Abstract
In recent years several of the key tenets of the original cytokine-STAT signaling paradigm have had to be revised. First, that nonphosphorylated “inactive” STATs are present in the cytoplasm as free monomers which dimerized only subsequent to Tyr-phosphorylation has been replaced by the understanding that nonphosphorylated STATs in the cytoplasm exist largely as dimers and high molecular mass “statosome” complexes. Second, the notion that phosphorylation, either of Tyr or Ser residues or both, in STAT species is required for transcriptional activation has been replaced by the realization that nonphosphorylated STATs can be transcriptionally active albeit with respect to sets of target genes distinct from phosphorylated STATs. Third, the notion that it is the activation by phosphorylation of STATs at the plasma membrane that then leads to their import into the nucleus has been replaced by the recognition that even nonphosphorylated STATs shuttle between the cytoplasm and nucleus at all times in a constitutive manner. Fourth, the notion that the trans-cytoplasmic transit of STATs from the plasma membrane to the nuclear import machinery takes place exclusively as a free cytosolic process has been replaced by the understanding that at least a portion of this trans-cytoplasmic transit is mediated via membrane-associated caveolar and endocytic trafficking (the “signaling endosome” hypothesis). Fifth, the targeting and sequestration of activated STAT3 to long-lived endosomes in the cytoplasm requires consideration of STAT3-mediated “signal transduction” from the plasma membrane to cytoplasmic membrane destinations potentially for function(s) in the cytoplasm. Indeed, in tissue sections many discrete histologic cell types display PY-STAT3 almost exclusively in the cytoplasm with little, if any, in the nucleus. New challenges include determining the structural bases for the recruitment of nonphosphorylated dimeric STAT species to the cytosolic face of membranes including at the cytoplasmic tails of respective receptor complexes, the conformational changes subsequent to phosphorylation and the structural bases for the targeting and functions of STAT proteins within the cytoplasm per se.
Keywords: statosome complexes, intracytoplasmic trafficking, signaling endosomes, sequestering endosomes, mitosis
1. Introduction
The focus of this review is the cell biology of “signal transduction” in the JAK/STAT pathway with particular emphasis on the trafficking and function of STAT species within and among different subcellular compartments in mammalian cells. “Signal Transducer and Activator of Transcription” (STAT)-family proteins are classically viewed as transducing cytokine- and growth factor-activated signals from the plasma membrane to the cell nucleus for the purpose of activating transcription [1-4]. The original STAT-signaling paradigm formulated in the early 1990s [1, 5] envisaged that monomeric nonphosphorylated STAT species in the cytosol were recruited to respective ligand-activated cytokine and growth factor receptors at the plasma membrane, Tyr- and/or Ser-phosphorylated with departure from the receptor complexes, subsequent dimerization of the phosphorylated STAT species, diffusional transit through the cytoplasm to the nuclear pore, import into the nucleus and transcriptional activation of target genes. Cessation of signaling involved dephosphorylation of DNA-bound STAT dimers within the nucleus and then export back to the cytoplasm [2, 3]. This formulation of the STAT signaling paradigm, particularly the representation that inactive STATs were all free monomers in the cytoplasm which dimerize only subsequent to phosphorylation followed by nuclear import and transcriptional activation by the phosphorylated dimers in the nucleus has had a seductive hold on the STAT signaling field. Though data showing that the original suggestion [5] that inactive nonphosphorylated STAT1 was monomeric in the cytoplasm was incorrect appeared almost a decade ago [6, 7], the simplicity of elocuting this “monomer to dimer transition” model has been hypnotic [see Fig. 1 in ref. 8 for one recent example].
Fig. 1.
Superose-6 FPLC analyses of STAT proteins in rat liver cytosol. An aliquot (200 μl) of rat liver cytosol (the 100,000 x g supernatant) was fractionated through Superose-6 FPLC (approximately 1 ml per fraction), and the elution of various STAT proteins was evaluated by SDS-PAGE and Western blotting of 100-μl aliquots of each elutae fraction. Panels A-D show the elution of STAT3, STAT1, STAT5A and STAT5B. Panel E shows the elution of murine anti-STAT3 mAb (IgG) of mass approximately 150-160 kDa off the same column in a separate run. Fr. No., fraction number; void volume of the column is in fractions 7-8. From ref. 7.
Extensive work by cell biologists over the last several decades shows that the cytoplasm is not simply an empty space between the plasma membrane and the nucleus for signal transduction purposes. Cell biologists have defined the “signaling endosome” compartment which mediates vesicular trafficking from the plasma membrane to the nucleus for productive transcriptional activation [9-14]. However, the trans-cytoplasmic traverse of “activated” STAT species is largely discussed as soluble and diffusion-driven process [1-4, 15]. The phosphorylation of STAT proteins (either Tyr-P or Ser-P or both) has itself been thought to be obligatorily required for nuclear import [1, 2, 15]. Indeed, despite book-keeping analyses to the contrary [16], it has been more-or-less assumed that the bulk of the pool of say a particular Tyrphosphorylated STAT species is automatically destined in its entirety for nuclear import and a transcriptional function [1, 2, 8, 15, 17].
It is the purpose of this essay to briefly revisit some of the considerations upon which the original STAT signaling paradigm was based, discuss some of the shifts in the original paradigm, and perhaps indulge a bit in the crystal ball. This essay builds upon two earlier discussions of adjustments to the STAT signaling paradigm [18, 19]. As the hyperactivation of STAT species, especially of STAT3, in localized tissues such as epithelia, endothelium and smooth muscle cells is increasingly implicated in disease processes such as cancer, gastrointestinal diseases and pulmonary arterial hypertension [20-24], and hypoactivity due to mutations in STAT3 implicated in immunological diseases [25, 26] it seems an appropriate moment to revisit some of the basic cell biology of this signaling pathway.
2. The classical STAT signaling paradigm: the “monomer to dimer transition” model
In the classical STAT signaling model inactive latent nonphosphorylated STAT species in the cytoplasm were posited to be free monomers each of mass approximately 80-90 kDa [1, 2, 5]. Cytokine binding to cognate cell-surface receptors led to the auto- or cross-Tyrphosphorylation of one or more members of JAK family kinases preassociated with the receptor complex. The activated JAKs phosphorylated Tyr residues on the cytoplasmic tails of respective cytokine receptor chains and served as docking sites for SH2-domain containing STAT monomers. Receptor bound STATs were then Tyr-phosphorylated on conserved Tyr residues (e.g. Tyr701 for STAT1 or Tyr705 for STAT3) by JAKs followed by departure of the phosphorylated STATs from the cytosolic face of the plasma membrane and their subsequent dimerization in the cytoplasm. Translocation to the nucleus of such STAT dimers, with or without additional Ser-phosphorylation, then positioned the “activated” STATs to transcriptionally regulate target genes. It was only the phosphorylated STAT homodimer or heterodimer species, which had DNA-binding capability, and which, in concert with co-activator proteins, led to transcriptional regulation. Moreover, the report in 1989 that “all” of the cytoplasmic DNA-shift activity characterized as ISGF3 (a protein complex now known to contain activated STAT1, STAT2 and interferon regulatory factor 9) was recovered in the S100 cytoplasmic fraction led to the suggestion that these proteins “are not associated with particles or membranes” [27]. Thus the trans-cytoplasmic transit of STAT signals was considered to be exclusively a cytosolic process, even though we (7, 14) and others (see Figs. 5A and 5B in ref. 28 for a recent dramatic example) reported punctuate localization of STAT3 and PY-STAT3 in the cytoplasm.
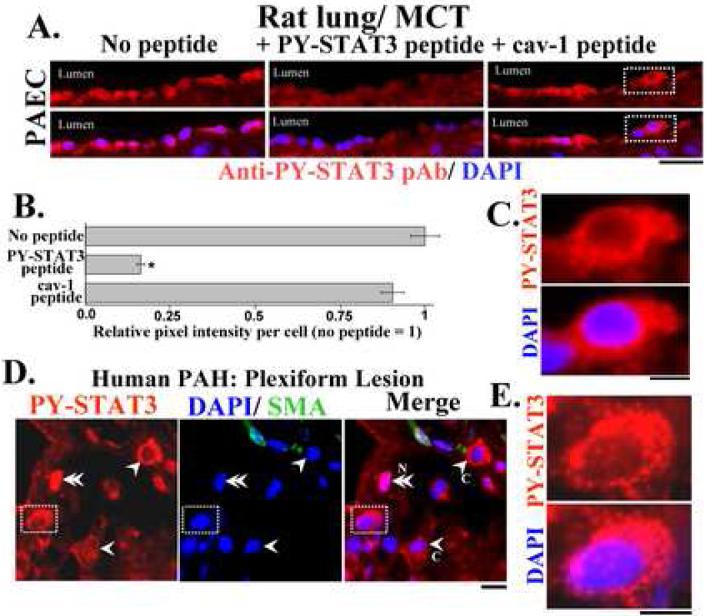
Fig. 5.
