Abstract
MafA is a basic leucine zipper transcription factor expressed within the beta cells of the pancreas and is required to maintain normal glucose homeostasis as it is involved in various aspects of beta cell biology. MafA protein levels are known to increase in response to high glucose through mechanisms that have yet to be fully characterized. We investigated whether discrete intracellular signaling events control mafA expression. We found that the general kinase inhibitor staurosporine induces mafA expression without altering the stability of the protein. Inhibition of the MAP-kinase JNK mimics the effects of staurosporine on the expression of mafA. Calmodulin kinase and calcium signaling are also important in stimulating mafA expression by high glucose. However, staurosporine, JNK, and calmodulin kinase have different effects on the induction of insulin expression. These data reveal that MafA levels are tightly controlled by the coordinated action of multiple kinase pathways.
Keywords: Diabetes, Glucose, Beta Cells, Insulin, Transcription, MafA, JNK, CaMK
INTRODUCTION
Insulin gene transcription is regulated by many cis-acting elements within the 350 base pair promoter region of the insulin gene. The glucose-dependent induction of insulin transcription is mainly driven by the A3, E1, and C1 elements of the insulin promoter [1–3], which are recognized by the transcription factors Pdx-1, NeuroD1, and MafA (formerly known as RIPE-3b1), respectively [1–4]. MafA is a major activator of insulin transcription [5] and consistent with this, insulin transcript levels are reduced in the mafA knockout mice [6]. Knockdown of MafA by siRNA also decreases insulin promoter activity and insulin mRNA levels [5, 7]. In addition to its importance in insulin transcription, MafA is also required for proper insulin secretion and maintenance of islet/beta cell integrity [6]. mafA knockout mice develop diabetes due to comprised beta cell function [6].
mafA mRNA and protein levels are increased in high glucose-grown beta cells [5, 8, 9] via increased flux through the hexosamine biosynthetic pathway and O-linked glycosylation [10, 11]. MafA protein is also constitutively phosphorylated by glycogen synthase kinase (GSK) leading to its ubiquitination and subsequent proteasomal degradation [12, 13].
Glucose metabolism stimulates several signal transduction events within beta cells, including those that rely on calcium influx [14]. Intracellular calcium metabolism activates multiple kinases including calmodulin kinase (CaMK) [15–17], the extracellular signal-regulated protein kinase 1/2 (ERK1/2) [18], and protein kinase C (PKC) [16, 17]. The signaling pathways associated with these kinases coordinate a variety of events including both insulin secretion and insulin transcription [15–19].
In this report, we investigated the role of kinases in the regulation of mafA expression. We provide evidence that mafA expression increases in response to the general kinase inhibitor staurosporine in a glucose-independent manner. This suggests that a kinase-dependent signaling event suppresses mafA expression irrespective of glucose concentration. The inhibition of the JNK MAP-kinase pathway mimics the effects induced by staurosporine on mafA expression. We also show that calmodulin kinase and calcium signaling influence the expression of mafA. These data reveal new pathways by which beta cells ultimately control the total levels of MafA.
MATERIALS AND METHODS
Chemicals
D-glucose, EGTA, and nifedipine were obtained from Sigma (St. Louis, MO). SP600125, JNK inhibitor V, staurosporine, and ionomycin were obtained from Calbiochem (San Diego, CA). Cell culture media were obtained from Atlanta Biologicals (Lawrenceville, GA).
Cell culture
Mouse insulinoma 6 (MIN6) cells of passages 20–30 were cultured in Dulbecco’s modified Eagle media (DMEM) containing 25 mM glucose, 10% (v/v) fetal bovine serum, 1% (v/v) penicillin/streptomycin, 2 mM glutamine, and 100 μM β-mercaptoethanol [20].
Western blotting
Western blotting was preformed as described previously [10]. Proteins were separated by SDS-polyacrylamide gel electrophoresis and subsequently transferred onto nitrocellulose membranes (Osmonics Inc.). Membranes were blocked for 1 hour in 1X TTBS (20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton-X 100) supplemented with 5% Carnation non-fat dry milk. After blocking, membranes were incubated overnight at 4°C with either MafA (Calbiochem), TATA-binding protein, TBP, CaMKII or phospho-CaMKII (Santa Cruz Biotechnology Inc.) antibody. Membranes were washed four times for 10 minutes in TTBS and subsequently incubated for 1 hour at room temperature with a secondary horseradish peroxidase-conjugated antibody and proteins were visualized by the ECL chemiluminescent detection system (Amersham Biosciences).
