Abstract
DNA polymerase beta plays a key role in base excision repair. We have previously shown that the hydrophobic hinge region of polymerase beta, which is distant from its active site, plays a critical role in the fidelity of DNA synthesis by this enzyme. The I260Q hinge variant of polymerase beta misincorporates nucleotides with a significantly higher catalytic efficiency than the wild-type enzyme. In the study described here we show that I260Q extends mispaired primer termini. The kinetic basis for extension of mispairs is defective discrimination by I260Q at the level of ground state binding of the dNTP substrate. Our results suggest that the hydrophobic hinge region influences the geometry of the dNTP binding pocket exclusively. Because the DNA forms part of the binding pocket, our data are also consistent with the interpretation that the mispaired primer terminus affects the geometry of the dNTP binding pocket such that the I260Q variant has higher affinity for the incoming dNTP than wild-type polymerase beta.
DNA Polymerase beta (pol β), a member of the X-family of DNA polymerases, is a key enzyme in base excision repair (BER) (1). The well-characterized BER pathway removes DNA damage that is induced by reactive oxygen species (ROS) and alkylating agents (2, 3). BER is initiated by DNA glycosylase and the type of glycosylase determines the pathway of BER (4). Monofunctional DNA glycosylases such as Alkyladenine DNA glycosylase (AAG), remove base damage and leave an abasic (AP) site. APE1 cuts the DNA 5′ to the AP site, leaving a 3′OH and a 5′dRP. Pol β removes the 5′dRP and fills in the gap. Bifunctional DNA glycosylases such as 8-OxoG DNA glycosylase 1 (OGG1) remove base damage and leave a modified 3′end, which is modified by Apurinic/apyrimidinic endonuclease 1 (APE1) (5) or by polynucleotide kinase (6). In this pathway, pol β fills in a one to six base gap. During long patch BER, pol β is likely to initiate large gap filling by performing strand displacement synthesis (7). Finally, DNA ligase IIIα-XRCC1 seals the nick (8). Approximately 20,000 lesions/cell/day are channeled through the BER pathway (9) highlighting the importance of this pathway for genomic integrity.
Pol β synthesizes DNA with error frequencies ranging from < 10−3 to 10−5, suggesting it is a lower fidelity polymerase than replicative DNA polymerases. However, nucleotide misincorporation by pol β is 10–100-fold lower on a 5′-phosphorylated 1 nt gapped DNA substrate, suggesting that this is likely to be the physiological substrate of pol β (10).
We use pol β as a model polymerase to gain insight into mechanisms of polymerase fidelity, due to its small size and lack of proofreading activity. In addition, the structure of DNA polymerase beta is well conserved from parasitic protozoans to humans (11, 12). There are several X-ray and NMR structures available for pol β (13–15) that facilitate interpretation of biochemical studies within the context of the polymerase’s structure. Pol β has a 31 kD polymerase domain which is comprised of three subdomains that consist of the palm, the thumb and the fingers. This architecture represents the hand-like motif of polymerases. In addition, pol β also has an N-terminal 8kD domain that houses its dRP lyase activity (16).
We have shown that the hydrophobic hinge region is important for catalysis and fidelity (17–21). This hinge is comprised of the F272, 1174, L194, T196, I260, and Y265 residues and where the motion for rotation of the fingers from an open to closed form originates. It is not part of the active site of pol β. In a previous study we demonstrated that the I260Q variant of the hydrophobic hinge is a mutator polymerase (22). We have shown using presteady state kinetics that it is a global misincorporator due to its inability to discriminate dNTP substrates during the binding step of the polymerase catalytic cycle. Misincorporation would be mutagenic upon extension of the mispaired primer terminus. In our current study we have explored the ability of I260Q to extend mispairs and showed that this variant possesses robust mispair extension activity.
Experimental Procedures
Materials
Ultrapure deoxynucleoside triphosphates, ATP, and [-γ32P] ATP (>6000 Ci/mmol, 150 mCi/mL) were purchased from New England Biolabs, Sigma, and Amersham Biosciences, respectively. T4 polynucleotide kinase (M0201S)) was purchased from New England Biolabs.
Cloning, Expression and Purification of WT and I260Q variant of pol β
The I260Q variant was generated by the Stratagene Quick-Change Site-Directed Mutagenesis kit according to the protocol of the manufacturer using pET28a-WT as a template, followed by DNA sequencing at the WM Keck Facility at Yale University School of Medicine as described before (22). N-terminal hexahistidine-tagged WT and I260Q variant were purified by two- step column chromatography (Ni-NTA affinity HP column and SP HP column from GE Healthcare) using fast protein liquid chromatography as described previously (22). Concentrations of purified pol β were determined using ε280= 21,200 M−1 cm−1 and a molecular mass of 40kDa for his-tagged protein.
Preparation of DNA substrates
Oligonucleotides were synthesized by WM Keck Facility at Yale University. The substrates used are shown in Table 1. The primer oligonucleotide was labeled at the 5′-end using T4 polynucleotide kinase and γ-32P ATP. Other Oligonucleotides were 5′-end-phosphorylated with the kinase and cold ATP. After purification on a Biorad spin column to remove unincorporated dNTPs, annealing was performed by mixing phosphorylated template, radiolabeled primer and phosphorylated downstream oligos in 50 mM Tris-HCI, pH 8.0, containing 0.25 M NaCl. The mixture was incubated sequentially at 95 °C (5 min), slowly cooled to 50 °C (for 30 min) and 50 °C (for 20 min), and immediately transferred to ice. To verify proper hybridization, the product was analyzed on an 18% native polyacrylamide gel followed by autoradiography to assess the quality of annealing.