Marked cytoplasmic provenance of PY-STAT3 in distinct cell types in lung tissue in pulmonary arterial hypertension assayed in formalin-fixed paraffin-block sections.
Panels A and B. Double-label immunofluorescence analyses using anti-PY-STAT3 pAb and DAPI showing PY-STAT3 in the cytoplasm of luminal pulmonary arterial endothelial cells in rats administered monocrotaline 4 wk earlier. Scale bar = 25 μm. Analyses included a peptide competition assay confirming the specificity of the PY-STAT3 pAb used. Per cell integrated pixel intensities were obtained using NIH Image J and expressed in terms of the section stained with PY-STAT3 pAb without any blocking peptide. * P < 0.05 in comparison to the section stained with PY-STAT3 pAb without any peptide, n = 50-60 cells per variable.
Panel C. High magnification panels of indicated area in Panel A. Scale bar = 5 μm.
Panel D. Plexiform lesion in idiopathic pulmonary arterial hypertension in man showing cells with both cytoplasmic (single arrowheads) and nuclear (double arrowheads) PY-STAT3. Scale bar = 5 μm.
Panel E. High magnification panels of indicated area in Panel D. Scale bar = 5 μm.
Adapted from ref. 24.
There is now extensive literature on considerations of Tyr-phosphorylation or Ser-phosphorylation of respective STAT proteins alone one at a time or both together in a stimulus and cell-type dependent manner [2, 15, 29]. There is also now extensive literature on the mechanisms for regulating STAT signaling through the activity of specific phosphatases, the PIAS and the SOCS family of proteins [2, 4, 15, 29], topics which are discussed elsewhere in this volume. The focus of this essay is the basic cell biology of the STAT signaling paradigm as it plays out in mammalian cells.
3. Limitations of techniques in use to investigate STAT signaling
In looking back (and forward) several limitations of the techniques used to investigate STAT signaling need to be kept in mind. Sucrose-gradient sedimentation methods to size proteins and their complexes are only as good as the sedimentation behavior of the size marker proteins, their replicability and the effect of detergents in such experiments. We suspect that these issues led Shuai et al [5] to infer that inactive STAT1 in the cytoplasm was all monomeric which dimerized only subsequent to Tyr-phosphorylation. Gel-filtration chromatography to correctly demonstrated that native STAT species in the cytoplasm and in solution as recombinant proteins are at least dimers and in higher molecular mass complexes - with little detectable monomers in the cytoplasm (Fig. 1)[6, 7, 30-32].
The use of immunofluorescence microscopy of fixed cells to illustrate the bulk transfer of the cellular pools of respective STAT species upon cytokine stimulation from the cytoplasm to the nucleus [33, 34] is open to several issues: (a) selective leaching and loss of soluble STAT proteins during fixation preferentially from the cytoplasm (we have observed loss of approximately half of STAT3 from cell cultures as a result of fixation, ref. 14); (b) different immunological reactivity of STAT proteins within the cytoplasmic or nuclear compartments in a manner that can depend upon the fixative used (Fig. 2); (c) the cell-culture state dependent behavior of STAT species within a heterogeneous population of cells even within the same culture vessel; (d) the ability of modern microscopy software to set “dark” thresholds so as to slice off lower intensity signals and represent these as completely negative; and, (e) the logarithmic nature of pixel intensity dependence upon analyte concentration allows the cytoplasm to appear devoid of a STAT signal because of a focus on the most intense pixels in the nucleus. Cross-comparisons between immunofluorescence and cell fractionation and Western blotting data reveal discrepancies between the two - usually in the direction that cytoplasmic STAT species were underrepresented in the immunofluorescence images [14, 35, 36]. Indeed in book-keeping studies using metabolic labeling of STAT species or Western blotting procedures, it is only the minority fraction (perhaps 15-20% at best) of cellular STAT species that is observed in the nucleus in cell-types such as HeLa or Hep3B [14, 16, 37]. From our perspective, the typical immunofluoresence images (Fig. 2)[14, 36] represent “etched” images of the residual STAT proteins associated with subcellular structures subsequent to leaching of at least a part of the soluble cytoplasmic pool. Additionally, different fixation protocols have distinctly different abilities to reveal nuclear versus cytoplasmic STAT species (Fig. 2)[14]. In the data in Fig. 2 we also draw attention to the presence of STAT3 immunofluorescence in the cytoplasm in clearly etched puncta.
Fig. 2.
Immunofluorescence analyses for STAT3 in Hep3B cells in culture showing punctuate cytoplasmic immunostaining and dependence of observations on specific fixation protocol used. Analyses for STAT3 were carried out in untreated and IL-6-treated (10 ng/ml for 30 min) Hep3B cells using the C20 anti-STAT3 rabbit IgG from Santa Cruz, Inc, Santa Cruz, CA using cold methanol-acetone, cold paraformaldehyde-Triton X-100 or formaldehyde (37°C)-Triton X-100 fixation methods. Respective peptide competition controls using relevant and irrelevant (cav-1) peptides were included in the analyses. Scale bar = 25 μm. From ref. 14.
While the use of fluorescence tagged STAT species (GFP, YFP or CFP) in live-cell imaging studies has been considered a better approach to investigate the dynamics of STAT signaling [35, 38-42] there are technical limitations to these approaches. This fluorescence can be markedly quenched by binding proteins, even by proteins that bind next to and at the fluorescent tag itself [43, 44]. A further limitation is the known ability of the fluorescent tag to itself affect trafficking - directing it to aberrant compartments, directing it to the correct compartment but to a structure segregated from the native molecules or altering kinetics [41, 45, 46]. Thus, controls based upon independent techniques become necessary in order to reliably interpret such tagged-protein studies and to extend the observations to the endogenous untagged copies [46].
The DNA-binding competence of STAT species using the well-known m67 SIE DNA probe has been often taken to quantitatively reflect the amounts of the respective Tyr-phosphorylated dimers in cytoplasmic or nuclear extracts (see ref. 17 for one example). However, in cross-comparisons of DNA-shift competence and Western-blottable Tyr-phosphorylated STAT1 and STAT3 species in cytoplasmic and nuclear extracts prepared from Hep3B cells treated with IFN-γ or IL-6 or both in the presence or absence of vanadate we observed up to a 30-fold reduced DNA shift competence compared to Western blotting data especially in cells treated with vanadate [16 and unpublished data]. Wenta et al [32] have recently shown that non-phosphorylated STAT1 dimers can also bind the m67 SIE DNA probe in gel-shift assays.
The question of the relevance of cell culture-derived trafficking data to cells located in distinct histologic cell types in intact tissues has rarely been raised. Most investigations of STAT trafficking have used cell culture systems in which the nuclear pool of activated STAT proteins is usually quite evident. However, the situation in vivo in cells in tissues can be quite different with, in certain instances, almost all the PY-STAT3 retained in cytoplasmic vesicles with very little detectable in the nucleus even when the same histologic cell type maintained in culture shows clear nuclear accumulation [24].The sustained long-term residence of activated STATs in the cytoplasm (for a particularly dramatic example of the imaging of PY-STAT3 exclusively in the cytoplasm see Fig. 2B in ref. 47) is little discussed. In our hands, STAT3 trafficking in vivo in the lung can be such that in specific histologic types of cells all of the PY-STAT3 remains cytoplasmic (see Fig. 6 below).
Fig. 6.
Predominant nuclear or cytoplasmic provenance of PY-STAT3 in cells of distinct histologic phenotypes in the human lung.
Panel A. Predominantly nuclear PY-STAT3 in cells lining thickened alveolar septa in a patient with pulmonary arterial hypertension.
