Abstract
In all animals, the initial events of embryogenesis are controlled by maternal gene products that are deposited into the developing oocyte. At some point after fertilization, control of embryogenesis is transferred to the zygotic genome in a process called the maternal to zygotic transition (MZT). During this time many maternal RNAs are degraded and transcription of zygotic RNAs ensues1. A longstanding question has been, what factors regulate these events? The recent findings that microRNAs2,3 and Smaugs4 mediate maternal transcript degradation have shed new light on this aspect of the problem. However, the transcription factor(s) that activate the zygotic genome remain elusive. The discovery that many of the early transcribed genes in Drosophila share a cis-regulatory heptamer motif, CAGGTAG and related sequences5,6, collectively referred to as TAGteam sites5 brought up the possibility that a dedicated transcription factor could interact with these sites to activate transcription. Here we report that the zinc-finger protein, Zelda (Zld; Zinc-finger early Drosophila activator), binds specifically to these sites, and is capable of activating transcription in transient transfection assays. Mutant embryos lacking zld are defective in cellular blastoderm formation, and fail to activate many genes essential for cellularization, sex determination, and pattern formation. Global expression profiling confirmed that Zld plays a key role in the activation of the early zygotic genome, and suggests that Zld may also regulate maternal RNA degradation during the MZT.
In Drosophila, an initial wave of zygotic gene transcription occurs between 1-2 hours of development during mitotic cleavage cycles 8-13. This is followed by a major burst of activity between 2-3 hours (cycle 14) when the embryo is undergoing cellular blastoderm formation. Many pre-cellular genes contain TAGteam sites in their upstream regulatory regions including several direct targets of Bicoid, Dorsal, and other key regulators of patterning5-7. ten Bosch et. al.5 demonstrated that TAGteam sites are required for the early expression of the dorsoventral (DV) gene zen, and the sex determination genes sisB and Sxl. To isolate the TAGteam binding factor, we performed a yeast one-hybrid screen with a 91 bp fragment (Fig. 1a, sequences in uppercase) from the zen cis-regulatory region8,9, which contains four TAGteam sites5 (Fig. 1a in red, the first two are the reverse complement). zld (CG12701 on the X-chromosome) was selected as the only candidate of the 11 recovered that had the potential to bind specific DNA sequences since it encoded a protein with six C2H2 zinc fingers (represented as green boxes in Fig. 1b). Oligonucleotides (Fig. 1a, underlined sequences) with different TAGteam sites were tested in gel shift assays with the 357 amino-acid C-terminal region of Zld fused to GST (GST-ZldC; Fig. 1b, stippled region). They all formed complexes with GST-ZldC, though with different affinities (Fig. 1c lanes 1-9), while mutations (Fig. 1a, in purple) in the heptanucleotide sequence abolished binding (Fig. 1c, lanes 10-12). Interestingly, the site with the strongest affinity, CAGGTAG, is the site most over represented in regulatory elements of pre-blastoderm genes versus postblastoderm genes5. A plasmid expressing full-length Zld protein promoted transcriptional activation of a zen91-lacZ reporter but not a mutated zen91m-lacZ reporter after co-transfection in Drosophila S2 cells (Fig. 1d). Taken together, these data strongly suggest that Zld activates transcription of zen, and likely other TAGteam containing genes.
Figure 1. TAGteam sites bind Zld and mediate transcriptional activation.
