Abstract
Background
Previous studies of brain and peripheral tissues in schizophrenia patients have indicated impaired energy supply to the brain. A number of studies have also demonstrated dysfunction of the microvasculature in schizophrenia patients. Together these findings are consistent with a hypothesis of blood-brain barrier dysfunction in schizophrenia. In this study, we have investigated the cerebral vascular endothelium of schizophrenia patients at the level of transcriptomics.
Methodology/Principal Findings
We used laser capture microdissection to isolate both microvascular endothelial cells and neurons from post mortem brain tissue from schizophrenia patients and healthy controls. RNA was isolated from these cell populations, amplified, and analysed using two independent microarray platforms, Affymetrix HG133plus2.0 GeneChips and CodeLink Whole Human Genome arrays. In the first instance, we used the dataset to compare the neuronal and endothelial data, in order to demonstrate that the predicted differences between cell types could be detected using this methodology. We then compared neuronal and endothelial data separately between schizophrenic subjects and controls. Analysis of the endothelial samples showed differences in gene expression between schizophrenics and controls which were reproducible in a second microarray platform. Functional profiling revealed that these changes were primarily found in genes relating to inflammatory processes.
Conclusions/Significance
This study provides preliminary evidence of molecular alterations of the cerebral microvasculature in schizophrenia patients, suggestive of a hypo-inflammatory state in this tissue type. Further investigation of the blood-brain barrier in schizophrenia is warranted.
Introduction
Despite decades of research and numerous competing hypotheses, our understanding of the pathophysiology of schizophrenia remains unclear, with adverse consequences for both diagnosis and treatment. In recent years an increasing body of evidence has pointed towards altered glucose metabolism in schizophrenic patients. In addition to the major findings of hypofrontality in patients obtained using brain imaging methods [1], numerous post mortem studies have shown alterations in the expression of genes and proteins involved in major energy metabolism pathways [2]–[6], and studies of peripheral tissues have also detected metabolic alterations in first onset and drug naïve patients [7], [8]. Examination of these data suggests an abnormality in glucose utilization in the brains of patients which may arise from impaired supply of energy substrates such as glucose and lactate [5], [7]. Such findings are consistent with an hypothesis of blood-brain barrier impairment in schizophrenia [9]. This hypothesis proposes that disruption in the coupling of cerebral blood flow to neuronal metabolic needs may be upstream of all conceivable functional neuronal abnormalities in schizophrenia.
A small but growing body of evidence points towards dysfunction of the microvasculature in schizophrenia. The niacin skin flush response has been widely reported to be abnormal in schizophrenic patients [10]–[14] and may be explained by abnormal vasodilatation. Furthermore, some studies have shown a decreased resting cerebral blood flow [15], [16] and decreased cerebral vascular volume in schizophrenic patients [17], with other data suggesting an increase in blood volume in certain brain regions [18], consistent with abnormalities of the cerebral microvasculature. A recent stereological study of capillary length density in schizophrenia brain tissue failed to find differences between schizophrenic and control subjects [19], but the authors reconcile these two apparently conflicting findings by proposing dysfunction of the cerebral microvasculature at a molecular, rather than structural level. However, few if any molecular studies of the cerebral vasculature in schizophrenia have been attempted, and existing quantitative molecular studies based on tissue homogenate or sections are unlikely to include a signal from the relevant cells as vasculature accounts for only 0.1% of whole brain tissue [20].
In this study we have attempted to characterise the cerebral microvasculature of schizophrenia patients by using laser microdissection to isolate cells from post mortem prefrontal cortex tissue. Laser microdissection has been widely touted as a major advance in molecular brain research [21], but has rarely [2], [22] been applied to the human postmortem brain due to the technical challenges raised by working with small amounts of tissue and the variability that may be introduced at various stages of the analytical process. Thus as a first step to check the integrity of the data, we compared data from endothelial cells to data from neurons in order to demonstrate whether the predicted differences between these two cells types could be detected. We then proceeded to investigate gene expression differences in these cell types in schizophrenia using microarrays. Although microarray technology has been demonstrated to be robust and reproducible in careful hands, validation of the results at the technical level is essential to increase confidence in a study. Quantitative real-time PCR, the most commonly used validation tool, is limited in that only a small number of genes can practically be measured within a single study, and the necessity to normalize to a so-called “housekeeping” gene introduces high levels of experimental noise. Thus in the present study we have taken the approach of using two array platforms, each possessing different probe design, synthesis and attachment strategies and different hybridization kinetics and lab procedure. Using this approach not only can mRNA levels be validated, but also differences in microarray methodology, normalization and data processing methods.