Panel B. Exclusively cytoplasmic PY-STAT3 in the tracheobronchila epithelium in normal lung in cells apparently of the basal cell progenitor phenotype.
Panel C. Low and high magnification panels of a peptide competition assays using sequential serial sections of the same block to verify the detection of PY-STAT3 as in panel B.
Scale bars = 25 μm except in the low magnification panel in C in which scale bar = 10 μm. (formalin-fixed paraffin-block sections of human lung specimens were kindly provided by Dr. Rubin M. Tuder, Department of Pathology, Johns Hopkins University School of Medicine as in ref. 24).
4. Higher-order structure of nonphosphorylated STAT species in the cytoplasm
Lackmann and colleagues [6] reported almost a decade ago that by gel-filtration sieving chromatography inactive nonphosphorylated STAT1 in the HeLa cell S100 cytosol prepared using detergent-free methods was not the expected 80-90 kDa free monomer but was at least a dimer of mass approximately 160-200 kDa. These investigators were unable to detect any free STAT1 monomer. Even more telling, these investigators showed that the nonphosphorylated STAT1 species eluting off a gel-filtration which was recruited to the cytoplasmic tail peptide of the IFN-γ-receptor in an immobilized-binding detection assay was in the 160-200 kDa range and thus at least dimeric. Importantly, there was no STAT1 in the monomer region of the FPLC eluate that bound the IFN-γ receptor fragment. Moreover, cytosol prepared from HeLa cells exposed to IFN-γ showed that the gel-filtration properties of Tyr-phosphorylated STAT1 detected by Western blotting were identical to those of unphosphorylated STAT1 from cells not treated with the cytokine. Thus there was no size shift of STAT1 upon phosphorylation as would have been predicted by the “inactive monomer becomes an active dimer upon Tyr-phosphorylation” paradigm. Also in 1998 Stanley and colleagues [48] reported their gel-filtration chromatography studies showing the presence of PY-STAT3, PY-STAT5A and PY-STAT5B isolated from the cytoplasm of a colony stimulating factor-1-treated macrophage cell line to be in complexes in the size range 160-600 kDa.
We reported in 1999 our failure to detect free monomers of STAT1, STAT3, STAT5A or STAT5B in the S100 cytoplasm of Hep3B cells and rat liver hepatocytes (Fig. 1)[7]. Nonphosphorylated STAT species were found to be in structures at least as large as dimers or in complexes larger than dimers (Fig. 2 and ref. 7). Nonphosphorylated STAT proteins in the S100 cytosol of Hep3B and rat liver hepatocytes eluted in a broad continuum in the range 200-400 kDa (“statosome I”) with an additional peak in the void volume around 1-2 MDa (“statosome II”)[7]. The latter larger complexes included clathrin heavy chain [14].
To keep the literature in perspective, cross-immunoprecipitation studies beginning in 1996 had revealed the interaction between and among non-phosphorylated STAT1 and STAT3 species in the cytoplasm [49, 50]. However, such cross-immunoprecipitation studies cannot exclude the presence of free STAT monomers in the cytoplasm. The resolution of the latter question derived solely from the use of gel-filtration chromatography techniques by Lackmann et al [6] and by Ndubusi et al [7] for native nonphosphorylated STAT1, STAT3, STAT5A and STAT5B species.
Subsequently, Ota et al [30] reported that recombinant nonphosphorylated STAT1, STAT3, STAT4, STAT5A, STAT5B and STAT6 formed dimers in solution as assayed by gel-filtration chromatography as well as in yeast two-hybrid assays. Moreover, nonphosphorylated STAT4 dimers formed in vivo before cytokine receptor-driven activation. Braunstein et al [30] also confirmed that STAT1 and STAT3 were dimeric in the cytoplasm of unstimulated cells and that the respective recombinant proteins were also dimeric in solution. Little or no free STAT1 or STAT3 monomer was detected in the cytoplasm. That nonphosphorylated STAT1 is dimeric and can even be tetrameric has now been additionally confirmed by the Darnell group [51, 52] and the Vinkemeir group [32]. The available data has led Darnell and colleagues and Vinkemeir and colleagues to now enunciate a conformational change model (switch from antiparallel dimer to a parallel dimer upon Tyr-phosphorylation)[3, 32, 51, 52]. Nevertheless, even today [8, 15] models of the recruitment of STAT species to the receptor cytoplasmic tails depict the docking of the free STAT monomer despite the lack of any experimental evidence that it is indeed the free monomer that binds the receptor. The mechanism by which the non-phosphorylated dimer is recruited to the receptor complex on the cytosolic face of the plasma membrane remains an enigma. At the moment, a conceptual way out has been to envisage some sort of a dimermonomer equilibrium [3, 32] with the monomer actually doing the docking. However there are no data one way or the other, except for those of Lackmann et al [6] showing that it is the nonphosphorylated dimer of STAT1 that docks with the IFN-γ-receptor PY-440-containing fragment.
The existence of STAT3 dimers in the cytoplasm of live cells has been confirmed using fluorescence resonance energy transfer (FRET) methods [53, 54]. Moreover, fluorescence relaxation spectroscopy approaches in analyzing the physical nature of STAT3-GFP in the cytoplasm of live Hep3B cells has provided evidence for inactive STAT3 to be in complexes of rotational radii consistent with a molecular mass of 200-400 kDa with the IL-6-induced generation of high molecular mass statosome complexes of STAT3 >1 MDa [40].
It is now clear that, in addition to their dimeric nature, cytoplasmic STAT species associate with a variety of proteins in the cytoplasm (as “dimer-plus” complexes). The associated proteins include regulatory proteins, chaperones, proteins involved in membrane trafficking, proteins ordinarily present in organelles such as mitochondria, and proteins involved in the trafficking of STAT species to the nucleus [2, 15, 19, 29]. From the point of view of endocytic membrane-associated trafficking of STAT proteins through the cytoplasm the association with clathrin heavy chain, caveolin-1 and the chaperones heat shock protein 90 and glucose regulated protein 58 [14, 55, 56](see below).
5. Constitutive nucleocytoplasmic shuttling of STATs
Early immunofluorescence studies showed an increase in the nuclear pool of respective STAT species when cells in culture were exposed to relevant cytokines and growth factors [2, 15, 33, 34]. The published illustrations were particularly dramatic and led to the inference that for example almost all of bulk cellular STAT1 accumulated in the nucleus of IFN-γ treated HeLa cells. With the availability of antibodies to Tyr-phosphorylated STAT species, immunofluorescence microscopy illustrations again represented that virtually all of the respective PY-STAT species was in the nucleus [1]. Moreover, assays of DNA-binding competent STAT1, and thus by inference of PY-STAT1 dimers, in IFN-γ-treated HeLa cells during a prolonged staurosporine chase were interpreted to show that the entirety of the cytoplasmic pool of say PY-STAT1 eventually transited to the nucleus [17]. These data led to the inferences (a) that it was only upon activation and phosphorylation that STAT proteins transited to the nucleus, and (b) that there was a bulk transit of STAT proteins, particularly of the phosphorylated pool, from the cytoplasm to the nucleus upon such activation.
Nevertheless, experiments using cell fractionation methods showed the discrepant results such as that at most 15-20% of cellular STAT1 was in the nuclear pool when assayed by Western blotting or by metabolic labeling [7, 14, 37]. Additional discrepant results using cell fractionation and Western blotting approaches showed that the majority of say PY-STAT1 in IFN-γ-treated Hep3B cells or of PY-STAT3 in IL-6-treated Hep3B cells remained cytoplasmic even during a prolonged staurosporine chase [16]. In contrast, other investigators highlighted their observations that nonphosphorylated STAT3 was nuclear in provenance in cultured Hep3B cells [35].
Upon the availability of expression constructs for fluorescently-tagged STAT species several investigators confirmed the cytokine or growth factor-driven increased accumulation of STAT proteins in the nucleus [38-41, 53]. The illustrations evident in the literature show an almost complete bulk transit of respective STAT proteins to the nucleus (in Fig. 3A note the upper cell showing almost all the STAT3-GFP in the nucleus). The nuclear import pathway for activated STAT species was delineated as the binding of the respective nuclear localization signals in STAT dimers to specific importins (for example STAT1 to importin α5 and β1, STAT3 to importin α3 or α6 and β1) followed by traverse through the nuclear pore [8]. Additional requirements for Rac1 and the MgcRacGAP GTPase have been reported for nuclear import of STAT5A and STAT3 [57, 58]. The export of dephosphorylated STAT species from the nucleus was delineated to be dependent upon the nuclear export signal in STAT proteins, the adapter CRM1 (thus inhibited by leptomycin B) and regulated by the DNA binding-competence of respective STAT complexes [8, 41].