Real-time qRT-PCR
Real-time qRT-PCR analysis was performed as previously described [10]. Briefly, RNA was isolated from MIN6 cells using the RNeasy® Mini Kit (Qiagen). First-strand cDNA synthesis was performed using the Enhanced Avian HS RT-PCR Kit (Sigma), after treatment with DNase I (Sigma) to remove contaminating genomic DNA. Real-time amplification of the cDNA was performed using the Brilliant SYBR Green QPCR Master Mix (Stratagene). The oligonucleotide primers used in this study are listed in Supplemental Table 1. Real-time PCR reactions were performed on an Mx4000 instrument (Stratagene) and the data were analyzed using the 2−ΔΔCt method as we have previously described [10].
RESULTS
Staurosporine Induces mafA Expression
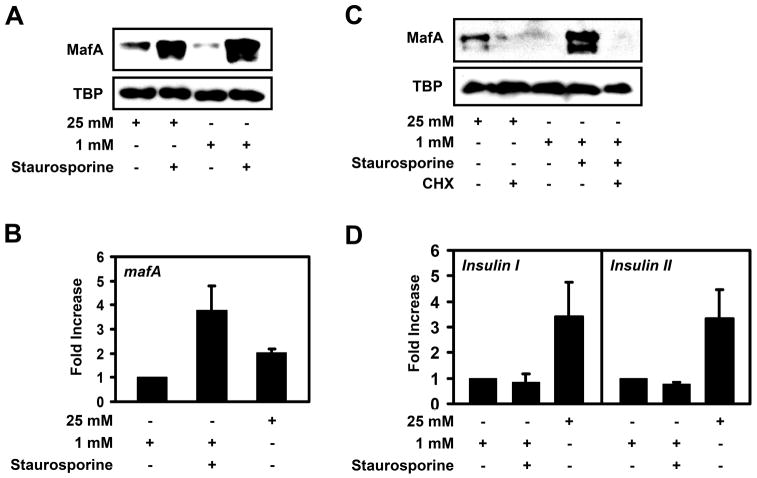
To test whether MafA levels are regulated by a cellular kinase(s), we treated high or low glucose-grown MIN6 beta cells with the general protein kinase inhibitor staurosporine. The mouse insulinoma MIN6 cell line is a widely used model system to study glucose metabolism and signaling pathways regulating insulin production and beta-cell function. MIN6 cells are responsive to physiological concentration of glucose and display glucose induced insulin secretion and insulin transcription [20]. Incubation of MIN6 cells with staurosporine dramatically increased MafA levels in a glucose-independent manner without altering the levels of TBP (Fig. 1A). Real-time RT-PCR analysis revealed that the increase in MafA protein levels after staurosporine treatment is due to increased mafA mRNA levels. We found that mafA mRNA levels were about 2-fold increased in high glucose incubated MIN6 cells as compared to low glucose alone (Fig. 1B). Staurosporine induced mafA expression nearly 4-fold above that within cells treated with low glucose alone (Fig. 1B). Treatment of MIN6 cells with the protein phosphatase inhibitor okadaic acid blocked the induction of MafA protein expression by high glucose (Supplemental Fig. 1). Overall, these data indicate that a phosphorylation event(s) suppresses mafA expression in both low and high glucose incubated MIN6 cells.
Figure 1. Staurosporine induces mafA, but not insulin expression.
A, MIN6 cells were cultured in the presence of 25 mM or 1 mM glucose +/− 100 nM staurosporine overnight (16 to 18 hours) and MafA and TBP protein levels were analyzed by Western blotting (n > 3). B, mafA mRNA levels, in MIN6 cells cultured overnight on 25 mM, 1 mM, or 1 mM glucose plus 100 nM staurosporine, were analyzed by real-time RT-PCR (n=4). C, Western blot analysis of the stability of MafA and TBP in MIN6 cells incubated with 25 mM glucose +/− 40 μM cycloheximide (CHX), 1 mM glucose, and 1 mM glucose and 100 nM staurosporine +/− 40 μM cycloheximide (n > 4). D, Real-time RT-PCR analysis of insulin mRNA levels in MIN6 cells cultured in 25 mM, 1 mM, or 1 mM glucose plus 100 nM staurosporine overnight (n=4).