Methods
Tissue collection
Consent: Human brain tissue was obtained from the Array collection of the Stanley Medical Research Institute (Bethesda, USA). Tissue was collected post mortem from patients and controls with full informed consent obtained from a first degree relative after death in compliance with the Declaration of Helsinki. The consent was obtained by questionnaires conducted over the phone and signed by two witnesses. All patient data are anonymised. Exemption from IRB approval was granted by the Uniformed Services University of Health Sciences IRB on the grounds that specimens were obtained via informed donation from cadaveric material in accordance with federal and state regulations, the research did not encompass genetic linkage studies, and all samples were de-identified and personal information anonymised.
Fresh-frozen gray matter tissue from dorsolateral prefrontal cortex (Brodmann area 9) of 12 schizophrenia patients and 12 matched control individuals was used in the study. The cell type analysis also included 12 subjects with bipolar disorder (see Supplementary Information S1).
Laser capture microdissection
Cells were visualised using a rapid fluorescence immunostaining method designed for optimal RNA preservation. Examples of immunostaining are shown in Fig 1. 15 µm tissue sections were cut onto polyethylene naphthalate membrane slides (Zeiss) and fixed for 10 minutes in acetone. After air drying, sections were incubated with either rabbit anti-von Willebrand factor (Chemicon) or mouse anti-neurofilament-160/200 kD (Cambridge Bioscience) for 5 minutes, followed by brief washing in RNase-free phosphate buffered saline (PBS) and incubation in secondary antibody for 5 minutes (Cy3-conjugated goat anti-rabbit IgG or Cy2-conjugated goat anti-mouse IgG (Jackson Immunoresearch). All antibodies were used at 1∶20 dilution with 1 unit/ml RNase inhibitor (GE Healthcare) in RNase-free PBS (Ambion). These incubation conditions were found to give the best staining for cell identification together with optimal RNA preservation. Following antibody incubation and brief washing in PBS, sections were then dehydrated through ethanol series and cell capture was initiated immediately. Laser capture microdissection was carried out using the PALM microlaser system [23]. 1000 neurons were captured from each subject, in two batches of 500, or an equivalent area of vascular endothelium (approximately 40 0000 µm2). Pyramidal neurons were selected based on staining and morphology. Following capture, RNA was extracted from cells using the PALM RNA extraction kit (Zeiss) and amplified through 2 rounds using the RiboAmp HS kit (Arcturus). The resulting aRNA was assessed on an Agilent Bioanalyser Nanochip to determine length of RNA transcripts in the samples. aRNA profiles with jagged curves or pronounced skews to the left, indicating degradation of the RNA, were eliminated from the analysis.
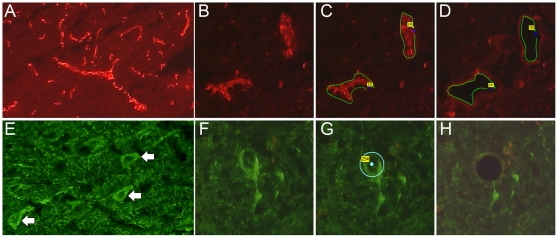
Figure 1. Examples of fluorescence immunostaining.
A rapid immunostaining method was developed to identify endothelial cells (A–D) and neurons (E–H) in human prefrontal cortex tissue, whilst maintaining optimal RNA preservation. (A, E) low power (×10) images of cell staining in prefrontal cortex tissue; (B, F) high power (×40) image of cells before capture; (C, G) cells selected ready for capture (D, H) tissue sections post-capture.
Microarray hybridisation
Amplified RNA was converted to cDNA using Round 2 components of the RiboAmp HS kit, labelled by in vitro transcription in the presence of biotinylated UTP (Codelink Expression Assay kit, GE Healthcare), and purified using YM-30 columns (Microcon).