Fig. 3.
Live-cell imaging of the sequestration of fluorescently tagged STAT3 to cytoplasmic sequestering endosomes.
Panels A and B. STAT3-GFP (green) transfected Hep3B cells were exposed to IL-6 for 30 min and imaged together with labeling for the lysosomal (LysoTracker in red), endoplasmic reticulum (ER-Tracker in red) or mitochondrial compartments (MitoTracker in blue).
Panel C. Sequestration of endogenous native PY-STAT3 in cytoplasmic membrane structures. Replicate Hep3B cells were exposed to IL-6 for 30 min and then sequentially to digitonin (50 μg/ml) in ice-cold 0.25 M sucrose-phosphate-buffered saline (sucrose buffer) and to Brij 58 (0.5% vol/vol in sucrose buffer), fixed with cold paraformaldehyde and immunostained for PY-STAT3.
Panels D and E. Cotransfection with an expression construct for the K44A dominant-negative mutant of dynamin II leads to marked accumulation of STAT3 in cytoplasmic vesicles with depletion from the nucleus.
All scale bars = 25 μm; adapted from ref. 41.
In a seminal discovery Vinkemeir and colleagues [38] used fluorescence recovery after photobleaching (FRAP) approaches to show that nonphosphorylated STAT1 traffics from the cytoplasm to the nucleus and out again in a constitutive manner. This constitutive import of nonphosphorylated STAT1 through the nuclear pore was found to be independent of the karyopherin (importin) mechanisms. On the other hand phosphorylated STAT1 was obligatorily dependent upon the karyopherin mechanism. Export required dephosphorylation and a CRM1-dependent mechanisms [38, 41]. The nucleocytoplasmic shuttling of nonphosphorylated STAT3 and of STAT3 persistently activated by src kinase is also now clear [35, 39, 59]. Indeed, Pranada et al [39] pointed out that the apparent IL-6-induced accumulation of STAT3 and PY-STAT3 in the nucleus was largely due to decreased export from the nucleus. In careful analyses of the dynamics of the nuclear import and export of nonphosphorylated and phosphorylated STAT1 species, Vinkemeir and colleagues [41] showed that the GFP tag itself altered the transit kinetics with a greater retardation of the kinetics of the transit of nonphosphorylated STAT1 than of phosphorylated STAT1. The slowing in the kinetics of nuclear transit due to the GFP tag was reflected in concomitant changes in the reduction of the respective STAT1 species to elicit a transcriptional response. Thus, not only is there constant nucleocytoplasmic shuttling of STAT proteins, its kinetics can be altered by the fluorescent tag itself. The nuclear import mechanisms applicable for the respective STAT proteins can be different for the nonphosphorylated and phosphorylated species [33, 41].
6. Transcriptional functions of non-phosphorylated STAT species
Even before the discovery of constitutive nucleocytoplasmic shuttling of STAT1, Stark and colleagues [60] pointed out that nonphosphorylated STAT1 must have a function in cells. STAT1-null cells in culture showed resistance to apoptosis by TNF. Replacement of STAT1 restored TNF-induced apoptosis and the expression of the caspases Ice, Cpp32 and Ich-1 [60]. Mutations in STAT1 which otherwise reduce cytokine-induced transcriptional capability also restored sensitivity to apoptosis. Thus constitutive STAT1 displayed biological activities different from those that mediated cytokine-induced gene expression.
Subsequently several investigators have highlighted the ability of nonphosphorylated forms of STAT1, STAT3 and STAT6 to transcriptionally activate target genes, albeit different from those activated during cytokine- or growth factor-mediated induction. Stark and colleagues have pioneered the demonstration that nonphosphorylated STAT1 and STAT3 have transcriptional effects in the nucleus through complexes formed with IRF1 or other transcription factors [61, 62]. There is also now evidence that unphosphorylated STAT6 contributes to the constitutive expression of cyclooxygenase-2 in a human cancer cell line [63].
7. STAT signaling along raft/caveolar and endocytic pathways (signaling endosome hypothesis)
Immunofluorescence analyses of STAT species, particularly STAT3, in the cytoplasm of Hep3B hepatocytes in culture fixed using the methanol-acetone procedure had a clearly punctuate appearance [36](see Fig. 2). Thus we asked whether at least a portion of cytoplasmic STAT3 might be associated with various membrane and organellar fractions in the cytoplasm. Cell fractionation, membrane rafting, double- and triple-label immunofluorescence studies disclosed the association of STAT species and cytokine receptor chains in plasma membrane rafts/caveolae and in association with cytoplasmic with vesicular elements of the caveosome/endosome pathways [14, 42, 55, 56]. Significantly, the disruption of plasma membrane rafts using β-methylcyclodextrin or filipin III reversibly affected not only the generation of cytokine-induced Tyr-phosphorylation of STAT1 or STAT3 but also productive transcriptional signaling [55, 56, 64]. Independently, in a seminal report in 2002 Jove and colleagues showed the association of STAT3 and PY-STAT3 in cells exposed to EGF with clearly defined cytoplasmic vesicles which colocalized with the endocytic marker α-adaptin [65]. Critically, these investigators showed that overexpression of proteins which have a dominant-negative effect on endocytic trafficking inhibited productive growth factor/STAT3 transcriptional signaling. Also independently, Scoles et al [66, 67] showed that the endocytosis regulators HRS (hepatocyte growth factor-regulated tyrosine kinase substrate) and schwanomin (product of the NF2 neurofibromatosis gene) inhibited STAT3 and STAT5 phosphorylation and transcriptional function in human schwannoma cell lines exposed to insulin-like growth factor-1 or epidermal growth factor.
Shah et al [14] confirmed and extended the observations of Jove and colleagues [65] to IL-6-induced trafficking of activated PY-STAT3 in the Hep3B cytoplasm. Cell fractionation methods revealed that the majority (60-75%) of the pool of cytoplasmic PY-STAT3 at 15 min after IL-6 exposure was associated with the early endosome fraction [14]. In terms of productive transcriptional signaling, overexpression of proteins which have a dominant negative effect on endocytic trafficking (the dynamin II K44A mutant deserves special note) reduced activity of STAT3-driven reporter constructs while overexpression of an active dynamin species (MxA) enhanced transcriptionally productive signaling [14]. Lamaze and colleagues [68] showed the dichotomy of IFN-α and IFN-γ induced STAT signaling along the endocytic pathway. Upon cytokine stimulation both IFN-α and IFN-γ receptors were internalized by a classical clathrin- and dynamin-dependent pathway and inhibition of clathrin-dependent endocytosis blocked the uptake of both receptors. However, this inhibition affected only IFN-α-induced transcriptional signaling involving STAT1 and STAT2 but not that by IFN-γ involving STAT1. Nevertheless, activated IFN-γ receptors rapidly became enriched in plasma membrane raft microdomains. The complexities of the interdependence between raft/caveolar and intracellular endocytic trafficking in the response of cells to IFN-γ in terms of STAT1 localization had been previously pointed out by Johnson and colleagues [69]. In the case of JAK/STAT signaling in Drosophila Ziedler and colleagues have identified several proteins including the early endosome marker Rab5 as able to modulate the strength of the productive transcriptional signal [70, 71] and Devergne et al [72] have provided evidence using genetic approaches for the critical dependence of productive JAK/STAT signaling on clathrin heavy chain, rab5, Hrs or deep orange mutations in all of which block endocytic trafficking.