Since MafA has been shown to be degraded via the proteasome [12, 13], we next tested whether staurosporine also prevents MafA degradation. For this, we treated MIN6 cells with low glucose and cycloheximide and with or without staurosporine. While the addition of staurosporine to low glucose incubated MIN6 cells drastically increased MafA protein levels, it did not prevent the degradation of MafA in the presence of cycloheximide (Fig. 1C). Proteasome inhibition even in the presence of cycloheximide prevents MafA degradation (data not shown) as previously reported [12, 13]. These data suggest that while staurosporine induces mafA expression, it has no effect on MafA protein stability.
Since MafA is a major regulator of insulin expression [5, 8, 9, 21–24], we next tested whether staurosporine treatment causes induction of insulin transcription. As illustrated in Fig. 1D, insulin I or insulin II mRNA levels in MIN6 cells treated with 1 mM glucose plus staurosporine were similar to low glucose only grown cells. However, as expected, 25 mM glucose induced both insulin I and insulin II expression (Fig. 1D). These data are consistent with previous work, demonstrating that staurosporine inhibits insulin promoter activity [25].
Inhibition of JNK Induces mafA Expression
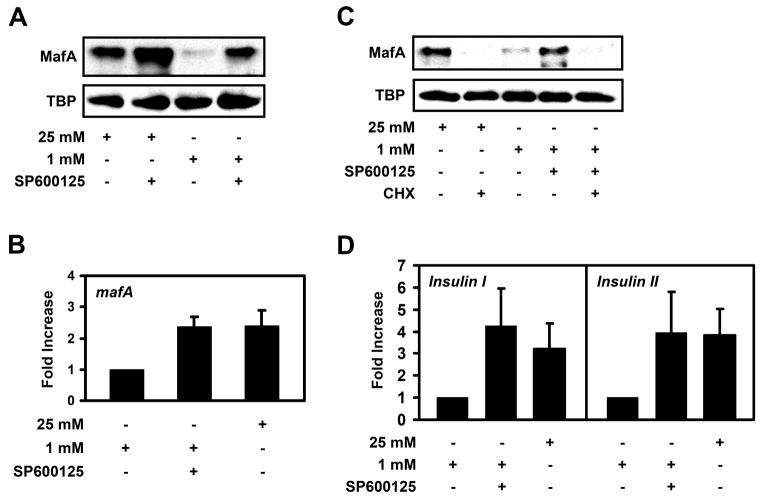
After screening many kinase inhibitors, we found that only the inhibition of JNK mimicked the effect staurosporine produced on mafA expression. To test whether JNK regulates MafA levels, we treated MIN6 cells with the JNK inhibitor SP600125. As shown in Supplemental Fig. 2A, phosphorylation of JNK was inhibited by treatment with SP600125. MafA protein levels increased in both high and low glucose-grown MIN6 cells treated with SP600125 (Fig. 2A). Similar effects on MafA expression levels were observed when a different JNK inhibitor was used (Supplemental Fig. 2B). Real-time RT-PCR analysis indicated that low glucose-grown MIN6 cells treated with SP600125 expressed about 2-fold more mafA than cells incubated in low glucose alone (Fig. 2B). Furthermore, like staurosporine, JNK inhibition did not alter the stability of MafA after the addition of cycloheximide, suggesting that JNK does not regulate MafA degradation (Fig. 2C). Overall, these data are consistent with the idea that JNK has an inhibitory effect on the expression of mafA.
Figure 2. Inhibition of JNK induces mafA and insulin expression.
A, MIN6 cells were cultured in the presence of 25 mM or 1 mM glucose +/− 20 μM SP600125 overnight. MafA and TBP protein levels were analyzed by Western blotting (n=3). B, MIN6 cells were cultured in 1 mM glucose, 1 mM glucose plus 20 μM SP600125, or 25 mM glucose overnight and real-time RT-PCR was utilized to measure mafA mRNA levels (n=3). C, Western blot analysis MafA and TBP protein levels in MIN6 cells incubated with 25 mM glucose +/− 40 μM cycloheximide (CHX), 1 mM glucose, and 1 mM glucose and 20 μM SP600125 +/− 40 μM cycloheximide (n=3). D, Real-time RT-PCR analysis of insulin expression in MIN6 cells incubated with 25 mM glucose, 1 mM glucose, or 1 mM glucose plus 20 μM SP600125 overnight (n=3).