Table 1.
DNA substrates used for Kinetic assays.
| Substrate | Sequence |
|---|---|
| CII | 5′ TTGCGACTTATCAACGCCCACA AGCTGTCTTCTCAGTTTC 3′ |
| 3 ′ AACGCTGAATAGTTGCGGGTGTAGTCATCGACAGAAGAGTCAAAG 5′ | |
| CII5T | 5′ TTGCGACTTATCAACGCCCACAX GCTGTCTTCTCAGTTTC 3′ |
| 3′ AACGCTGAATAGTTGCGGGTGTAGTCATCGACAGAAGAGTCAAAG 5′ | |
| CII5C | 5′ TTGCGACTTATCAACGCCCACAX GCTGTCTTCTCAGTTTC 3′ |
| 3′ AACGCTGAATAGTTGCGGGTGTAGCCACCGACAGAAGAGTCAAAG 5′ | |
| 45AG5 | 5′ GCCTCGCAGCCGTCCAACCAAY CTCGATCCAATGCCGTCC 3′ |
| 3′ CGGAGCGTCGGCAGGTTGGTTGAGTTGGAGCTAGGTTACGGCAGG 5′ |
X=T for correct paired termini and A for mispaired termini; Y= C for correct pair and G for mispaired end. The templating base is underlined.
Qualitative missing base primer extension assay
Purified WT and I260Q (750 nM) were incubated with radiolabeled DNA substrate (50 nM). Two types of primer extension assays were performed. The ”Missing Base Primer Extension” used three of the four dNTPs, while “One at a Time Primer Extension” used only one of the four dNTPs. The missing substrates used in each reaction are indicated in the respective Figure. The reactions were incubated for 20 min at 37°C and terminated by adding 0.3M EDTA. The products resulting from incorporation of nucleotides into the primer were resolved on a 20% denaturing polyacrylamide gel and visualized on a Phosphorimager using Imagequant software analysis program.
Single turnover mispair extension assay
The rate of next correct nucleotide incorporation of either correctly paired or mispaired termini of a five base pair gapped DNA substrate was measured under single turnover conditions. The single turnover conditions were determined empirically by titration of increasing concentrations of DNA substrate with enzyme and a ratio of 15:1 (750nM enzyme/50nM DNA) was chosen for each reaction. Kinetics of extension from a correctly paired primer-terminus were determined using the Rapid Quench-Flow apparatus with concentrations of dNTP ranging from 1–500uM depending on the substrate employed. Mispair extension kinetics were carried out manually with dNTP concentrations ranging from 1–500uM.
Data were analyzed by Kaleidagraph software (synergy software) using the appropriate equations. To determine kpol, the rate of maximum polymerization and Kd, the dissociation constant for dNTP binding, the single turnover kinetic data were fit to the single exponential equation: , where A is the amplitude; t is the time; and kobs is the observed rate constant. Observed rate constants were then plotted against [dNTP], and the data were fit to the hyperbolic equation: , where kpol is the maximum rate of polymerization, and Kd is the equilibrium dissociation constant for dNTP. Relative extension frequency values were calculated using the relationship: fidelity = ((kpol/Kd)cp + (kpol/Kd)mp)/((kpol/Kd)mp), where cp and mp represent the correct and mispaired termini, respectively.
DNA binding assay
Various concentrations of WT and I260Q pol β protein (0.1–1000 nM) were incubated with 0.1 nM radiolabeled gapped DNA substrate (CII5T; Table 1) in buffer containing 50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 10% glycerol, and 0.1% Nonidet P-40 at room temperature (23°C) for 15 min. Samples were loaded onto a 6% native polyacrylamide gel with the current running at 300 V at 4°C. After the sample was loaded, the voltage was reduced to 150 V. Bound protein was quantified using Imagequant software, after scanning the gel using a Molecular Dynamics Phosphorimager. Protein bound to DNA resulted in a shift of the DNA on the gel when compared to DNA without bound protein. Fraction bound is the ratio of the intensity of all shifted species divided by the total. The dissociation constant for DNA (KD) was estimated from fitting the bound protein (Y) versus protein concentration (x) with the equation: , where m is a scaling factor and b is the apparent minimum Y value.
Results
In our previous study we have shown that the I260Q variant is a misincorporation mutator (22). The mechanistic basis of misincorporation is due to loss of discrimination during dNTP ground state binding. Extension of a mispair that was produced during misincorporation could lead to fixation of a mutation. Therefore, in this study we characterized the ability of the I260Q variant to extend mispaired primer-termini.