Labelled RNA was hybridised to both Affymetrix and Codelink chips according to manufacturers' recommendations. Balanced numbers of patient and control samples were included in each hybridisation batch.
Data analysis
All datasets (endothelial and neuronal, disease and control samples) were subjected to normalisation and quality control measures together, within each array platform. Only samples which passed quality control on both platforms were included in the final dataset, to facilitate cross-platform validation. An outline of the data analysis workflow is shown in Fig 2.
Figure 2. Diagram showing overview of experimental workflow.
Affymetrix arrays
Data pre-processing
Quality control protocols for Affymetrix microarray data derived from human postmortem brains were applied as previously described [24], and samples which did not pass were removed from the dataset. Expression measures were computed for each of the probesets on each of the GeneChips in the dataset using the robust multichip average (RMA) method [25], which is implemented in the BioConductor package ‘Affy’ [26]. The ‘Affy’ package was also used to generate RNA digestion plots which allow any 5′ to 3′ trend to be visualized. A linear regression of expression values on the logarithm (base 2) of slope of the RNA digestion plots for each probeset was performed and the residuals from the regression were assigned as expression values for further analysis. This transformation effectively corrected for any systematic error in the data introduced by 3′ signal bias and significantly improved the quality of the data (see Supplementary Information S1). Following transformation, all 54647 probes were included in the analysis. The data have been submitted to GEO (www.ncbi.nlm.nih.gov/geo/), accession numbers GSM318410-GSM318441; series record GSE12679.
Demographics
Demographic variables for the disease-based analysis are shown in Tables 1 & 2. No significant differences between patients and controls were found for pH, PMI or age in either cell type. The distribution of demographic variables for the cell-type analysis can be found in Supplementary Information S1.
Table 1. Endothelial sample demographics.
| Group (n) | Control (7) | Schizophrenia (9) | p value |
| Age (yrs; mean±SD) | 42.7±7.8 | 43.2±10.6 | 0.917 |
| Gender (n; F/M) | 1/6 | 3/6 | n/a |
| pH (mean±SD) | 6.6±0.2 | 6.5±0.3 | 0.363 |
| PMI (hours; mean±SD) | 20.3±10.4 | 29.6±10.1 | 0.094 |
| Diagnosis (n; paranoid/undifferentiated) | n/a | 2/7 | n/a |
| Medicated at TOD (n; yes/no) | 0 | 8/1 | n/a |
| Lifetime medication (fluphenazine mg equiv; mean±SD) | 0 | 112838±154662 | n/a |
Demographic variables for endothelial samples included in the disease-based analysis (F: female; M: male; PMI: post mortem interval; SD: standard deviation; TOD: time of death).
Table 2. Neuronal sample demographics.
| Group (n) | Control (6) | Schizophrenia (5) | p value |
| Age (yrs; mean±SD) | 41.2±7.2 | 45.0±5.4 | 0.353 |
| Gender (n; F/M) | 2/4 | 2/3 | n/a |
| pH (mean±SD) | 6.6±0.2 | 6.6±0.1 | 0.682 |
| PMI (hours; mean±SD) | 30.0±18.6 | 28.4±7.9 | 0.862 |
| Diagnosis (n; paranoid/undifferentiated) | n/a | 1/4 | n/a |
| Medicated at TOD (n; yes/no) | 0 | 4/1 | n/a |
| Lifetime medication (fluphenazine mg equiv; mean±SD) | 0 | 44010±35056 | n/a |
Demographic variables for neuronal samples included in the disease-based analysis (F: female; M: male; PMI: post mortem interval; SD: standard deviation; TOD: time of death).
LIMMA
A Bayesian moderated t-test was applied to identify differentially expressed genes as implemented in the LIMMA (linear models for microarray analysis) package [27] from Bioconductor. Firstly, pre-processed Affymetrix datasets were collapsed to the probeset with the maximum expression value for each gene using the Gene Set Enrichment Analysis software (details below). Differentially expressed genes were then identified using the LIMMA package and raw p-values were adjusted for multiple hypothesis testing using the false discovery rate (FDR) method of Benjamini and Hochberg [28].
Functional profiling
Two complimentary approaches to gene set analysis were employed. Both methods investigated all “biological process” categories as defined by the Gene Ontology consortium (GO). The default GSEA significance threshold of q<0.25 (after controlling the false discovery rate) was used for all functional analyses.