Because JNK inhibition induced mafA expression similar to staurosporine treatment, we next tested whether JNK inhibition had any effect on the expression of insulin in low glucose-grown cells. In contrast to staurosporine treatment, both insulin I and insulin II mRNA levels were increased in MIN6 cells treated with 1 mM glucose plus SP600125 (Fig. 2D). Therefore, the expression of mafA correlates with an increase in insulin expression in cells treated with the JNK inhibitor SP600125. These data are consistent with previous reports indicating JNK suppresses insulin expression [26, 27].
Calmodulin kinase and calcium signaling regulate mafA expression on high glucose
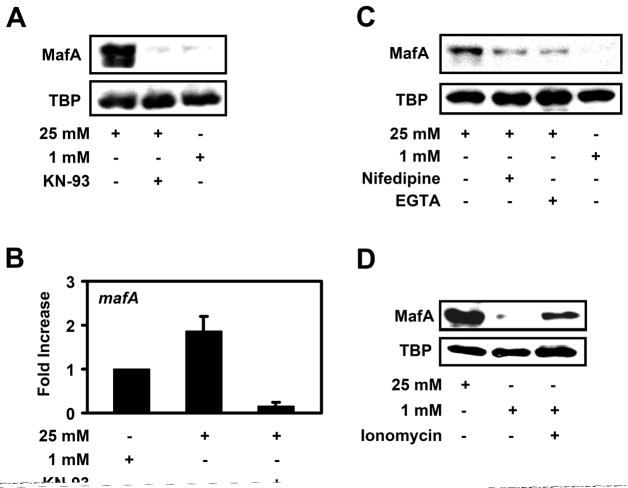
Staurosporine is known to inhibit many kinases, including calmodulin kinase (CaMK). Our screen for various kinase inhibitors revealed a role for CaMK in regulating MafA levels. We treated MIN6 cells with the CaMK inhibitor KN-93, which blocks CaMKII activation (Supplemental Fig. 3A). Treatment of MIN6 cells with KN-93 lead to a decline in MafA protein levels in high glucose-grown cells (Fig. 3A), while there was no effect on MafA levels on low glucose treated MIN6 cells (Supplemental Fig. 3B). This suggests that CaMK activity is required to induce mafA expression under high glucose conditions. Real-time RT-PCR analysis revealed that KN-93 decreased mafA expression below that observed in MIN6 cells grown in low glucose alone (Fig. 3B). These data are consistent with the idea that CaMK plays a role in mediating the high glucose-dependent expression of mafA.
Figure 3. CaMK and calcium signaling regulate mafA expression.
A, MIN6 cells were cultured in the presence of 25 mM glucose +/−- 10 μM of the CaMK inhibitor KN-93 or 1 mM glucose overnight and MafA and TBP protein levels were analyzed by Western blotting (n > 3). B, MIN6 cells were cultured in 1 mM glucose, 25 mM glucose +/− 10 μM KN-93 overnight and mafA mRNA levels were measured by real-time RT-PCR (n=4). C, MIN6 cells were cultured overnight in 25 mM glucose, 25 mM glucose plus 10 μM nifedipine, 25 mM glucose plus 1 mM EGTA, or 1 mM glucose. MafA and TBP protein levels were analyzed by Western blotting. D, After overnight incubation of MIN6 cells in 25 mM glucose or 1 mM glucose +/− 1 μM ionomycin, Western blotting was utilized to measure MafA and TBP levels.
Glucose metabolism is known to induce an increase in intracellular calcium levels leading to CaMK activation [15, 28]. Thus, we next determined whether calcium levels may affect mafA expression. We treated high glucose-grown MIN6 cells with the calcium channel blocker nifedipine or with the calcium chelating agent EGTA. Both nifedipine and EGTA blunted, but did not completely inhibit MafA protein expression induced by high glucose (Fig. 3C). However, the calcium ionophore ionomycin led to an increase in MafA levels in low glucose-grown MIN6 cells (Fig. 3D). These data strongly suggests that CaMK and calcium signaling mediate the high glucose-dependent expression of mafA in beta cells.