The immunofluorescence observations of Jove and colleagues [65] showing activated STAT3 along the endocytic pathway have received strong independent confirmation in recent studies of STAT3-GFP and STAT3-YFP trafficking [41, 73]. Ng and colleagues [73] were the first to draw attention to the constitutive targeting of an N-terminally truncated STAT3β-GFP to “vesicular-like structures in the cytoplasm [of dengue virus NS1 transfected BHK cells] that may be of endosomal or lysosomal origin” (see Fig. 3 in ref. 73). In our hands [41], Fig. 3A shows one example in which two IL-6-treated Hep3B cells growing side-by-side show either the classical phenotype (almost all evident STAT3-GFP in the nucleus) or a phenotype in which considerable STAT3-GFP is localized to cytoplasmic endosome structures. Figs. 3A and 3B together show that the cytoplasmic structures were distinct from the lysosomal, endoplasmic and mitochondrial compartments. Fig. 3C confirms that endogenous PY-STAT3 in IL-6-treated Hep3B cells localized to the endosomal compartment as was first shown by Jove and colleagues [65]. Moreover Xu et al [42] used the fluorescence protease protection assay to determine that the STAT3 was on the surface of such sequestering endosomes and went on to isolate purified endosomal fractions and verified the presence of STAT3 on the surface of such vesicles by whole-mount immunoelectron microscopy.
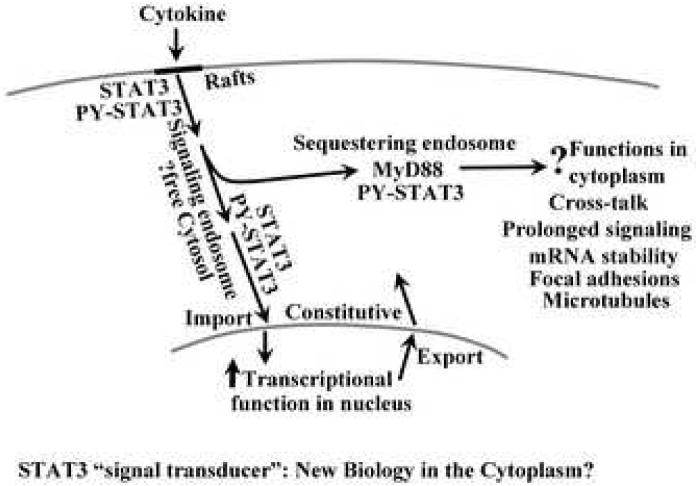
The key question remains whether membrane-associated trafficking of activated STATs is obligatorily required for productive transcriptional signaling. Thus far we are not able to answer this question in mammalian cells and it is likely that the answer will be STAT specific and cell-type specific. As an example, Devergne et al [72] showed using genetic approaches that that productive JAK/STAT signaling in the Drosophila embryo required intact endocytic trafficking. In a second clear example, O'Brien and Nathanson [74] used neuronal cell cultures with long dendrites to physically separate the location of application of the cytokine LIF and investigated STAT3 activation in the neuronal cell body at a distance. In this elegant system, they were able to show that application of LIF at a distal location led to activation of gp130 which was associated with an endosome which then traveled upstream to the cell body and there activated STAT3. Thus, in this cell type it was the activated gp130 complex resident on a “signaling endosome” that traveled towards the cell body. It is noteworthy that the cytokine-induced internalization of gp130 and the receptor chains for other cytokine and growth factors along the caveolar/endocytic pathway with continued signaling from within the cytoplasmic endocytic compartments has been well characterized [15, 19, 66, 71]. Indeed the notion of the “signaling endosome” has been in the literature for the last two decades in a variety of different contexts [9-13](Fig. 4).
Fig. 4.
Signal transduction through the cytoplasm: the signaling and sequestering endosome mechanisms. Adapted from ref. 24.
A host of mechanistic issues remain unresolved in terms of the association and trafficking of STAT proteins with the caveolar/endocytic signaling pathways. The molecular mechanisms of membrane-targeting are unknown except for the hint that clathrin heavy chain is somehow involved [14]. There appears to be an inverse relationship between levels of caveolin-1 (cav-1) and STAT3 signaling, at least in certain cell types [24, 75]. Reduced cav-1 is accompanied by upregulation of PY-STAT3 and PY-STAT3 DNA-binding activity in the cytoplasm and the nucleus in pulmonary arterial endothelial cells [24, 75]. Overexpression of cav-1 inhibits transcriptional cav-1 signaling [24]. Yet, raft/caveolar disruption also inhibits IL-6/STAT3 signaling [55, 56, 64]. For the moment it is not clear what the role of cav-1 is at the level of the plasma membrane and what its function is at the level of the targeting of cytoplasmic endosomes to the lysosomal compartment (Fig. 4; see ref. 24). In other contexts such as in myeloid cells, cav-1 has a positive-going effect on IL-6/gp130 signaling [76]. Moreover, the functional dependence of IL-6/STAT3 signaling on cholesterol-enriched plasma membrane rafts is evident even in cells with little cav-1 [55, 56, 76, 77]. It is well known that cav-1 is only one of many proteins that contribute to the structure of plasma membrane localized raft microdomains so cells with little or no cav-1 can continue to display detergent-resistant functional rafts [19, 77]. Although “preformed” membrane complexes containing JAKs, cytokine receptors and STAT proteins have been observed [55, 56, 76, 77], the mechanisms by which STAT proteins cycle on and off intracellular membranes remain unknown.
8. Activated STAT3 in sequestering endosomes: function(s) in the cytoplasm?
Until recently all discussions of the activation and trafficking of STAT proteins took place exclusively in the context of the eventual transcriptional regulatory function of these proteins. However there were already hints in the literature that activated PY-STAT3 may not always find their way into the nucleus. Silver et al [78] reported that activated STAT3 localized not only to the nuclei but also to focal adhesions in ovarian cancer cells in culture. Mukhopadhyay et al [24] have confirmed the localization of PY-STAT3 to focal adhesions in human pulmonary arterial smooth muscle cells in culture. Cao and colleagues [79] have implicated activated STAT3 in the regulation of microtubule function by antagonizing the depolymerization activity of stathmin. Impressively, Mitsuyama et al [47] published an illustration showing PY-STAT3 and CD4 double-label immunostaining of lymphoid cell collections in the gut of genetically altered SAMP1/Yit mice with ileitis showing virtually all of the PY-STAT3 in the cytoplasm of CD4+ cells with none detectable in the nucleus (Fig. 2B in ref. 47) but did not comment upon this paradigm-breaking observation.
Figs. 3A and 3B showed the IL-6-induced targeting of STAT3-GFP to cytoplasmic endosomes. Figs. 3D and 3E show the massive accumulation of bulk fluorescently-tagged STAT3 in cytoplasmic vesicular structures in Hep3B expressing the K44A dynamin II dominant-negative mutant with depletion of the nuclear pool. This DN mutant inhibits not merely the budding of endosomes at the plasma membrane but also a variety of intracellular trafficking events. This bulk accumulation of STAT3 in cytoplasmic vesicular structures in K44A-expressing cells demonstrates that the membrane-associated station in the cytoplasm is a major locale for constitutive trafficking of STAT3 within the cell. In staurosporine chase experiments Xu et al [42] discovered that the STAT3-GFP-sequestering endosomes were long-lived (t1/2 approx 15 min), did not physically move towards the nucleus and did not colocalize with early endosome markers. We suggested that these entities represented PY-STAT3 “sequestering endosomes” and may serve a function(s) in the cytoplasm per se [42].
The discovery that PY-STAT3 is almost entirely sequestered in the cytoplasm in pulmonary arterial endothelium (PAEC) in rats which develop pulmonary hypertension (PAH) subsequent to a single administration of the plant alkaloid monocrotaline (MCT) and in some of the cells in the plexiform lesions in the lungs of patients with PAH [24](Fig. 5) pointed to a function of PY-STAT3 in the cytoplasm. Indeed, in quantitative analyses, almost 70-80% of a particular cell type can display PY-STAT3 exclusively in the cytoplasm (see Table 1 in ref. 24). From a technical standpoint it is reassuring that in these analyses clear nuclear provenance of PY-STAT3 was also observed in discrete cells in the same sections (Fig. 5). In an even more dramatic demonstration, Fig. 6A illustrates sections of lungs of a patient with PAH showing the clear nuclear localization of PY-STAT3 in the nuclei of epithelial cells lining the thickened alveolar septa. In contrast Figs. 6B and 6C identify a specific cell type in the normal tracheobronchial epithelium - likely to be the basal cell (which is a progenitor cell to the other differentiated goblet, columnar and Clara cells) - which has PY-STAT3 exclusively in the cytoplasm. It is noteworthy that in both Fig. 5 and Fig. 6 the PY-STAT3 immunofluorescence was confirmed to be specific by showing that it was competed by the relevant PY-STAT3 peptide but not by the irrelevant cav-1 peptide.