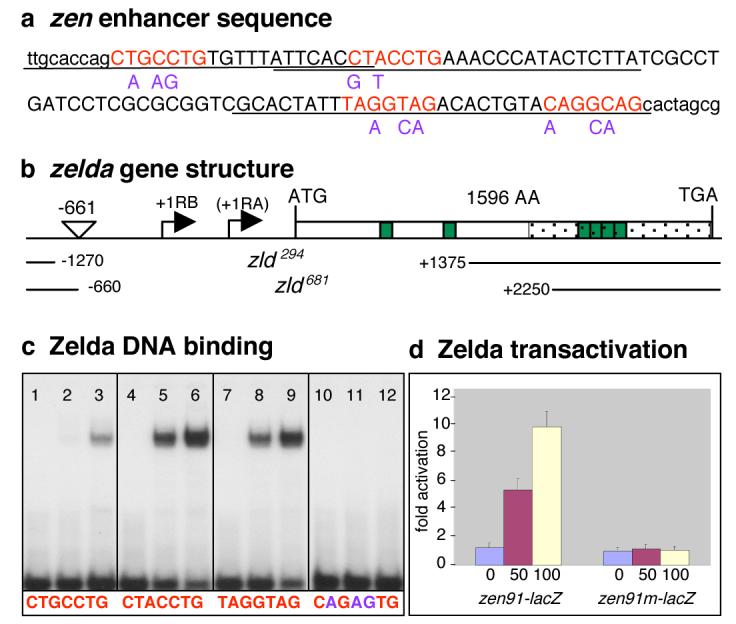
(a) DNA sequence of the 91 bp zen enhancer (uppercase) plus surrounding sequences (lowercase). Base substitutions are in purple. (b) Schematic organization of the zld locus (CG12701; Flybase) with the two predicted transcription start sites, RB and RA. The P{RS3}UM8171-3 insertion site is between -661 and -660. The nucleotides deleted in zld294 and zld681 are indicated as blank space between solid lines. (c) Zld binding to oligonucleotides containing different TAGteam sites (denoted beneath each section of the gel). The first lane in each section contains free probe. The second lane contains probe plus 10ng GST-ZldC, the third 30 ng GST-ZldC. (d) S2 cells were transfected with 0 ng (blue bar), 50 ng (red bar), or 100 ng (yellow bar) of plasmid expressing zld under control of the inducible metallothionein promoter, the zen91-lacZ or zen91m-lacZ reporter plasmids, and the luciferase control. Error bars, s.e.m.; n=3.
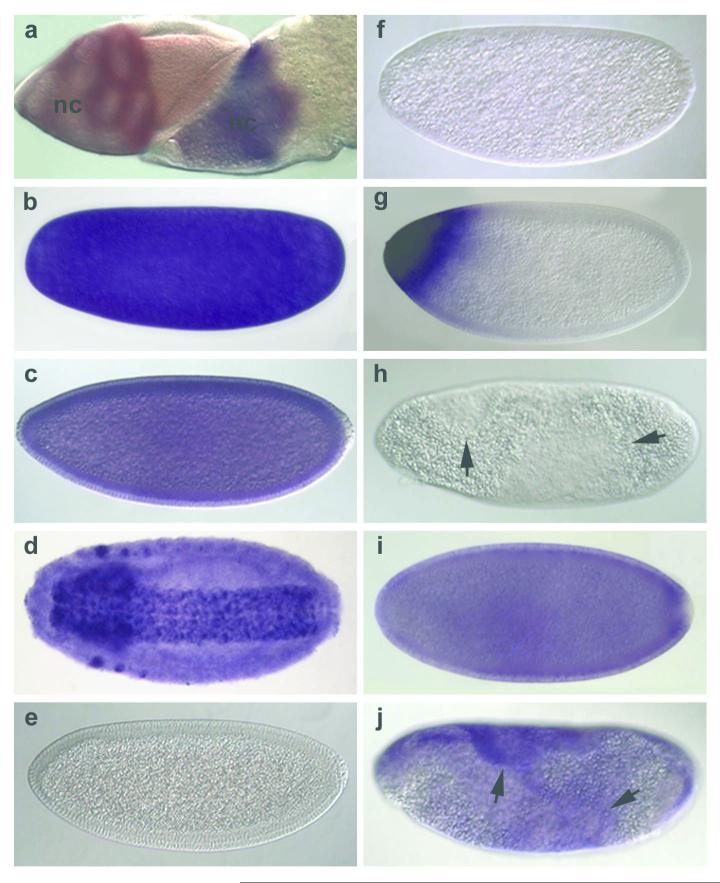
zld transcripts were detected in the germline cells of the ovary (Fig. 2a), in unfertilized eggs (Fig. 2b), and throughout early development (Fig. 2c). Later zld becomes restricted to the nervous system and specific head regions (Fig. 2d), as previously shown10. To analyze zld function, we generated deletion alleles of zld by imprecise excision (schematized in Fig. 1b). Hemizygous embryos showed abnormal CNS and head development (data not shown), consistent with previous reports of CG12701 lethal P-insertion phenotypes10,11. zld transcripts were not observed in these embryos after cycle 14 (Fig. 2e). However, younger embryos had high levels of maternal zld transcripts (data not shown), indicating that maternally loaded zld transcripts are degraded during cellularization, and replaced with zygotic zld.