I260Q misincorporates and extends mispairs in the missing base primer extension assay and in the one at a time primer extension assay
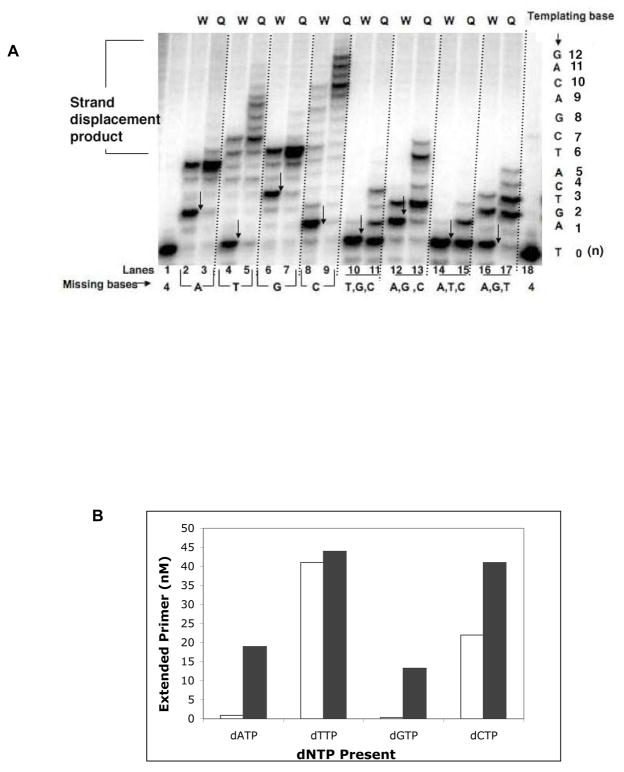
As a first screen, we employed a missing base primer extension assay and used a 5-bp gapped substrate, CII (Table 1), as described previously, with slight modifications (23). This assay was used to assess the ability of I260Q to misincorporate and extend mispairs in a qualitative manner. When the correct base is not present in the reaction mixture, a low fidelity polymerase could both misincorporate nucleotides and extend the resulting mispair. Figure 1A shows that the I260Q variant extended the primer (“Q” lanes) to a greater degree or farther than the WT enzyme (“W” lanes) in primer extension assays, in which one (lanes 2–9) or three dNTPs (lanes 10–17) were missing from the reaction. For example, when only dATP is missing, WT and I260Q both misincorporate and extend a mispair to a certain extent. For I260Q, we observe a greater amount of mispair extension (lane 3) compared to WT (lane 2). When only dTTP is missing most of the primers were not extended beyond template A by WT pol β (lane 4) but I260Q misincorporates and extends the mispair to a greater extent (lane 5). When only dCTP is missing (lanes 8– 9), I260Q extends the primer past template G (lane 9), whereas very little extension occurs with the WT polymerase (lane 8).
Figure 1. A. I260Q misincorporates and extends mispairs in the missing base primer extension assay.
On the left side of the gel image are results from experiments in which one base is deleted from the reaction mixture. On the right side of the gel image are results from “one at a time primer extension” experiments in which only one base is present in the reaction mixture. These primer extension assays were carried out with 50 nM CII, a 5bp-gapped substrate and 750nM enzyme. “W” lanes are for WT and “Q” lanes are for I260Q variant. “Missing nucleotide(s)” represents nucleotide(s) that were excluded from the reaction. The arrows point to the expected pause sites in the absence of the complementary base. B. Quantification of “one at a time primer extension” assay. Empty bars represent the amount of primer extended by WT and filled bars represent the amount of primer extended by I260Q.
The “one at a time” primer extension assay confirms similar misincorporation and mispair extension characteristics of I260Q. When only dATP Is present (lanes 10 and 11), I260Q misincorporates opposite template A and a significant fraction of these mispaired primer-termini (A:A) appear to be extended (lane 11). When dTTP is the sole source of dNTP in the mixture (lanes 12 and 13), WT and I260Q both correctly incorporate dTTP opposite template A, then misincorporate dTTP opposite G (lane 12 and 13). Whereas WT cannot extend mispaired G:T (lane 12), I260Q extends the mispaired end (G:T) (lane 13). When either dGTP or dCTP is present in the mixture, I260Q shows more misincorporation followed by mispair extension (lanes 15 and 17) compared to WT (lanes 14 and 16). We have quantified the data obtained from the “one at a time primer extension” assay (Figure 1B). In general, I260Q appears to be able to misincorporate nucleotides and extend the resulting mispair much more extensively than WT. In the reaction where only dATP is present, I260Q has a much greater propensity to misincorporate dATP opposite template A and then continue DNA synthesis.
I260Q and WT pol β have similar affinities for their DNA substrates
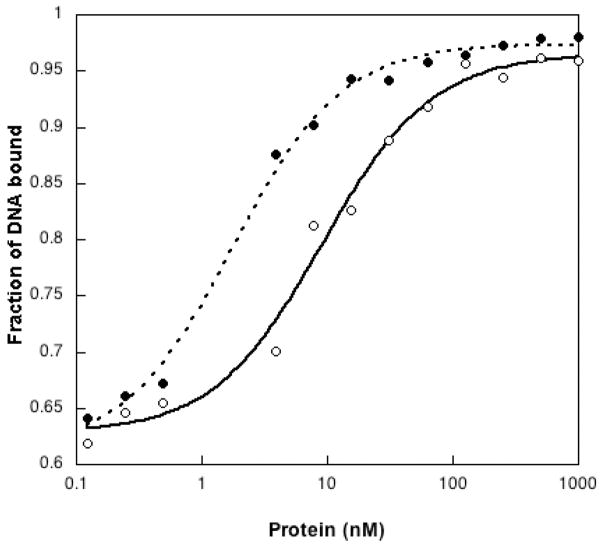
We characterized the binding affinities of I260Q and WT pol β for the 5bp correctly paired (A:T) gapped substrate that we employed in our missing base primer extension assay and initial kinetics experiments (Table 2), using a gel mobility shift assay. As shown in Figure 2, WT and I260Q both have similar sigmoidal profile of DNA binding. The KD DNA for WT is 9.3± 1.5 nM and for I260Q is 1.8±0.2 nM. This suggests that I260Q has a slightly higher affinity for this DNA substrate than WT pol β.