Tissue-derived data such as in Figs. 5 and 6 compel us to ask about the function of PY-STAT3 in the cytoplasm in a manner separate from that in the nucleus i.e. in a manner not secondary to nuclear events (Fig. 7). Candidate possibilities include regulation of the duration of signaling to the nucleus or functions in the cytoplasm including cross-talk with other signaling pathways. One hint about the latter possibility comes from the observation that the adapter protein MyD88 colocalized with the STAT3-GFP sequestering endosomes as in Fig. 3, particularly in dynamin II K44A expressing cells [42]. This adapter has been shown to mediate the stabilization target mRNA species through the MEKK6/p38 pathway. Indeed there has been literature around for the last two decades that IL-6 not only transcriptionally activates acute phase plasma protein genes but also increases the stability of several of the acute phase protein mRNA species [42]. However the mechanisms for the latter have not yet been delineated. It could well be that IL-6, via activation of STAT3, might not only transcriptionally activate acute phase plasma protein genes at the level of the nucleus but also stabilize the respective mRNA species at the level of the cytoplasm (Fig. 7). The role of activated STAT3 in regulating mitochondrial energy generation has also emerged in recent years [80], although it is not yet clear whether that function depends on the direct transit of PY-STAT3 to the mitochondria per se or is an indirect consequence of transcriptional effects in the somatic nucleus.
Fig. 7.
Signal transduction by STAT3 to cytoplasmic destinations.
9. STAT3 in mitosis
Despite the intense focus in the STAT signaling field on mechanisms of promoting or inhibiting cell proliferation by these proteins [15, 20-23], extant literature is largely silent about what happens to STAT proteins during cell division per se. Metge et al [81] reported that STAT3 activity was required for centromere duplication during cell division and Cao and colleagues [79] showed that STAT3 affected microtubule function by antagonizing the depolymerization activity of stathmin. Although during mitosis the Golgi organelle and the nuclear membrane break up the fate of STAT proteins during this process has not been investigated. Fig. 8 shows the dramatic colocalization of STAT3 and PY-STAT3 with clathrin heavy chain and with the membrane protein LAMP-1 along elements of the mitotic spindle during mitosis [82]. These data are reflective of the growing recognition that proteins otherwise thought to be involved in endocytosis (such as dynamin II, clathrin and the endocytic coat protein ARH) have additional functional roles during mitosis [83-85]. Clearly elucidation of the biology and function of STAT proteins during mitosis per se remains ahead of us.
Fig. 8.
STAT3 and PY-STAT3 during mitosis. Double-label confocal images of cells in mitosis in cultures of Hep3B or of bovine pulmonary arterial endothelial cells were captured au naturel showing the co-localization of STAT3 with clathrin heavy chain (CHC) or with LAMP1 (Hep3B cells) or of PY-STAT3 with CHC (endothelial cells). Scale bar = 10 μm. Adapted from ref. 82.
10. Concluding remarks
Almost a decade after the reports of Lackmann et al [6] and Ndubuisi et al [7] it is time that the STAT signaling field moved away from the mindset of the original “monomer to dimer” transition paradigm. Structural mechanisms of recruitment of nonphosphorylated dimers to the cytokine receptors and their “activation” following phosphorylation likely involve novel conformational rearrangements. Moreover, nonphosphorylated STAT proteins traffic constitutively into and out of the nucleus and exert transcriptional activation functions in the nucleus. Cytokine- or growth-factor mediated activation of STAT species by virtue of Tyr- or Ser-phosphorylation events or both adds to the ability of these signal transducer proteins to transcriptionally activate distinct classes of “induced” genes. The IL-6-treated Hep3B cell illustrated in Fig. 3A showing STAT3-GFP almost exclusively in the nucleus represents a continuum with the past literature. However, the same illustration showing an adjacent cell with STAT3-GFP sequestered in endosomes in the cytoplasm represents a step ahead of extant literature. The schematic in Fig. 4 attempts to place recent discussions of the trans-cytoplasmic transit of activated STAT species within the larger context of discussions of the signaling endosome pathways in cell biology in diverse experimental systems. Whether membrane-associated trafficking of activated STATs is obligatorily required for productive transcriptional signaling in mammalian cells remains open. The best answer at the moment comes from genetic analyses in Drosphila showing defective productive STAT signaling in vivo in mutants in the endocytic pathway [72]. The tissue level-derived data from this lab (Figs. 5 and 6 and Table 1; ref. 24) and other investigators [46] evidencing the almost exclusive localization of PY-STAT3 in the cytoplasm in discrete histological cell types under specific biological conditions pose new questions about the mechanisms and functions of “activated” STAT species in the cytoplasm (Fig. 7). We suspect that novel aspects of the biology of STAT proteins in the cytoplasm, including during mitosis (Fig. 8), remain to be discovered.
Acknowledgements
I thank Drs. McKevin I. Ndubuisi, Gary Guo, Mehul Shah, Somshuvra Mukhopadhyay, Fang Xu and Mr. Kirit Patel for their careful experimental contributions to work in my laboratory in recent years. Supported by Research Grant HL073301 from the NIH-NHLBI.
Abbreviations
- cav-1
caveolin-1
- CHC
clathrin heavy chain
- GFP
green fluorescent protein tag (YFP and CFP represent analogous yellow and cyan tags)
- HRS
hepatocyte growth factor-regulated tyrosine kinase substrate
- IFN
interferon
- IL
interleukin
- IRF
interferon regulator factor
- JAK
Janus kinase
- LIF
leukemia inhibitory factor
- PAEC
pulmonary arterial endothelial cell
- STAT
signal transducer ad activator of transcription protein family
- TNF
tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Darnell JE, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- [2].Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- [3].Mertens C, Darnell JE. Snapshot: JAK-STAT signaling. Cell. 2007;131:612. doi: 10.1016/j.cell.2007.10.033. [DOI] [PubMed] [Google Scholar]
- [4].Sehgal PB, Levy DE, Hirano T, editors. Signal Transducers and Activators of Transcription. Activation and Biology, Kluwer Academic Press; Dordrecht, The Netherlands: 2003. pp. 1–746. [Google Scholar]
- [5].Shuai K, Horvath CM, Huang LH, Qureshi SA, Cowburn D, Darnell JE., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH-2 phosphotyrosyl peptide interactions. Cell. 1994;76:821–8. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- [6].Lackmann M, Harpur AG, Oates AC, Mann RJ, Gabriel A, Meutermans W, et al. Biomolecular interaction analysis of IFN gamma-induced signaling events in whole-cell lysates: prevalence of latent STAT1 in high- molecular weight complexes. Growth Factors. 1998;16:39–51. doi: 10.3109/08977199809017490. 1998. [DOI] [PubMed] [Google Scholar]
- [7].Ndubuisi MI, Guo GG, Fried VA, Etlinger JD, Sehgal PB. Cellular physiology of STAT3: where's the cytoplasmic monomer? J Biol Chem. 1999;274:25499–509. doi: 10.1074/jbc.274.36.25499. [DOI] [PubMed] [Google Scholar]
- [8].Reich NC, Liu L. Tracking STAT nuclear traffic. Nature. 2006;6:602–12. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- [9].Beattie EC, Zhou J, Grimes ML, Bunnet NW, Howe CL, Mobley WC. A signaling endosome hypothesis to explain NGF actions: potential implications for neurodegeneration. Cold Spring Harb. Symp. Quant. Biol. 1996;61:389–406. [PubMed] [Google Scholar]
- [10].Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–14. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- [11].Saxena S, Howe CL, Cosgaya JM, Steiner P, Hirling H, Chan JR, et al. Differential endocytic sorting of p75NTR and TrkA in response to NGF: a role for late endosomes in TrkA trafficking. Mol. Cell. Neurosci. 2005;28:571–87. doi: 10.1016/j.mcn.2004.11.011. [DOI] [PubMed] [Google Scholar]