Figure 2. Maternal zld transcripts are lost as zygotic zld is activated in cycle 14.
Wild-type (wt; a-d) and zld294 (e-j) ovaries (a) and embryos (b-j) were hybridized with zld (all but g) or bcd (g) RNA probes. (a) mid (left) and late-stage (right) egg chambers with zld transcripts in the nurse cells (nc) but not the columnar follicle cells that overlay the oocyte. (b) Unfertilized egg. (c) cycle 14 embryo undergoing cellularization. (d) late-stage embryo. (e) M+Z- zld cycle 14 embryo. Maternal zld transcripts have disappeared. (f) M- zld cycle 10-11 embryo. (g) M- zld cycle 14 embryo has a normal bcd pattern. (h) M-Z- zld late cycle14 embryo showing anomalous distribution of cytoplasm (arrows). (i) M-Z+ zld early cycle 14 embryo showing onset of zygotic zld expression. (j) M-Z+ zld late cycle14 embryo showing abnormalities (arrows).
To eliminate maternal zld from embryos, we induced clones of zld294 mutant germ cells in the adult female. All resulting embryos were null for maternal zld (M- zld), and the male embryos were also null for zygotic zld (M-Z- zld). All early M- zld embryos lacked zld transcripts (Fig. 2f), but had normal patterns of other maternally deposited factors such as bicoid RNAs (Fig. 2g) and the Dorsal protein gradient (data not shown). M-Z- zld embryos never expressed zld (Fig. 2h) unlike M-Z+ zld embryos, which began to express zld ubiquitously in cycle 14 (Fig. 2i). However, regardless of their zygotic genotypes, all M- zld embryos showed a severe abnormal morphology after cycle 14 (Fig. 2h,j), and did not survive to make cuticle.
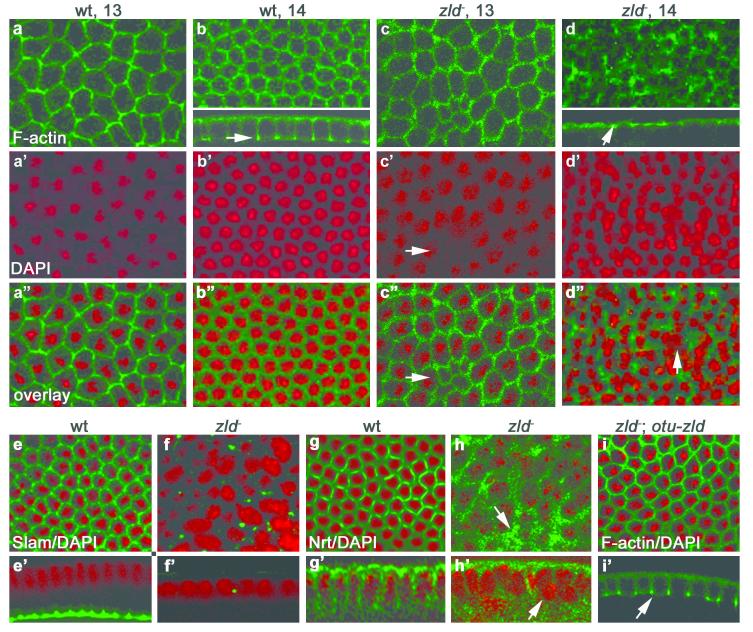
Before cycle 14, M zld embryos are similar to wild-type, except for sporadic nuclear fallout (Fig. 3c’). However, at early cycle 14 the hexagonal-actin network becomes disorganized (Fig. 3c) and begins to degenerate (Fig. 3d) resulting in a multinucleated phenotype (Fig. 3d”, arrow) resembling nullo12 and serendipity-α13 (sry-α) mutants. Celllarization does not proceed as furrow canals never move inward (Fig. 3d, arrow) like in wild-type (Fig. 3b, arrow), and Neurotactin (Nrt) accumulates abnormally in the apical cytoplasm (Fig. 3h, arrow), reminiscent of the slam mutant phenotype14,15. Staining with α-Slam antibody confirmed that Slam protein is mostly absent by mid-cycle 14 (Fig. 3f) while in wild-type, Slam has moved basally (Fig. 3e’). In addition, nuclei do not elongate but instead become rounded, enlarged, and clump together (Fig. 3f and h’ arrow). Regions of higher nuclear density were observed (data not shown), a phenotype similar to that obtained by injection of dsRNA against CG1270110, which we noticed resembles a frühstart (frs) phenotype16. Despite their aberrant morphology, M- zld embryos attempt to form a ventral furrow (Fig. S1b,c,e) but soon become highly disorganized with only pole cells recognizable (Fig. S1f). We rescued the M- zld cellularization defects by driving a wild-type copy of zld in the germline using the ovarian tumor (otu) promoter17. The cytoskeleton becomes well structured (Fig. 3i) and furrow canal ingression are normal (Fig. 3i’) as Slam protein is restored (data not shown).
Figure 3. Maternal zld is required for cellularization.
Confocal images of wt, zld- (M- zld294) and rescued (M- zld294; otu-zld) embryos (as indicated) stained with phalloidin to detect F-actin, α-slam, or α-Nrt antibodies (green), and DAPI to detect DNA (red). In M- zld embryos the cytoskeletal network is disorganized and quickly degenerates in early cycle 14 (d) accompanied by nuclear fallout (d’, arrow). Slam protein disappears in cycle 14 (f, f’), while Nrt accumulates apically (h, arrow). In M- zld294; otu-zld embryos the cytoskeleton is organized (i) and cellularization proceeds (i’, arrow).
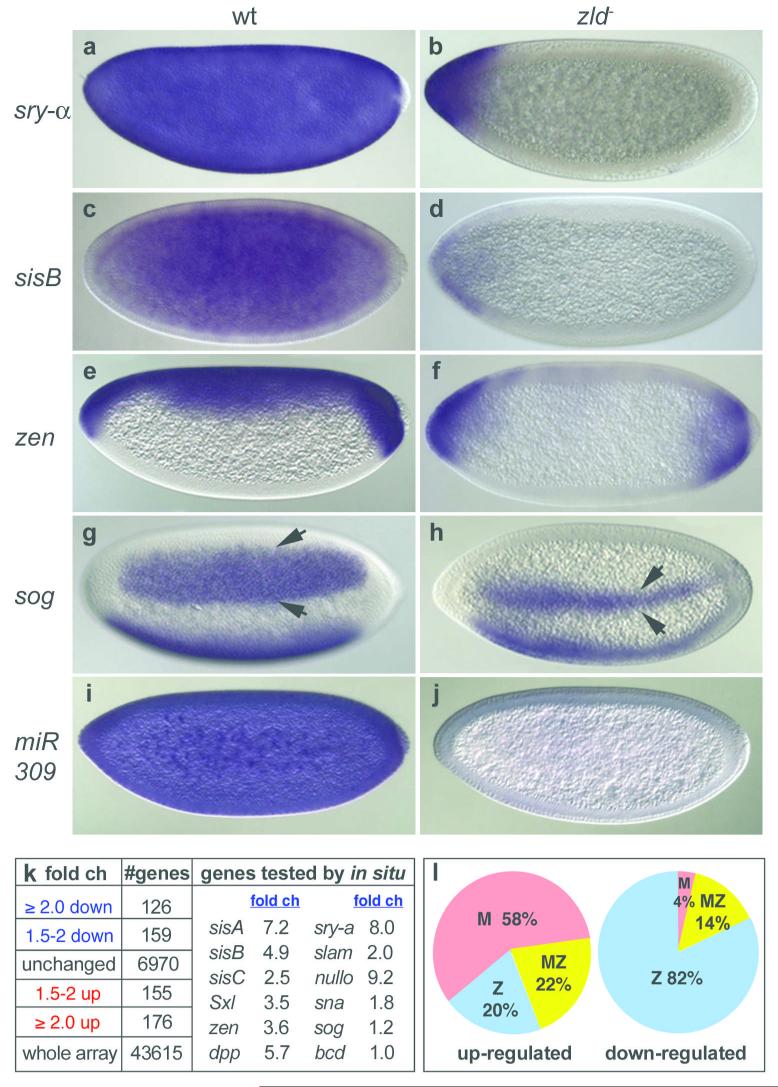
The broad range of phenotypes strongly indicated that M- zld embryos do not express genes essential for cellular blastoderm formation. We assayed the expression of sry-α, slam, and nullo, as well as sisA, sisB, sisC, Sxl, zen and dpp. None of these genes were activated in M- zld embryos (data shown for sry-α, sisB , and zen in Fig. 4b, d, f, respectively), except at the poles in some cases. However, sna and sog, which are activated by Dorsal18, were not absent but delayed in expression by at