Table 2.
I260Q extends an A:A mispair.
| Terminal base pair | Enzyme | kpol (sec−1) | Kd(μM)a | Kpol(cp)/kpol(mp)b | Kd(mp)/Kd(cp)c | kpol/Kd M−1 sec−1 | Relative extension frequency ×103 |
|---|---|---|---|---|---|---|---|
| A:dTTP | WT | 27.4±3.2 | 157±35 | 721.0 | 0.08 | 17.4×104 | 64 |
| I260Q | 10.8±1.5 | 165±40 | 450.0 | 0.0036 | 6.5 ×104 | 2.6 | |
| A:dATP | WT | 0.038±0.003 | 13±3 | 2. 75×103 | |||
| I260Q | 0.024±0.0008 | 0.6±0.1 | 4.0×104 |
The correct pair is bold.
The kpol for correct pair(cp) is divided by kpol for mispair (mp) end substrate.
The Kd for correct pair(cp) is divided by Kd for mispair (mp) end substrate. Fidelity is calculated as described in Materials and Methods. The DNA substrate is CII5T.
Figure 2. Plots of the fraction of DNA (CII5T) bound vs the concentration of enzyme. WT (●), I260Q (○).
The Kaleidagraph graphics program was used to plot the data, and the data were fit as described in Experimental Procedures to obtain the KD. KD for WT is 9.3 ±1.5 nM and for I260Q=1.8±0.2nM.
We also determined if I260Q and WT pol β differed in their affinity for a mispaired DNA substrate (CII5T, A:A) using a gel mobility shift assay. We found that the apparent KD for I260Q is 2.0±0.2 nM and for WT it is 6.0±1.3 nM. Thus, the affinity of I260Q for the DNA substrate is not much different than that of WT pol β and cannot explain its ability to extend mispaired primer-termini.
Mispair extension is due to higher binding affinity for dNTP
In our missing base primer extension assay, I260Q exhibits remarkable mispair extension when the A:A mispair is formed by misincorporation. To obtain mechanistic insight into the A:A mispair extension ability of I260Q we employed a 5-bp gapped substrate (CII5T). Presteady state kinetic assays were carried out to compare the efficiency of extension of a correctly paired primer-terminus (A:T) versus a mispaired primer-terminus (A:A). An example of single turnover kinetics of I260Q with the CII5T substrate is shown in Figure 3. In Table 2, we have summarized our results. WT and I260Q exhibit a similar binding affinity (Kd) for correct dNTP when a correctly paired primer-terminus was present (157uM and 165uM respectively). In contrast, with a mispaired primer-terminus, I260Q has a much higher affinity (0.6 uM) for the next correct dNTP compared to WT (13.8 uM). This results in a catalytic efficiency (kpol/Kd) for insertion of the next correct base that is 15-fold higher for I260Q than wild-type pol β.
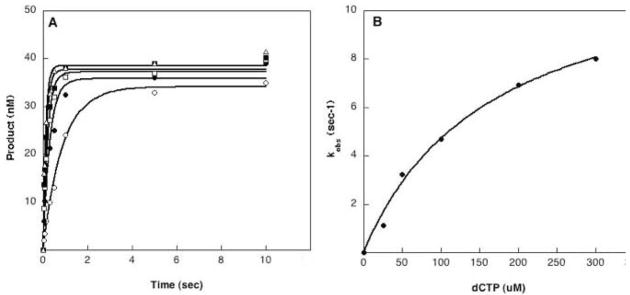
Figure 3. Single turnover experiments of correctly paired CII5T substrate using rapid quench apparatus.
A. Incorporation of dCTP opposite template G (substrate CII5T) for I260Q at 37°C. A preincubated solution containing 750nM enzyme and 50nM substrate was mixed with 10mM MgCl2 and 25(○),50(●),100(□),.200 (△) and 300 (■) μM dCTP. The reactions were quenched by EDTA at different time points, and the products were resolved by denaturing sequencing gel electrophoresis. The data were fit to the single exponential equation to obtain the kobs. B. The secondary kinetic plot of kobs versus the dCTP concentration for I260Q (●). The solid line represents the best fit of the data to the hyperbolic equation. The kpol is 10.8 sec−1 and Kd is 165.8 uM.