- [12].Leof EB. Growth factor receptor signaling: location, location, location. Trends Cell Biol. 2000;10:343–8. doi: 10.1016/s0962-8924(00)01795-5. [DOI] [PubMed] [Google Scholar]
- [13].Howe CL. Modelling the signaling endosome hypothesis: why a drive to the nucleus is better than a (random) walk. Theor Biol Med Model. 2005;2:43. doi: 10.1186/1742-4682-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shah M, Patel K, Mukhopadhyay S, Xu F, Guo G, Sehgal PB. Membrane associated STAT3 and PY-STAT3 in the cytoplasm. J Biol Chem. 2006;281:7302–8. doi: 10.1074/jbc.M508527200. [DOI] [PubMed] [Google Scholar]
- [15].Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin-6 cytokine signalling and its regulation. Biochem J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sehgal PB, Kumar V, Guo G, Murray WC. Different patterns of regulation of Tyr-phosphorylated STAT1 and STAT3 in human hepatoma Hep3B cells by the phosphatase inhibitor orthovanadate. Arch Biochem Biophys. 2003;412:242–50. doi: 10.1016/s0003-9861(03)00050-x. [DOI] [PubMed] [Google Scholar]
- [17].Haspel RL, Darnell JE. A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc Natl Acad Sci USA. 1999;96:10188–93. doi: 10.1073/pnas.96.18.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sehgal PB. STAT-signalling through the cytoplasmic compartment: consideration of a new paradigm. Cell Signal. 2000;12:525–35. doi: 10.1016/s0898-6568(00)00098-x. [DOI] [PubMed] [Google Scholar]
- [19].Sehgal PB. Plasma membrane rafts and chaperones in cytokine/STAT signaling. Acta Biochemica Polonica. 2003;50:583–94. [PubMed] [Google Scholar]
- [20].Darnell JE. Validating Stat3 in cancer therapy. Nat Med. 2005;11:623–9. doi: 10.1038/nm0605-595. [DOI] [PubMed] [Google Scholar]
- [21].Yu H, Jove R. The STATs of cancer - new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- [22].Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, Waring P, et al. Hyperactivation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-β signaling. Nat Med. 2005;11:845–1097. doi: 10.1038/nm1282. [DOI] [PubMed] [Google Scholar]
- [23].Gao SP, Mark KG, Leslie K, Pao W, Motoi N, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–56. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mukhopadhyay S, Shah M, Patel K, Tuder RM, Sehgal PB. Cytoplasmic provenance of STAT3 and PY-STAT3 in the endolysosomal compartments in pulmonary arterial endothelial and smooth muscle cells: implications in pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2008;294:L449–68. doi: 10.1152/ajplung.00377.2007. [DOI] [PubMed] [Google Scholar]
- [25].Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007:448, 1058–62. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- [26].Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. New Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- [27].Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes and Develop. 1989;3:1362–71. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- [28].O'Hara KA, Vaghjiani RJ, Nemec AA, Klei LR, Barchowsky A. Cr(VI)-stimulated STAT3 tyrosine phosphorylation and nuclear translocation inhuman airway epithelial cells requires Lck. Biochem J. 2007;402:261–69. doi: 10.1042/BJ20061427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. 2007;282:20059–63. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- [30].Ota N, Brett TJ, Murphy TL, Fremont DH, Murphy KM. N-domain-dependent nonphosphorylated STAT4 dimers required for cytokine-driven activation. Nat Immunol. 2004;5:208–15. doi: 10.1038/ni1032. [DOI] [PubMed] [Google Scholar]
- [31].Braunstein J, Brutsaert S, Olson R, Schindler C. STATs dimerize in the absence of phosphorylation. J Biol Chem. 2003;278:34133–40. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- [32].Wenta N, Staruss H, Meyer S, Vinkemeir U. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc Natl Acad Sci. 2008;105:9238–43. doi: 10.1073/pnas.0802130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–13. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- [34].Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–12. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- [35].Liu L, McBride KM, Reich NC. SAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc Natl Acad Sci. 2005;102:8150–55. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rayanade RJ, Patel K, Ndubuisi M, Sharma S, Omura S, Etlinger JD, et al. Proteosome- and p53-dependent masking of signal transducer and activator of transcription (STAT) factors. J Biol Chem. 1997;272:4659–62. doi: 10.1074/jbc.272.8.4659. [DOI] [PubMed] [Google Scholar]
- [37].Haspel RL, Salditt-Georgieff M, Darnell JE. The rapid inactivation of nuclear turosine-phosphorylated Stat1 depends upon a protein tyrosine phosphatase. EMBO J. 1996;15:6262–8. [PMC free article] [PubMed] [Google Scholar]
- [38].Vinkemeier U. Getting the message across, STAT! Design principles of a molecular signaling event. J Cell Biol. 2004;167:197–201. doi: 10.1083/jcb.200407163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pranada AL, Metz S, Herrmann A, Heinrich PC, Muller-Newen G. Real time analysis of STAT3 nucleocytoplasmic shuttling. J Biol Chem. 2004;279:15114–23. doi: 10.1074/jbc.M312530200. [DOI] [PubMed] [Google Scholar]
- [40].Watanabe K, Saito K, Matsuda T, Tamura M, Kon S, Miyazaki T, Uede T. Molecular dynamics of STAT3 on IL-6 signaling pathway in living cells. Biochem Biophys Res Commun. 2004;324:1264–73. doi: 10.1016/j.bbrc.2004.09.187. [DOI] [PubMed] [Google Scholar]
- [41].Meyer T, Begitt A, Vinkemeier U. Green fluorescent protein-tagging reduces the nucleocytoplasmic shuttling specifically of unphosphorylated STAT1. FEBS J. 2007;274:815–26. doi: 10.1111/j.1742-4658.2006.05626.x. [DOI] [PubMed] [Google Scholar]
- [42].Xu F, Mukhopadhyay S, Sehgal PB. Live-cell imaging of interleukin-6-induced targeting of “transcription factor” STAT3 to sequestering endosomes in the cytoplasm. Am J Physiol Cell Physiol. 2007;293:C1374–82. doi: 10.1152/ajpcell.00220.2007. [DOI] [PubMed] [Google Scholar]
- [43].Deo SK, Daunert S. Green fluorescent protein mutant as label in homogeneous assays for biomolecules. Anal Biochem. 2001;289:52–9. doi: 10.1006/abio.2000.4909. [DOI] [PubMed] [Google Scholar]
- [44].Chun W, Waldo GS, Johnson GV. Split GFP complementation assay: a novel approach to quantitatively measure aggregation of tau in situ: effects of GSK3beta activation and caspase 3 cleavage. J Neurochem. 2007;103:2529–39. doi: 10.1111/j.1471-4159.2007.04941.x. [DOI] [PubMed] [Google Scholar]
- [45].Falcón-Pérez JR, Nazarian R, Sabatti C, Dell'Angelica EC. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J Cell Sci. 2005;118:5243–55. doi: 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- [46].Sobota JA, Ferraro F, Bäck N, Eipper BA, Mains RE. Not all secretory granules are created equal:partitioning of soluble contents proteins. Mol Biol Cell. 2006;17:5035–53. doi: 10.1091/mbc.E06-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mitsuyama K, Matsumoto S, Rose-John S, Suzuki A, Hara T, Tomiyasu N, et al. STAT3 activation via interleukin 6 trans-signalling contributes to ileitis in SAMP1/Yit mice. Gut. 2006;55:1263–9. doi: 10.1136/gut.2005.079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yeung Y-G, Wang Y, Einstein DB, Lee PSW, Stanley ER. Colony-stimulating factor-1 stimulates the formation of multimeric cytosolic complexes of signaling proteins and cytoskeletal components in macrophages. J Biol Chem. 1998;273:17128–37. doi: 10.1074/jbc.273.27.17128. [DOI] [PubMed] [Google Scholar]
- [49].Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB. Preassociation of STAT1 with STAT2 and STAT3 in separate signaling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134–7. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- [50].Haan S, Kortylewski M, Behrmann I, Muller-Esterl W, Heinrich PC, Schaper F. Cytoplasmic STAT proteins associate prior to activation. Biochem J. 2000;345:417–21. [PMC free article] [PubMed] [Google Scholar]