least 2 cycles (data not shown), suggesting that Zld facilitates the onset of early-gene transcription. Furthermore, the lateral stripes of sog were greatly reduced in width (Fig. 4h), indicating that in regions where Dorsal protein levels are low, a combinatorial mechanism involving both Dorsal and Zld establishes the broad sog domain. Interestingly, the two TAGteam sites in the 393bp sog enhancer19 lie close to four Dorsal binding sites.
Figure 4. Zld plays a role in zygotic gene activation and maternal RNA degradation during the MZT.
wt (left) and M-Z- zld294 (right) mitotic cycle 12-14 embryos were hybridized as indicated (309 = pri-miR-309). (k) Summary of expression profiles of 1-2 hr wt and M-zld294 embryos. fold ch = fold change with respect to wt (genes absent in the array data are not included). (l) Percentage of genes for which there is expression data3,5,19 described as maternal (M), zygotic (Z) or both (MZ) in the down- (≥ 2 fold) and up-regulated (≥ 1.5 fold) gene sets.
Our results hinted that Zld is a global activator of early genes. To test this directly we compared the expression profiles of wild-type and M- zld embryos in mitotic cycles 8-13, a time point presumably enriched in genes that are direct Zld targets. 120 genes were down-regulated at least 2-fold, and surprisingly 176 genes were up-regulated, in the absence of Zld (p<0.05; Fig. 4k). The down-regulated set (Table S1) was strongly enriched in genes that are zygotically expressed (Fig. 4l) and involved in early developmental processes (Fig. S2), including most of the genes we assayed by in situ (Fig. 4k). For example, 12 genes involved in cellular blastoderm formation (nullo, slam, sry-α, bnk, frs, btsz, halo and 5 halo-like genes20), 6 sex determination genes (sisA, sisB, sisC, run, Sxl, dpn), and 8 DV genes (dpp, tld, tok, tsg, tsg-like, scw, zen, zen-2) are in our down-regulated dataset. Overall, 75% of the early genes previously described as pre-cellular5,6,21 are included. This number may be an underestimate since there may be many genes like sna and sog that did not make the 2-fold cut-off (Fig. 4k), but are indeed regulated by Zld.