GSEA
The GSEA algorithm examines a ranked list of all genes on the chip, and identifies whether members of a gene category are enriched at either the top or bottom, using a modified Kolmogorov-Smirnov statistic. GSEA was carried out following the recommendations of the authors [29], [30]. Pre-processed expression data was inputted to the GSEA software and collapsed to the probe with the maximum expression level for each gene prior to analysis. Genes were ranked by fold change calculated using the “difference of class means” metric implemented in the GSEA software, such that genes ranked towards the top of the list are considered enriched in one sample group and genes ranked at the bottom are considered enriched in the other. Enrichment scores were calculated using the weighted enrichment statistic, and significance levels calculated by permuting phenotype labels 1000 times. Gene set size filters were set to exclude gene sets containing fewer than 25, or greater than 500 members. All other parameters were set to GSEA defaults.
In the schizophrenia versus control analysis the gene sets investigated comprised the complete list of human biological process categories present on the U133 Plus 2.0 array as defined by the GO consortium (subject to filters as described above; around 3000 categories in total). Some categories represent closely related functions and in addition, multifunctional genes may be annotated in more than one category. GSEA examines each gene set independently and hence multiple categories annotated to the same or similar genes can arise due to the hierarchical nature of the GO database. We therefore used the leading edge analysis tool within GSEA to identify related sets, i.e. those in which the significance is driven by an overlapping subset of genes (the “leading edge”).
OntoExpress
The OntoExpress software uses an algorithm which examines a predetermined list of significant genes and identifies categories of genes which are over or under represented in this list relative to their representation on the entire chip. Following LIMMA analysis genes were ranked in order of t statistic. A list of the top 2% of genes most significantly up- and down-regulated (this included 402 genes from the Affymetrix dataset corresponding to a p value cutoff of 0.027 among the upregulated genes and 0.047 among the downregulated genes) were analysed separately using OntoExpress, using default settings (hypergeometric distribution and FDR (Benjamini-Hochberg) correction) and the Affymetrix human HG-U133 Plus 2.0 array as reference. Categories which had at least 3 members were considered in the analysis.
Codelink arrays
Data preprocessing
Image analysis and feature extraction was performed using the proprietary Amersham CodeLink Bioarray software (GE Healthcare). A flag-based noise filter was applied such that probes were retained for further analysis which had a “good” flag in a minimum number of arrays corresponding to the smallest sample group tested (eg where there were 12 control and 12 schizophrenia samples, the filter was set to retain probes which had a “good” flag in at least 12 samples). This step was carried out independently for the cell-type and disease analyses. The spot mean signal intensities for probes passing the filter were quantile normalized [31] to generate gene expression measures. Outlier removal was carried out based on a correlation matrix generated from all possible pairwise comparisons between arrays using Pearson's product-moment correlation coefficient as the metric. Poorly correlating arrays were removed from the analysis. After removing outliers, the flag-based noise filter and normalisation process were re-applied. The final dataset for the cell type analysis (all samples) contained 10487 probes. For the disease-based analysis endothelial and neuronal samples were normalised separately and the final dataset contained 8846 probes for the endothelial samples and 14262 probes for the neuronal samples. Due to the much reduced number of probes on the Codelink arrays compared to the Affymetrix arrays, Codelink arrays were used solely for validation purposes and not for the primary analysis.
Cross platform validation
Data were cross-validated between array platforms using GSEA and the method of Cheadle et al (2006) [32]. This method was developed to examine the entire dataset, taking into account the differences in absolute mRNA quantitation which often occur between array platforms. Following one microarray analysis, the most significantly altered genes are used to create a category. Data from an alternative microarray platform are then probed with this category. If the data are reproducible in the second platform, the category should be significantly enriched in the predicted direction. Datasets were collapsed to the maximum probe level per gene using GSEA, filtered to those genes which were present across both platforms and ranked using the GSEA metric “difference of class means”. The 200 top and bottom ranking genes from each array platform were each used to create a gene set. We then determined whether the top ranking genes from each platform showed enrichment in the same direction in the other platform, using the GSEA parameters described above.