Mispair extension is not due to misalignment of primer-template
In our mispair extension assay, the template sequence of the gap was –GTCAT-(Table 1). Thus, one possible mechanism of mispair extension would be that the A at the 3′end of the primer would pair with the T adjacent to the G of the gap, creating a paired primer-terminus via transient misalignment. In this case, we would not be observing true mispair extension, but rather misincorporation on a misaligned template. This could be a viable possibility given the fact that I260Q is prone to misincorporation of nucleotides (22). To test this possibility, we redesigned the substrate CII5T to CII5C, where the gap sequence is changed to –GCCAC-, so that mispaired primer has little chance to become misaligned in the presence of dTTP. We repeated the mispair extension kinetics experiments on this substrate and the results are tabulated in Table 3. The rate of extension (kpol) was slow for the mispaired primer-terminus compared to the correctly paired terminus similarly for WT and I260Q. As obvious from the catalytic efficiency values (kpol/Kd) shown in Table 3,1260Q has greater ability than WT to incorporate the next correct base, dCTP, with the mispaired primer-terminus. The mechanistic basis for this is that I260Q has a much higher affinity than WT for dNTP substrate (Kd= 0.98uM) in the presence of a mispaired primer-terminus. The binding affinity of WT for the dNTP substrate was similar for both correct or mispaired primer-terminus. The net result is a 15-fold higher catalytic efficiency for mispair extension by I260Q versus WT pol β. Thus, I260Q appears to be proficient at mispair extension.
Table 3.
Mispair extension by I260Q is not due to misalignment of template.
| Terminal base pair | Enzyme | kpol(sec−1) | Kd(μM)a | Kpol(cp)/kpol(mp)b | Kd(mp)/Kd(cp)c | kpol/Kd M−1 sec−1 | Relative extension frequency ×103 |
|---|---|---|---|---|---|---|---|
| A:dTTP | WT | 3.9±0.11 | 21.8±3.2 | 108 | 1.0 | 17.8×104 | 112 |
| I260Q | 6.6±0.26 | 70.6±10.2 | 275 | 0.013 | 9.3×104 | 4.8 | |
| A:dATP | WT | 0.036±0.001 | 21.3±1.8 | 1.6×103 | |||
| I260Q | 0.024±0.0005 | 0.98±0.21 | 2.4×104 |
The correct pair is bold.
The kpol for correct pair(cp) is divided by kpol for mispair (mp) end substrate.
The Kd for correct pair(cp) is divided by Kd for mispair (mp) end substrate. Fidelity is calculated as described in Materials and Methods. The DNA substrate is CIISC.
I260Q can extend a G:G mispair
Joyce et al. previously reported for Klenow fragment that pur:pur mismatches are less efficiently extended and that extension is likely to be dependent upon local sequence context (24). To test the possibility that mispair extension by I260Q is sequence context dependent, we performed the mispair extension kinetics experiments on a DNA substrate with a different sequence context than CII5T and CII5C. Since we have already shown that I260Q extends an A:A mispair, we characterized mispair extension for a G:G mispair (45AG5). In Table 4 we show that I260Q is much more proficient than WT pol β in extending a G:G mispair. Again, I260Q is less able than WT pol β to discriminate during ground state binding in the presence of a mispaired end. The catalytic efficiency (kpol/Kd) of mispair extension is almost 8-fold higher for I260Q with a G:G mispair at the primer-terminus of the DNA substrate. These results suggest that the abilities of I260Q to extend mispairs relative to wild-type is not sequence context dependent.
Table 4.
Mispair extension of I260Q is not sequence context dependent.
| Terminal base pair | Enzyme | kpol(sec−1) | Kd(μM)a | Kpol(cp)/kpol(mp)b | Kd(mp)/Kd(cp)c | kpol/Kd M−1 sec−1 | Relative extension frequency ×103 |
|---|---|---|---|---|---|---|---|
| G:dCTP | WT | 9.6±1.2 | 130±10 | 505.0 | 2.4 | 7.38 ×104 | 1251 |
| I260Q | 8.4±1.3 | 145±15 | 420.0 | 0.3 | 5.7 ×104 | 129 | |
| G:dGTP | WT | 0.019±0.0009 | 320±10 | 5.9×101 | |||
| I260Q | 0.020±0.0002 | 45±2.2 | 4.44×102 |
The correct pair is bold.
The kpol for correct pair(cp) is divided by kpol for mispair (mp) end substrate.
The Kd for correct pair(cp) is divided by Kd for mispair (mp) end substrate. Fidelity is calculated as described in Materials and Methods. Substrate is 45AG5.