CaMK inhibition decreases insulin expression
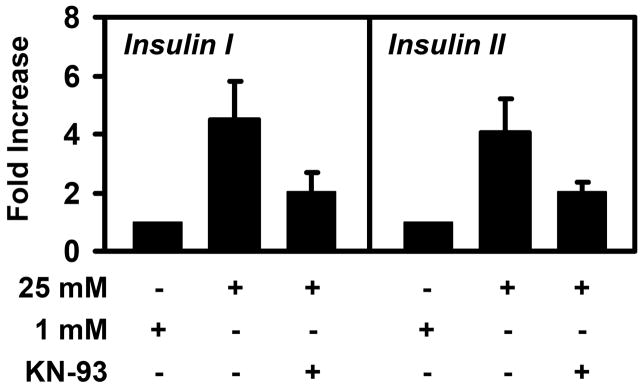
Calcium signaling and CaMK is known to regulate insulin secretion and insulin transcription [15–17, 28–30]. To establish a correlation between the effects of KN-93 on mafA expression and the involvement of CaMK in insulin expression, we measured insulin mRNA levels in high glucose-grown MIN6 cells treated with KN-93. Consistent with previous work [30–32], the high glucose-dependent expression of insulin I and insulin II were blunted, but not completely inhibited by the inhibition of CaMK (Fig. 4). These data indicate that calcium signaling and CaMK affect both mafA and insulin expression.
Figure 4. Inhibition of CaMK suppresses insulin expression.
MIN6 cells were cultured in 1 mM glucose or 25 mM glucose +/− 10 μM KN-93 overnight and insulin levels were measured by real-time RT-PCR. The data shown are averages of three to four measurements +/− standard deviations.
DISCUSSION
Glucose metabolism induces mafA expression via flux through the hexosamine biosynthetic pathway followed by an unknown O-linked glycosylation event [10]. MafA protein levels are also controlled through ubiquitin-mediated proteasomal degradation after phosphorylation of MafA by GSK [12, 13]. Other signaling events controlling MafA levels are unknown.
Here we demonstrate that the kinase inhibitor staurosporine increases mafA expression independent of glucose concentration. Staurosporine has previously been shown to inhibit insulin transcription and insulin secretion [25, 33–35]. Consistent with these data, we observed that despite the fact that staurosporine induced mafA expression in low glucose-grown MIN6 cells, insulin expression was not increased. This is likely because stauroporine inhibits many kinases that may be required for proper insulin expression.
Staurosporine inhibits the expression of many genes including several transcription factors, such as c-Jun [36]. It is possible that staurosporine blocks the expression of a transcriptional regulator(s) that normally suppresses mafA expression, thereby increasing expression of mafA. On the other hand, staurosporine also activates some transcription factors like NF-κB. As such, staurosporine increases the synthesis of some NF-κB target genes [37, 38]. Therefore, perhaps staurosporine activates a transcription factor necessary for inducing mafA expression.
The inhibition of the MAP-kinase JNK mimics the effects of staurosporine on mafA expression while the inhibition of other MAP-kinases like ERK do not affect the expression of mafA (data not shown). There are several putative binding sites for JNK-sensitive transcription factors such as c-Jun within the mafA promoter (data not shown). Beyond the expression of mafA, JNK also plays a role in inhibiting insulin expression under oxidative stress conditions [26, 27, 39]. We found that the inhibition of JNK promotes insulin expression in low glucose-grown beta cells, suggesting a role for JNK in the suppression of insulin expression under low glucose conditions.
We have also shown that KN-93, nifedipine, and EGTA blocked the glucose-dependent increase in mafA expression, while the calcium ionophore ionomycin induces mafA expression in low glucose-grown beta cells. This suggests that CaMK and calcium signaling are important for the induction of mafA by high glucose. CaMKs are known to regulate gene expression via the activation of the cAMP response element-binding protein (CREB). CREB regulates the expression of many genes via cAMP response elements (CREs) within target gene promoters [40, 41]. There are potential binding sites for CREB and for several other calcium-sensitive transcription factors within the mafA promoter (data not shown).