- [51].Mao X, Ren Z, Parker GN, Snydermann H, Pastorello MA, McMurray JS, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–71. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- [52].Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, et al. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-I nactivation cycle. Proc. Natl. Acad. Sci. USA. 2005;102:3966–71. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Analysis of Stat3 (signal transducer and activator of transcription 3) dimerization by fluorescence resonance energy transfer in living cells. Biochem. J. 2004;377:289–97. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schroder M, Kroeger KM, Volk HD, Eidne KA, Grutz GJ. Preassociation of nonactivated STAT3 molecules demonstrated in living cells using bioluminescence resonance enery transfer: a new model of STAT activation? Leukocyte Biol. 2004;75:792–7. doi: 10.1189/jlb.1003496. [DOI] [PubMed] [Google Scholar]
- [55].Sehgal PB, Guo GG, Shah M, Kumar V, Patel K. Cytokine signaling: STATs in plasma membrane rafts. J Biol Chem. 2002;277:12067–74. doi: 10.1074/jbc.M200018200. [DOI] [PubMed] [Google Scholar]
- [56].Shah M, Patel K, Fried VA, Sehgal PB. Interactions of STAT3 with caveolin-1 and heat shock protein 90 in plasma membrane raft and cytosolic complexes. preservation of cytokine signaling during fever. J Biol Chem. 2002;277:45662–69. doi: 10.1074/jbc.M205935200. [DOI] [PubMed] [Google Scholar]
- [57].Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, et al. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J Cell Biol. 2006;175:937–46. doi: 10.1083/jcb.200604073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Tonozuka Y, Minoshima Y, Bao YC, Moon Y, Tsubono Y, Hatori T, et al. A GTPase-activating protein binds STAT3 and is required for IL-6-induced STAT3 activation and for differentiation of a leukemic cell line. Blood. 2004;104:3550–57. doi: 10.1182/blood-2004-03-1066. [DOI] [PubMed] [Google Scholar]
- [59].Herrmann A, Vogt M, Mönnigmann M, Clahsen T, Sommer U, Haan S, et al. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007;120:3249–61. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- [60].Kumar A, Commane M, Flickinger TW, Horvath CM, Stark GR. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitute levels of caspases. Science. 1997;278:1630–2. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- [61].Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–22. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, et al. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–47. [PubMed] [Google Scholar]
- [63].Cui X, Zhang L, Luo J, Rajasekaran A, Hazra S, Cacalano N, et al. Unphosphorylated STAT 6 contributes to constitute cyclooxygenase-2 expression in human non-small cell lung cancer. Oncogene. 2007;26:4253–60. doi: 10.1038/sj.onc.1210222. [DOI] [PubMed] [Google Scholar]
- [64].Jain S, Li Y, Kumar A, Sehgal PB. Transcriptional signaling from membrane raft-associated glucocorticoid receptor. Biochem Biophys Res Commun. 2005;336:3–8. doi: 10.1016/j.bbrc.2005.08.057. [DOI] [PubMed] [Google Scholar]
- [65].Bild AH, Turkson J, Jove R. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 2002;21:3255–63. doi: 10.1093/emboj/cdf351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Scoles DR, Nguyen VD, Qin Y, Sun CS, Morrison H, Gutmann DH, et al. Neurofibromatosis 2 (NF2) tumor suppressor schwannomin and its interacting protein HRS regulate STAT signaling. Hum Mol Genet. 2002;11:3179–89. doi: 10.1093/hmg/11.25.3179. [DOI] [PubMed] [Google Scholar]
- [67].Scoles DR, Qin Y, Nguyen V, Gutmann DH, Pulst DM. HRS inhibits EGF receptor signaling in the RT4 rat schwannoma cell line. Biochem Biophys Res Commun. 2005;335:385–92. doi: 10.1016/j.bbrc.2005.07.083. [DOI] [PubMed] [Google Scholar]
- [68].Marchetti M, Monier M-N, Fradagrada A, Mitchell K, Baychelier F, Eid P, et al. Stat-mediated signaling induced by Type I and Type II interferons (IFNs) is differentially controlled through lipid microdomain associated and clathrin-dependent endocytosis of IFN receptors. Mol Biol Cell. 2006;17:2896–09. doi: 10.1091/mbc.E06-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Subramaniam PS, Johnson HM. Lipid microdomains are required sites for the selective endocytosis and nuclear translocation of IFN-γ, its receptor chain IFN-γ receptor-1, and the phosphorylation and nuclear translocation of STAT1α. J Immunol. 2002;22:1959–69. doi: 10.4049/jimmunol.169.4.1959. [DOI] [PubMed] [Google Scholar]
- [70].Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signaling components by genome-wide RNA interference. Nature. 2005;436:871–5. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- [71].Mukherjee T, Schäfer U, Zeidler MP. Identification of Drosophila genes modulating Janus Kinase/signal transducer and activator of transcription signal transduction. Genetics. 2006;172:1683–97. doi: 10.1534/genetics.105.046904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Devergne O, Ghiglione C, Noselli S. The endocytic control of JAK/STAT signaling in Drosophila. J Cell Sci. 2007;120:3457–64. doi: 10.1242/jcs.005926. [DOI] [PubMed] [Google Scholar]
- [73].Chua JJ-E, Bhuvanakantham R, Chow VT-K, Ng M-L. Recombinant non-structural 1 (NS1) protein of dengue-2 virus interacts with human STAT3β protein. Virus Research. 2005;112:85–94. doi: 10.1016/j.virusres.2005.03.025. [DOI] [PubMed] [Google Scholar]
- [74].O'Brien JJ, Nathanson NM. Retrograde activation of STAT3 by leukemia inhibitory factor in sympathetic neurons. J Neurochemistry. 2007;103:288–302. doi: 10.1111/j.1471-4159.2007.04736.x. [DOI] [PubMed] [Google Scholar]
- [75].Mathew R, Huang J, Shah M, Patel K, Gewitz M, Sehgal PB. Disruption of endothelial-cell caveolin-1α/raft scaffolding during development of monocrotaline-induced pulmonary hypertension. Circulation. 2004;110:1499–1506. doi: 10.1161/01.CIR.0000141576.39579.23. [DOI] [PubMed] [Google Scholar]
- [76].Podar K, Tai YT, Cole CE, Hideshima T, Sattler M, Hamblin A, et al. Essential role of caveolae in interleukin-6 and insulin-like growth factor I-triggered Akt-1-mediated survival of multiple myeloma cells. J Biol Chem. 2003;278:5794–801. doi: 10.1074/jbc.M208636200. [DOI] [PubMed] [Google Scholar]
- [77].Dráber P, Dráberová L, Heneberg P, Šmíd F, Farghali H, Dráber P. Preformed STAT3 transducer complexes in human HepG2 cells and rat hepatocytes. Cell Signal. 2007;11:2400–12. doi: 10.1016/j.cellsig.2007.07.018. [DOI] [PubMed] [Google Scholar]
- [78].Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT)3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–8. doi: 10.1158/0008-5472.CAN-03-3959. [DOI] [PubMed] [Google Scholar]
- [79].Ng DC, Lin BH, Lim CP, Huang G, Zhang T, Poli V, et al. Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol. 2006;172:245–57. doi: 10.1083/jcb.200503021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Potla R, Koeck T, Wegrzyn J, Cherukuri S, Shimoda K, Baker DP, et al. Tyk2 tyrosine kinase expression is required for the maintenance of mitochondrial respiration in primary pro-b lymphocytes. Mol Cell Biology. 2006;26:8562–71. doi: 10.1128/MCB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Metge B, Ofori-Acquah S, Stevens T, Balczon R. Stat3 activity is required for centrosome duplication in Chinese hamster ovary cells. J Biol Chem. 2004;279:41801–6. doi: 10.1074/jbc.M407094200. [DOI] [PubMed] [Google Scholar]
- [82].Shah MB. Membrane-assosciated trafficking of STAT3 in the cytoplasm. New York Medical College; 2006. PhD Thesis. [Google Scholar]
- [83].Thompson HM, Cao H, Chen J, Euteneur U, McNiven MA. Dynamin 2 binds γ-tubulin and participates in centrosome cohesion. Nat Cell Biol. 2004;6:335–42. doi: 10.1038/ncb1112. [DOI] [PubMed] [Google Scholar]
- [84].Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–7. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lehtonen S, Shah M, Nielsen, Iino N, Ryan JJ, et al. The endocytic protein coat protein ARH associates with motor and centrosomal proteins and is involved in centrosome assembly and cytokinesis. Mol Biol Cell. 2008;19:2949–61. doi: 10.1091/mbc.E07-05-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]