I260Q extends a mispaired primer-terminus with incorrect dNTP
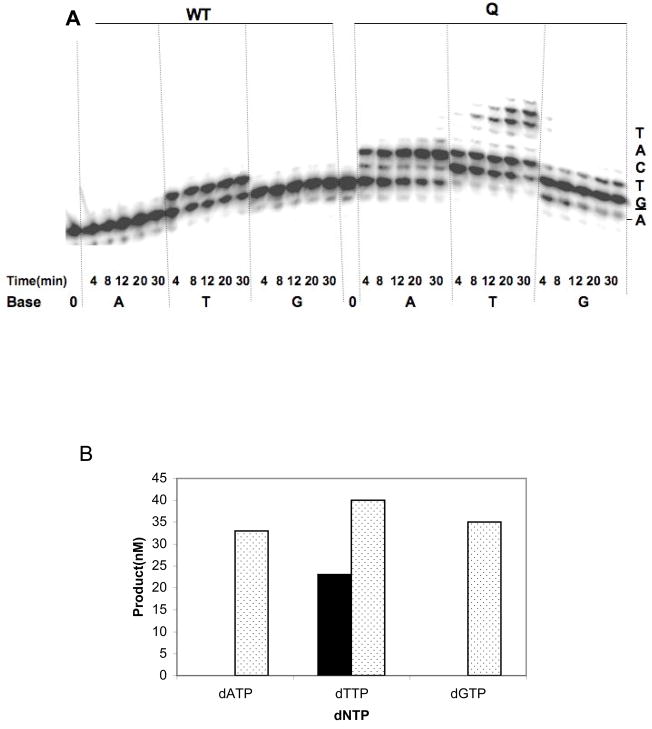
We asked whether I260Q could extend a mispaired primer-terminus in the presence of the incorrect nucleoside triphosphate. We employed the CII5T DNA substrate, in which A is mispaired with A and added one dNTP per reaction for various amounts of time. As shown in Figure 4A, WT pol β can extend this mispair in the presence of dTTP only. In contrast, the I260Q variant is able to extend the A:A mispair in the presence of dATP, dTTP, and dGTP (Figure 4A and 4B).
Figure 4. I260Q extends mispaired primer-termini with incorrect bases.
A missing base primer extension assay was carried out as described in Materials and Methods. The substrate employed was CII5T primer-template that contains a mispaired (A:A) at the 3′end of gthe 5 nucleotide gap. 50nM of mispaired substrate was incubated with either 750nM WT or I260Q protein. Reactions were conducted in presence of only one type of 100uM dNTP (dATP, dTTP or dGTP) at 37°C for different time intervals (4–30 min). A. A phosphorimager representation of sequencing gel of mispair extension by incorrect dNTP. The dNTP used was indicated at the bottom of the gel. B. A quantitative representation of mispair extension by WT and I260Q. The amount of mispair extension by each dNTP at 30 minutes was plotted against the respective dNTP. The filled bar represents WT and dotted bar is for I260Q.
Discussion
The goal of this study was to determine if the I260Q pol β variant was able to extend mispaired primer-termini. We found that I260Q can extend mispairs in the presence of the next correct and even the next incorrect dNTP substrate. In our first mispair extension study (Table 2), the I260Q variant exhibits a catalytic efficiency (kpol/Kd) that is only about two-fold lower than that of WT pol β when extending a correctly paired primer-terminus. Remarkably, the catalytic efficiency (kpol/Kd) of I260Q for extension of a mispaired primer-terminus is only two-fold lower than that for extension of a correctly paired primer-terminus whereas for WT it is 63-fold lower (Table 2). The kinetic basis for the extension of a mispaired primer-terminus by I260Q appears to be during the dNTP binding step. Surprisingly, I260Q exhibits a 276-fold increased dNTP binding affinity when extending from a (A:A) mispaired versus a correctly paired primer-terminus. We conclude that the dNTP binding pocket of I260Q has a geometric shape that is different from that of WT pol β, and that this altered shape likely facilitates mispair extension by this variant.
The Rate Of Mispair Extension Is Slow for I260Q
The maximum rate of polymerization in the presence of a mispaired primer-terminus is much slower for both WT and I260Q pol β than in the presence of a correctly paired DNA substrate. The simplest interpretation is that the 3′-OH of the primer strand in both the WT and the variant is greater than 3 Angstroms away from the alpha-phosphate of the incoming dNTP most of the time during the polymerase catalytic cycle. This idea is supported by the structures of pol β in the presence of mispairs (25). Krahn and colleagues showed that within the WT pol β active site, the 3′-OH of a mispaired primer-terminus is too far from the alpha-phosphate for an in line attack, likely due to the staggered nature of the terminal mispair. Importantly, because the reaction rate is slowed to a similar extent in both WT and I260Q, it is not likely that the alteration of the hydrophobic hinge affects the overall rate of DNA synthesis.
I260Q Has a Higher Affinity For The Incoming dNTP in the presence of a mispair
It is interesting that WT pol β exhibits increased or equal affinity for the incoming dNTP substrate in the presence of five nucleotide gapped DNA template-primer with a terminal mispair compared to a DNA substrate with a correctly paired primer terminus. This implies the presence of a terminal mispair influences the geometry of the dNTP binding pocket, as shown in Figure 5. We previously showed that I260Q misincorporates nucleotides due to its lack of discrimination at the level of dNTP binding. This variant has the same affinity for the correct and incorrect dNTP substrate during incorporation. Thus, in the presence of a correctly paired primer-terminus, alteration of residue 260 to Q results in a dNTP binding pocket with a geometry that is different than that of WT pol β, suggesting that the hinge influences dNTP binding pocket geometry. In the presence of a terminal mispair and within the same sequence context used in our previous studies, I260Q exhibits no discrimination at the level of dNTP binding. In fact, it has a much higher affinity for the incoming dNTP in the presence of a terminal mispair compared to WT pol β.