In summary, we report that the JNK MAP-kinase pathway suppresses mafA expression irrespective of glucose concentration. CaMK and calcium signaling also play a role in high glucose-dependent expression of mafA. Future studies will address the exact molecular mechanisms by which JNK and CamK regulate mafA expression and determine how this information can be utilized to control the levels and/or activity of MafA in beta cells.
Supplementary Material
Footnotes
ACKNOWLEDGMENTS: This work was supported by NIH/NIDDK grant 5R01DK067581 and the National Center for Research Resources Grant P20 RR20171 and a Pre-doctoral Fellowship from the American Heart Association, Great Rivers Affiliate, to NLV.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Melloul D, Marshak S, Cerasi E. Diabetologia. 2002;45:309–326. doi: 10.1007/s00125-001-0728-y. [DOI] [PubMed] [Google Scholar]
- 2.Ohneda K, Ee H, German M. Semin Cell Dev Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- 3.Poitout V, Hagman D, Stein R, Artner I, Robertson RP, Harmon JS. J Nutr. 2006;136:873–876. doi: 10.1093/jn/136.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aramata S, Han SI, Kataoka K. Endocr J. 2007 doi: 10.1507/endocrj.kr-101. [DOI] [PubMed] [Google Scholar]
- 5.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S. Mol Cell Biol. 2005;25:4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchiya M, Yoshida T, Taniguchi S, Yasuda K, Maeda A, Hayashi A, Tanaka J, Shigemoto M, Nitta K, Tsuchiya K. Biochem Biophys Res Commun. 2007;356:129–135. doi: 10.1016/j.bbrc.2007.02.105. [DOI] [PubMed] [Google Scholar]
- 8.Kajihara M, Sone H, Amemiya M, Katoh Y, Isogai M, Shimano H, Yamada N, Takahashi S. Biochem Biophys Res Commun. 2003;312:831–842. doi: 10.1016/j.bbrc.2003.10.196. [DOI] [PubMed] [Google Scholar]
- 9.Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Handa H. J Biol Chem. 2002;277:49903–49910. doi: 10.1074/jbc.M206796200. [DOI] [PubMed] [Google Scholar]
- 10.Vanderford NL, Andrali SS, Ozcan S. J Biol Chem. 2007;282:1577–1584. doi: 10.1074/jbc.M605064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrali SS, Sampley ML, Vanderford NL, Ozcan S. Biochem J. 2008;415:1–10. doi: 10.1042/BJ20081029. [DOI] [PubMed] [Google Scholar]
- 12.Han SI, Aramata S, Yasuda K, Kataoka K. Mol Cell Biol. 2007;27:6593–6605. doi: 10.1128/MCB.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rocques N, Abou Zeid N, Sii-Felice K, Lecoin L, Felder-Schmittbuhl MP, Eychene A, Pouponnot C. Mol Cell. 2007;28:584–597. doi: 10.1016/j.molcel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Mears D. J Membr Biol. 2004;200:57–66. doi: 10.1007/s00232-004-0692-9. [DOI] [PubMed] [Google Scholar]
- 15.Easom RA. Diabetes. 1999;48:675–684. doi: 10.2337/diabetes.48.4.675. [DOI] [PubMed] [Google Scholar]
- 16.Jones PM, Persaud SJ. Endocr Rev. 1998;19:429–461. doi: 10.1210/edrv.19.4.0339. [DOI] [PubMed] [Google Scholar]
- 17.Nesher R, Anteby E, Yedovizky M, Warwar N, Kaiser N, Cerasi E. Diabetes. 2002;51(Suppl 1):S68–73. doi: 10.2337/diabetes.51.2007.s68. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence M, Shao C, Duan L, McGlynn K, Cobb MH. Acta Physiol (Oxf) 2008;192:11–17. doi: 10.1111/j.1748-1716.2007.01785.x. [DOI] [PubMed] [Google Scholar]