Results
Cell type analysis
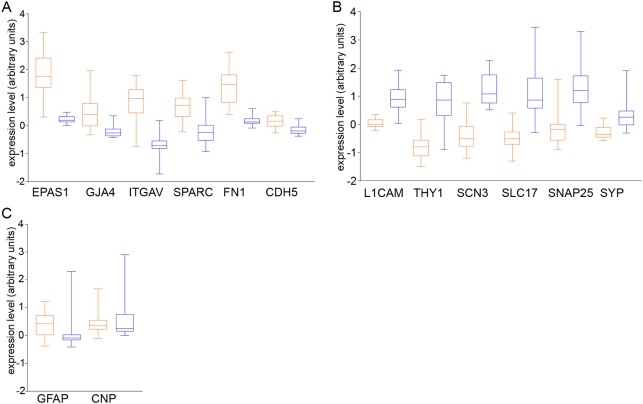
As an initial assessment of the biological validity of the data, we compiled a list of biological processes, as defined by the Gene Ontology Consortium, likely to be specific to one cell type or the other, using the search terms “neuron”, ”neurotransmitter” and “synapse” plus “endothelial”, and “angiogenesis”. Due to the small number of categories found to be endothelial-related, we also included cell proliferation as an endothelial-related category, as there is no published evidence for neurogenesis in the adult prefrontal cortex [33]. These categories were tested for enrichment in the Affymetrix data using GSEA. All endothelial categories were found to be significantly enriched in the endothelial data, and all neuronal categories were found to be significantly enriched in the neuronal data (q<0.25; Table 3). Additionally we chose a panel of six genes expected from the literature to be preferentially expressed in one cell type or the other in brain tissue, and investigated whether these were found differentially expressed between cell types using our methods. In the Affymetrix dataset, all six genes considered to differentiate endothelial cells from neurons (fibronectin (FN1), osteonectin (SPARC), integrin alpha5 (ITGAV), vascular endothelial cadherin (CDH5), endothelial PAS domain protein 1 (EPAS1), gap junction protein alpha4 (GJA4)) were significantly upregulated in our endothelial cell samples, and all six of those chosen to be neuronal differentiators (neural cell adhesion molecule L1 (L1CAM), synaptosomal protein 25 (SNAP25), synaptophysin (SYP), voltage gated sodium channel type IIIbeta (SCN3B), vesicular glutamate transporter (SLC17A7), Thy-1 cell surface antigen (THY1)) were significantly upregulated in our neuronal samples (Fig 3, Table 4). Furthermore, we examined the expression of markers of other cell types: GFAP, an astrocyte marker, and CNP, a marker of oligodendrocytes. These genes showed low expression values, and did not show differential expression between cell types. The method of Cheadle et al [32] was used to cross validate the entire dataset between chip platforms. In all cases both platforms reflected similar changes for genes with the greatest differential expression between endothelial cells and neurons (Table 5). Furthermore, four of each of the neuronal and endothelial markers were also detected in the Codelink dataset; only one failed to cross validate between datasets (Table 4).
Table 3. Results of cell type comparison–gene categories.
| GO ID | Category Name | Cell type | NES | FDR q value |
| GO:0001525 | angiogenesis | E | 1.49 | 0.028 |
| GO:0008283 | cell proliferation | E | 1.51 | 0.037 |
| GO:0045765 | regulation of angiogenesis | E | 1.40 | 0.046 |
| GO:0048666 | neuron development | N | −1.42 | 0.055 |
| GO:0030182 | neuron differentiation | N | −1.43 | 0.060 |
| GO:0019226 | transmission of nerve impulse | N | −1.45 | 0.061 |
| GO:0048699 | generation of neurons | N | −1.45 | 0.075 |
| GO:0048667 | neuron morphogenesis during differentiation | N | −1.46 | 0.092 |
| GO:0007269 | neurotransmitter secretion | N | −1.56 | 0.093 |
| GO:0007268 | synaptic transmission | N | −1.48 | 0.118 |
| GO:0042551 | neuron maturation | N | −1.23 | 0.166 |
| GO:0001505 | regulation of neurotransmitter levels | N | −1.26 | 0.169 |
| GO:0050808 | synapse organization and biogenesis | N | −1.24 | 0.173 |
| GO:0006836 | neurotransmitter transport | N | −1.13 | 0.241 |
GSEA results for categories characteristic of either endothelial cells (E) or neurons (N). Positive normalized enrichment scores (NES) indicate categories enriched in endothelial samples, negative NES indicate categories enriched in neuronal categories. All endothelial categories showed significant enrichment in the endothelial samples and all neuronal categories showed significant enrichment in the neuronal samples (q<0.25).
