doi: 10.1242/dev.02874. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005a;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Cordes R, Kopan R, Gossler A, Gridley T. The Notch ligands DLL1 and JAG2 act synergistically to regulate hair cell development in the mammalian inner ear. Development. 2005b;132:4353–4362. doi: 10.1242/dev.02002. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladher RK, Wright TJ, Moon AM, Mansour SL, Schoenwolf GC. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford PJ, Lan Y, Jiang R, Lindsell C, Weinmaster G, Gridley T, Kelley MW. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- Lee YS, Liu F, Segil N. A morphogenetic wave of p27Kip1 transcription directs cell cycle exit during organ of Corti development. Development. 2006;133:2817–2826. doi: 10.1242/dev.02453. [DOI] [PubMed] [Google Scholar]
- Lim DJ, Anniko M. Developmental morphology of the mouse inner ear. A scanning electron microscopic observation. Acta Otolaryngol Suppl. 1985;422:1–69. [PubMed] [Google Scholar]
- Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Morrison A, Hodgetts C, Gossler A, Hrabe de Angelis M, Lewis J. Expression of Delta1 and Serrate1 (Jagged1) in the mouse inner ear. Mech Dev. 1999;84:169–172. doi: 10.1016/s0925-4773(99)00066-0. [DOI] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–3335. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KL, Jacques BE, Kelley MW. Fibroblast growth factor signaling regulates pillar cell development in the organ of Corti. J Neurosci. 2002;22:9368–9377. doi: 10.1523/JNEUROSCI.22-21-09368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:REVIEWS3005. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert J, McConnell S, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol Suppl. 1967;220:1–44. [PubMed] [Google Scholar]
- Sher AE. The embryonic and postnatal development of the inner ear of the mouse. Acta Otolaryngol. 1971;285(Suppl):1–77. [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Hatch EP, Karabagli H, Karabagli P, Schoenwolf GC, Mansour SL. Expression of mouse fibroblast growth factor and fibroblast growth factor receptor genes during early inner ear development. Dev Dyn. 2003;228:267–272. doi: 10.1002/dvdy.10362. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage PI, Powell LM, Prentice DR, McLaughlin P, Jarman AP. EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev Cell. 2004;7:687–696. doi: 10.1016/j.devcel.2004.09.015. [DOI] [PubMed] [Google Scholar]