- 19.Zawalich WS, Bonnet-Eymard M, Zawalich KC. Front Biosci. 1997;2:d160–172. doi: 10.2741/a180. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, Yamamura K. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 21.Matsuoka TA, Artner I, Henderson E, Means A, Sander M, Stein R. Proc Natl Acad Sci U S A. 2004;101:2930–2933. doi: 10.1073/pnas.0306233101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Mol Cell Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olbrot M, Rud J, Moss LG, Sharma A. Proc Natl Acad Sci U S A. 2002;99:6737–6742. doi: 10.1073/pnas.102168499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A, Fusco-DeMane D, Henderson E, Efrat S, Stein R. Mol Endocrinol. 1995;9:1468–1476. doi: 10.1210/mend.9.11.8584024. [DOI] [PubMed] [Google Scholar]
- 25.Goodison S, Kenna S, Ashcroft SJ. Biochem J. 1992;285(Pt 2):563–568. doi: 10.1042/bj2850563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneto H, Xu G, Fujii N, Kim S, Bonner-Weir S, Weir GC. J Biol Chem. 2002;277:30010–30018. doi: 10.1074/jbc.M202066200. [DOI] [PubMed] [Google Scholar]
- 27.Kawamori D, Kaneto H, Nakatani Y, Matsuoka TA, Matsuhisa M, Hori M, Yamasaki Y. J Biol Chem. 2006;281:1091–1098. doi: 10.1074/jbc.M508510200. [DOI] [PubMed] [Google Scholar]
- 28.Wenham RM, Landt M, Easom RA. J Biol Chem. 1994;269:4947–4952. [PubMed] [Google Scholar]
- 29.Tabuchi H, Yamamoto H, Matsumoto K, Ebihara K, Takeuchi Y, Fukunaga K, Hiraoka H, Sasaki Y, Shichiri M, Miyamoto E. Endocrinology. 2000;141:2350–2360. doi: 10.1210/endo.141.7.7553. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Murao K, Sayo Y, Imachi H, Cao WM, Ohtsuka S, Niimi M, Tokumitsu H, Inuzuka H, Wong NC, Kobayashi R, Ishida T. Diabetes. 2004;53:1475–1481. doi: 10.2337/diabetes.53.6.1475. [DOI] [PubMed] [Google Scholar]
- 31.Ban N, Yamada Y, Someya Y, Ihara Y, Adachi T, Kubota A, Watanabe R, Kuroe A, Inada A, Miyawaki K, Sunaga Y, Shen ZP, Iwakura T, Tsukiyama K, Toyokuni S, Tsuda K, Seino Y. Diabetes. 2000;49:1142–1148. doi: 10.2337/diabetes.49.7.1142. [DOI] [PubMed] [Google Scholar]
- 32.Osterhoff M, Mohlig M, Schwanstecher M, Seufert J, Ortmann J, Schatz H, Pfeiffer AF. Cell Calcium. 2003;33:175–184. doi: 10.1016/s0143-4160(02)00227-0. [DOI] [PubMed] [Google Scholar]
- 33.Gray E, Muller D, Squires PE, Asare-Anane H, Huang GC, Amiel S, Persaud SJ, Jones PM. J Endocrinol. 2006;190:703–710. doi: 10.1677/joe.1.06891. [DOI] [PubMed] [Google Scholar]
- 34.Zawalich WS, Zawalich KC. Mol Cell Endocrinol. 2001;177:95–105. doi: 10.1016/s0303-7207(01)00422-1. [DOI] [PubMed] [Google Scholar]
- 35.Zawalich WS, Zawalich KC, Ganesan S, Calle R, Rasmussen H. Biochem J. 1991;279( Pt 3):807–813. doi: 10.1042/bj2790807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zayzafoon M, Stell C, Irwin R, McCabe LR. J Cell Biochem. 2000;79:301–310. doi: 10.1002/1097-4644(20001101)79:2<301::aid-jcb130>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 37.Chabot-Fletcher M, Breton JJ. Biochem Pharmacol. 1998;56:71–78. doi: 10.1016/s0006-2952(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 38.Mahon TM, Matthews JS, O’Neill LA. Biochem J. 1997;325( Pt 1):39–45. doi: 10.1042/bj3250039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawamori D, Kajimoto Y, Kaneto H, Umayahara Y, Fujitani Y, Miyatsuka T, Watada H, Leibiger IB, Yamasaki Y, Hori M. Diabetes. 2003;52:2896–2904. doi: 10.2337/diabetes.52.12.2896. [DOI] [PubMed] [Google Scholar]
- 40.Means AR. Mol Endocrinol. 2000;14:4–13. doi: 10.1210/mend.14.1.0414. [DOI] [PubMed] [Google Scholar]
- 41.Shaywitz AJ, Greenberg ME. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.