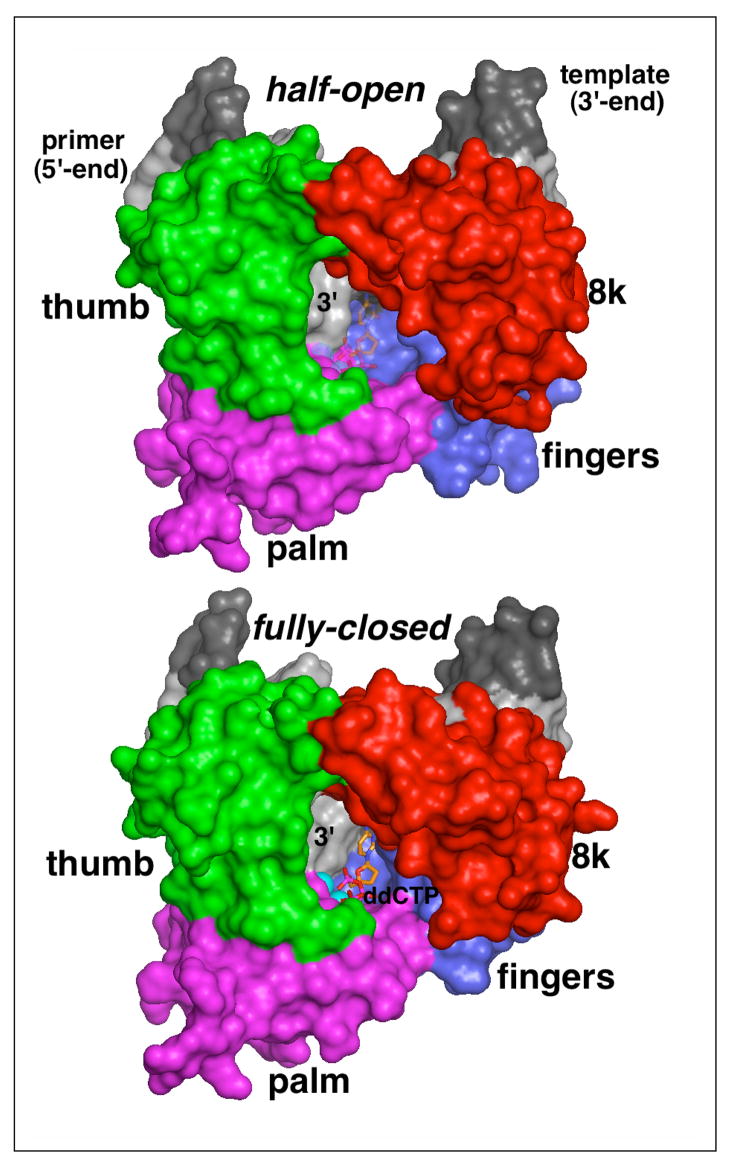
Figure 5. Surface representations of the half-open (top) and fully closed (bottom) form of wild-type polymerase β complexed with gapped DNA.
The surface of polymerase β is colored according to domains (red, 8K; green, thumb; magenta, palm; blue, fingers) and the gapped duplex is shown in shades of gray (dark, template; light, primer). The ddCTP in the half-open form (top), shown as transparent stick model, is modeled into the nucleotide binding pocket by superimposing three nucleotides at the 3′-end of the primer and critical active site residues from the closed conformation onto the respective residues in the half-open form. Note, that the left part of the nucleotide binding pocket is formed by the penultimate nucleotide in the primer strand. In the full-closed form, the fingers domain and the DNA primer have a tight grip around the incoming nucleotide while in the half-open form the geometry and the packing in the dNTP pocket is changed. The apo-polymerase structure of I260Q and computer modeling suggest that the hinge region affects the shape and size of the dNTP pocket, thereby likely leading to changes in the ground-state binding of a mismatched, incoming nucleotide. The turn-over rate of I260Q polymerase β is not affected as the alpha phosphate is still sufficiently close to the 3′-O′ on the primer.
Our results suggest that both the DNA and the hydrophobic hinge region influence the geometry of the dNTP binding pocket. Because alteration of residue 260 to Q only affects ground state binding, we suggest that the hinge region functions in ensuring proper dNTP binding pocket geometry. Molecular modeling studies suggest that the cavity of the hinge region becomes smaller in the presence of Q at position 260. This smaller cavity could restrict domain movements and affect the geometry of the dNTP binding pocket, likely by altering the snug fit that usually occurs upon interaction with the correct dNTP. The pocket geometry is unlikely to be affected directly by residues that are so far away from the binding pocket. Rather, we suggest that the shape of the binding pocket is a function of the conformational changes that originate with the hinge and precede chemistry.
Abbreviations and textual footnotes
- pol β
DNA polymerase beta
- BER
base excision repair
- ROS
reactive oxygen species
- AAG
alkyladenine DNA glycosylase
- AP
apurinic or apyrimidinic site
- OGG1
8-oxoguanine DNA glycosylase
- APE1
apurinic or aprimidinic exonuclease 1
- Kd
equilbrium dissociation constant for dNTP
- kpol
maximum rate of polymerization
- KD
equilibrium dissociation for DNA
Footnotes
Acknowledgement of financial aid: This work was funded by CA080830 from the National Cancer Institute.