About 80% of the down-regulated genes have TAGteam sites within 2 Kb upstream of the transcription start site (Table S2), and another 10% have TAGteam sites in introns, such as slam with two sites in its first intron, supporting the idea that most of our down-regulated genes are direct Zld targets. In addition, the TAGteam sites upstream of the down-regulated genes tend to be located very close to the transcription start site, within 200 bp (Table S2), consistent with the previous finding that early zygotic genes have a statistical overrepresentation of TAGteam sites close to the start site5,6.
In contrast to the down-regulated genes, the up-regulated set is strongly enriched in genes that are maternally expressed (Fig. 4l). We considered the possibility that Zld activates components of the RNA degradation machinery that in turn destabilize maternal RNAs. Since the miR-309 enhancer22 contains two TAGteam sites, we assayed for miR-309 primary transcripts in M- zld embryos (Fig. 4j), and indeed they were absent. It was recently shown that mature miR-309 miRs become abundant during cycle 14, and play a role in maternal transcript turnover in 2-4 hr embryos3. Not surprisingly, our 1-2 hr (cycles 8-13) dataset had no overlap with the 44 published miR-309 targets3, however 2-4 hr profiling experiments should reveal whether they are up-regulated in the absence of zld. We also compared our up-regulated genes to those affected by smaug4, another gene required for removal of maternally supplied RNAs. We found there was little overlap with the published Smaug targets4, suggesting that Zld is involved in a parallel pathway of maternal RNA degradation.
In summary, we have demonstrated that Zld functions as a key transcriptional activator during the MZT in Drosophila. This is the first demonstration of such an activator in any organism. We propose that the biological role of Zld in the preblastoderm embryo is to set the stage for key processes such as cellular blastoderm formation, counting of X chromosomes for dosage compensation and sex determination, and pattern formation, by ensuring the coordinated accumulation of batteries of gene products during the MZT. This early preparedness should allow sufficient time for the formation of molecular machines23 involved in these processes, and so are ready to spring into action during the prolonged interphase of cycle 14.
Methods Summary
Fly strains
The zld294 and zld681 alleles were generated by imprecise excision of the P{RS3}24 element UM-8171-3 (Flybase, Szeged stock center). The ovoD FRT19A stock was generated by transposition of P{mini w+, ovoD1-26}25 onto y w sn FRT19A, hsFLP122. Germ-line clones were induced in zld294 FRT 19A / ovoD1 FLP122 FRT19A by the Flp-FRT technique26. Virgin females were collected and mated to yw, FM7, or FM7c-ftz-lacZ males. The otu-zld construct was microinjected into w1118 embryos.
Yeast one-hybrid assay
The yeast one-hybrid screen was performed following the Matchmaker One-Hybrid System (Clontech) protocol with the 91 bp zen-promoter and a 0-6 hours Drosophila embryonic cDNA library fused to the Gal4 activation domain27 (gift from L. Pick).
Molecular Biology
DNA binding assays9 and Drosophila S2 cell transient transfection assays28 were performed as previously described. The fold activation was calculated as a ratio of the normalized (for transfection efficiency) lacZ activity in cells treated with 0.5 mM CuSO4 and untreated cells.
Analysis of phenotypes
Various RNA probes, antibodies and molecular probes were used to detect gene expression or to visualize the cytoskeleton and nuclei (further described in the online Methods). Embryos were viewed by fluorescence microscopy using a Nikon FX-A microscope for whole embryo views, or an Improvision Yokogawa CSU-10 spinning disk confocal system for grazing and sectional views, and by Nomarski optics using a Zeiss Axiophot microscope.
Microarray analysis
Total RNA was extracted from three independent collections of 1-2 hr yw and M- zld embryos by TRIzol (invitrogen). cDNA was prepared using the GeneChip® HT One-Cycle cDNA Synthesis Kit (Invitrogen), labeled with the BioArray™ HighYield™ RNA Transcript Labeling Kit (Enzo), and hybridized to Affymetrix Drosophila Genome 2 arrays and processed by a GeneChip Fluidics Station 400.