Figure 3. Characteristic endothelial and neuronal genes.
Boxplots showing the expression levels (arbitrary units, derived from Affymetrix Genechips) in endothelial (red) and neuronal (blue) samples of a panel of genes known to be preferentially expressed in either (a) endothelial cells or (b) neurons in brain tissue, plus (c) two markers known to be expressed in other cell types.
Table 4. Results of cell type comparison–individual genes.
| Gene symbol | Gene name | Cell type | Affymetrix | Codelink | ||
| Fold change | q value | Fold change | q value | |||
| EPAS1 | Endothelial PAS domain protein 1 | E | 3.09 | <0.0001 | 0.93 | 0.5568 |
| GJA4 | Gap junction protein alpha4 | E | 1.62 | 0.0116 | 1.18 | 0.0645 |
| ITGAV | Integrin alpha5 | E | 2.95 | <0.0001 | Not detected | |
| SPARC | Osteonectin | E | 1.87 | 0.0012 | 1.56 | 0.0003 |
| CDH5 | Vascular endothelial cadherin | E | 1.24 | 0.0124 | Not detected | |
| FN1 | Fibronectin | E | 2.36 | <0.0001 | 1.99 | <0.0001 |
| L1CAM | Neural cell adhesion molecule L1 | N | −1.84 | 0.0001 | Not detected | |
| THY1 | Thy-1 cell surface antigen | N | −2.94 | <0.0001 | −1.99 | 0.0045 |
| SCN3B | Voltage gated sodium channel type IIIbeta | N | −3.04 | <0.0001 | Not detected | |
| SLC17A7 | Vesicular glutamate transporter | N | −3.14 | <0.0001 | −1.32 | 0.0011 |
| SNAP25 | Synaptosomal protein 25 | N | −2.75 | 0.0007 | −1.32 | 0.0423 |
| SYP | Synaptophysin | N | −1.54 | 0.0121 | −1.20 | 0.0009 |
| GFAP | Glial fibrillary acidic protein | A | 1.22 | 0.4727 | Not detected | |
| CNP | 2′,3′-cyclic nucleotide 3′ phosphodiesterase | O | −1.07 | 0.8321 | Not detected | |
Differential expression of genes previously shown to be preferentially expressed in neurons (N), endothelial cells (E), astrocytes (A), or oligodendrocytes. Fold changes are calculated as endothelial cells relative to neurons. Of the genes which were present across both array platforms, only EPAS1 was not significant in both.
Table 5. Results of cross platform validation of the cell type comparison.
| CODELINK DATA | ||
| Categories tested | Enrichment score | FDR q-value |
| AFFY_ENDO | 1.63 | 0.001 |
| AFFY_NEURON | −1.32 | 0.148 |
Based on the Affymetrix data, the 200 genes whose expression was the highest in endothelial cells compared to neurons (AFFY_ENDO), and the 200 genes whose expression was the highest in neurons compared to endothelial cells (AFFY_NEURON) were used to create categories. These Affymetrix-derived categories were then investigated in the ranked gene list (endothelial cells vs neurons) generated using the Codelink platform. A positive enrichment score indicates enrichment in endothelial cells, and a negative score indicates enrichment in neurons. The categories were all found to be significantly enriched in the predicted direction in the Codelink data (q<0.25). The converse procedure was carried out testing categories generated from the Codelink data (CL_ENDO, CL_NEURON) on the Affymetrix platform, with comparable results.