References
- 1.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 2.Cabelof DC, Raffoul JJ, Yanamadala S, Guo Z, Heydari AR. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002;23:1419–1425. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- 3.Sobol RW, Wilson SH. Mammalian DNA beta-polymerase in base excision repair of alkylation damage. Prog Nucleic Acid Res Mol Biol. 2001;68:57–74. doi: 10.1016/s0079-6603(01)68090-5. [DOI] [PubMed] [Google Scholar]
- 4.Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem. 1999;274:15230–15236. doi: 10.1074/jbc.274.21.15230. [DOI] [PubMed] [Google Scholar]
- 5.Xu YJ, Kim EY, Demple B. Excision of C-4′-oxidized deoxyribose lesions from double-stranded DNA by human apurinic/apyrimidinic endonuclease (Ape1 protein) and DNA polymerase beta. J Biol Chem. 1998;273:28837–28844. doi: 10.1074/jbc.273.44.28837. [DOI] [PubMed] [Google Scholar]
- 6.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Podlutsky AJ, Dianova II, Wilson SH, Bohr VA, Dianov GL. DNA synthesis and dRPase activities of polymerase beta are both essential for single-nucleotide patch base excision repair in mammalian cell extracts. Biochemistry. 2001;40:809–813. doi: 10.1021/bi002064s. [DOI] [PubMed] [Google Scholar]
- 8.Tomkinson AE, Chen L, Dong Z, Leppard JB, Levin DS, Mackey ZB, Motycka TA. Completion of base excision repair by mammalian DNA ligases. Prog Nucleic Acid Res Mol Biol. 2001;68:151–164. doi: 10.1016/s0079-6603(01)68097-8. [DOI] [PubMed] [Google Scholar]
- 9.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 10.Chagovetz AM, Sweasy JB, Preston BD. Increased activity and fidelity of DNA polymerase beta on single-nucleotide gapped DNA. J Biol Chem. 1997;272:27501–27504. doi: 10.1074/jbc.272.44.27501. [DOI] [PubMed] [Google Scholar]
- 11.Chang LM, Plevani P, Bollum FJ. Evolutionary conservation of DNA polymerase beta structure. Proc Natl Acad Sci U S A. 1982;79:758–761. doi: 10.1073/pnas.79.3.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recupero AJ, Rein DC, Meyer RR. Structure-function analysis of DNA polymerase-beta using monoclonal antibodies: identification of a putative nucleotide binding domain. Biochemistry. 1992;31:7989–7997. doi: 10.1021/bi00149a033. [DOI] [PubMed] [Google Scholar]
- 13.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 14.Pelletier H, Sawaya MR, Woffle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 15.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 16.Maciejewski MW, Liu D, Prasad R, Wilson SH, Mullen GP. Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity. J Mol Biol. 2000;296:229–253. doi: 10.1006/jmbi.1999.3455. [DOI] [PubMed] [Google Scholar]
- 17.Clairmont CA, Narayanan L, Sun KW, Glazer PM, Sweasy JB. The Tyr-265-to-Cys mutator mutant of DNA polymerase beta induces a mutator phenotype in mouse LN12 cells. Proc Natl Acad Sci U S A. 1999;96:9580–9585. doi: 10.1073/pnas.96.17.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li SX, Vaccaro JA, Sweasy JB. Involvement of phenylalanine 272 of DNA polymerase beta in discriminating between correct and incorrect deoxynucleoside triphosphates. Biochemistry. 1999;38:4800–4808. doi: 10.1021/bi9827058. [DOI] [PubMed] [Google Scholar]
- 19.Opresko PL, Shiman R, Eckert KA. Hydrophobic interactions in the hinge domain of DNA polymerase beta are important but not sufficient for maintaining fidelity of DNA synthesis. Biochemistry. 2000;39:11399–11407. doi: 10.1021/bi000698t. [DOI] [PubMed] [Google Scholar]
- 20.Opresko PL, Sweasy JB, Eckert KA. The mutator form of polymerase beta with amino acid substitution at tyrosine 265 in the hinge region displays an increase in both base substitution and frame shift errors. Biochemistry. 1998;37:2111–2119. doi: 10.1021/bi9722711. [DOI] [PubMed] [Google Scholar]
- 21.Shah AM, Maitra M, Sweasy JB. Variants of DNA polymerase Beta extend mispaired DNA due to increased affinity for nucleotide substrate. Biochemistry. 2003;42:10709–10717. doi: 10.1021/bi034885d. [DOI] [PubMed] [Google Scholar]
- 22.Starcevic D, Dalal S, Jaeger J, Sweasy JB. The hydrophobic hinge region of rat DNA polymerase beta is critical for substrate binding pocket geometry. J Biol Chem. 2005;280:28388–28393. doi: 10.1074/jbc.M502178200. [DOI] [PubMed] [Google Scholar]
- 23.Starcevic D, Dalal S, Sweasy J. Hinge residue Ile260 of DNA polymerase beta is important for enzyme activity and fidelity. Biochemistry. 2005;44:3775–3784. doi: 10.1021/bi047956x. [DOI] [PubMed] [Google Scholar]
- 24.Joyce CM, Sun XC, Grindley ND. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment) J Biol Chem. 1992;267:24485–24500. [PubMed] [Google Scholar]
- 25.Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12:1823–1832. doi: 10.1016/j.str.2004.08.001. [DOI] [PubMed] [Google Scholar]