Supplementary Material
Acknowledgements
We thank the following people for generous gifts of RNA probes, plasmids, antibodies, and fly stocks: J. Erickson, R. Lehmann, R. Cinalli, R. Martinho, C. Navarro,J. Treisman, and L. Pick. We are indebted to J. Rhee, and A. Chung for help with isolating and characterizing zld null mutants. We thank S. Fu for help locating TAGteam sites in Zelda target genes. We are grateful to M. Siegel and K. Birnbaum for help with the microarray analysis, and C. Desplan, P. Struffi, S. Small, and R. Lehmann for critical reading of the manuscript. This work was supported by a grant from the National Institutes of Health (GM63024).
References
- 1.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 2.Giraldez AJ, et al. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 3.Bushati N, Stark A, Brennecke J, Cohen S. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 4.Tadros W, et al. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 5.ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 6.De Renzis SD, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:1036–1051. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6:365–388. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang J, Rushlow CA, Zhou Q, Small S, Levine M. Individual Dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 1992;11:3147–3154. doi: 10.1002/j.1460-2075.1992.tb05387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirov N, Zhelnin L, Shah J, Rushlow C. Conversion of a silencer into an enhancer: evidence for a co-repressor in dorsal-mediated repression in Drosophila. EMBO J. 1993;12:3193–3199. doi: 10.1002/j.1460-2075.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staudt N, Fellert S, Chung H, Jäckle H, Vorbrüggen G. Mutations of the Drosophila zinc finger-encoding gene vielfältig impair mitotic cell divisions and cause improper chromosome segregation. Mol. Biol. Cell. 2006;17:2356–65. doi: 10.1091/mbc.E05-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourbon HM, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 12.Simpson L, Wieschaus EF. Zygotic activity of the nullo locus is required to stabilize the actin-myosin network during cellularizatiron in Drosophila. Development. 1988;110:851–863. doi: 10.1242/dev.110.3.851. [DOI] [PubMed] [Google Scholar]
- 13.Schweisguth F, Lepesant JA, Vincent A. The serendipity alpha gene encodes a membrane-associated protein required for the cellularization of the Drosophila embryo. Genes. Dev. 1990;4:922–931. doi: 10.1101/gad.4.6.922. [DOI] [PubMed] [Google Scholar]
- 14.Lecuit T, Samanta R, Wieschaus E. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev. Cell. 2002;2:425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- 15.Stein JA, Broihier HT, Moor LA, Lehmann R. Slow as molasses is required for polarized membrane growth and germ cell migration in Drosophila. Development. 2002;129:3925–3934. doi: 10.1242/dev.129.16.3925. [DOI] [PubMed] [Google Scholar]
- 16.Grosshans J, Müller H, Wieschaus E. Control of cleavage cycles in Drosophila embryos by frühstart. Dev. Cell. 2003;5:285–294. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- 17.Robinson DN, Cooley L. Examination of the function of two kelch proteins generated by stop codon suppression. Development. 1997;124:1405–1417. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- 18.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev. Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross SP, Guo Y, Martinez JE, Welte MA. A determinant for directionality of organelle transport in Drosophila embryos. Curr. Biol. 2003;13:1660–1668. doi: 10.1016/j.cub.2003.08.032. [DOI] [PubMed] [Google Scholar]
- 21.Pilot F, Philippe JM, Lemmers C, Chauvin JP, Lecuit T. Developmental control of nuclear morphogenesis and anchoring by charleston, identified in a functional genomic screen of Drosophila cellularization. Development. 2006;133:711–723. doi: 10.1242/dev.02251. [DOI] [PubMed] [Google Scholar]
- 22.Biemar F, et al. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunsalus K, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 24.Ryder E, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- 26.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y, et al. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]
- 28.Kirkpatrick H, Johnson K, Laughon A. Repression of dpp targets by binding of brinker to mad sites. J. Biol. Chem. 2001;276:18216–18222. doi: 10.1074/jbc.M101365200. [DOI] [PubMed] [Google Scholar]
- 29.Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 30.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol. 2004;14:159–165. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.