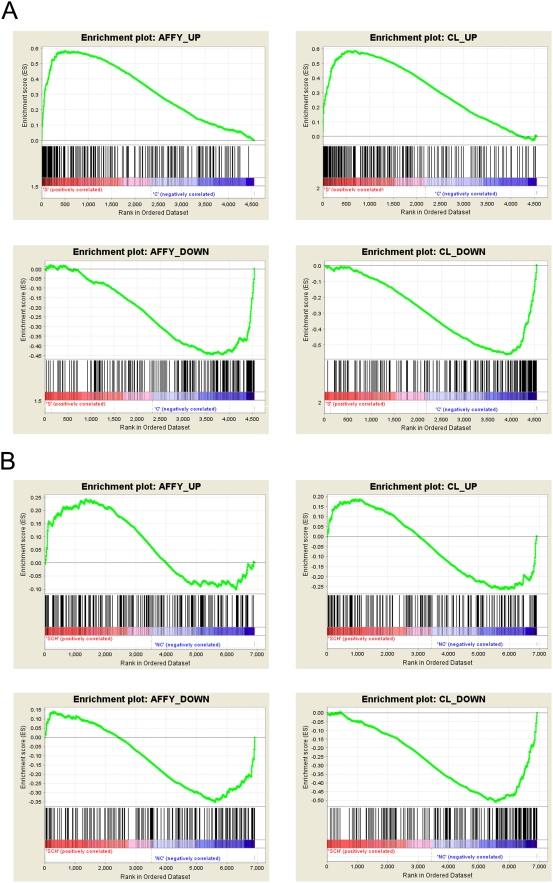
Having established that biologically valid differences between cell types could be detected in the dataset, we moved on to investigate whether differences could be detected between schizophrenia patients and controls in either cell type. LIMMA analysis revealed 1156 genes significantly differentially expressed in endothelial cells and 803 in neurons at p<0.05; however, when correction for multiple hypothesis testing was applied, no genes reached significance. This is likely to be explained by the small sample number and relative subtlety of the expected disease-related changes. In order to determine whether the disease signal could be validated using an alternative methodology, we applied the method of Cheadle et al [32] to validate across chip platforms. In the endothelial dataset, a set of genes whose expression was the most different between schizophrenia and control on one chip platform were found to be significantly enriched in the predicted direction on the other platform (Table 6, Fig 4), suggesting that a reproducible disease signal could be detected in the endothelial samples. However, in the neuronal dataset, alterations in gene expression between schizophrenia and control could not be validated across chip platforms (Table 7). This indicates that using this methodology, no technically robust disease-related alterations could be detected in neurons from the schizophrenia samples, in contrast to the endothelial cells.
Table 6. Results of cross platform validation of the schizophrenia vs control comparison for endothelial cells.
| CODELINK DATA | ||
| Categories tested | Enrichment score | FDR q-value |
| AFFY_UP (SZ) | 1.5 | 0.013 |
| AFFY_DOWN (C) | −1.44 | 0.044 |
Based on the Affymetrix data, the 200 genes whose expression was most upregulated in schizophrenia (AFFY_UP), and the 200 genes whose expression was the most downregulated in schizophrenia (AFFY_DOWN) were used to create categories for analysis in the ranked list (schizophrenia vs control) derived from the Codelink platform. A positive enrichment score indicates enrichment in schizophrenia samples, and a negative enrichment score indicates enrichment in control samples. These were all found to be significantly enriched in the expected direction in the Codelink data (q<0.25). The converse procedure was carried out testing categories generated from the Codelink data (CL_UP, CL_DOWN) on the Affymetrix platform, with comparable results.
Figure 4. Results of cross-platform validation of the schizophrenia vs control analysis for (a) endothelial cells and (b) neurons.
The figures were created by the GSEA software and plot the running enrichment score (ES) which reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. The score at the peak of the plot (the score furthest from 0.0) is the ES for the gene set. The position of individual members of the gene set in the ranked list is indicated by vertical lines.
Table 7. Results of cross platform validation of the schizophrenia vs control comparison for neurons.
| CODELINK DATA | ||
| Categories tested | Enrichment score | FDR q-value |
| AFFY_UP (SZ) | 0.77 | 0.734 |
| AFFY_DOWN (C) | −1.15 | 0.283 |
The same procedure as described in Table 6 (endothelial cells) was carried out on data from the neuronal samples. In only one of the four tests was the category derived from one platform significantly enriched in the expected direction on the other platform.
In order to further characterise the disease signal in endothelial cells, we investigated alterations in functional categories of genes, a statistically more powerful approach than considering individual gene changes. Numerous approaches to functional profiling of gene expression data exist, which rely on different computational approaches and assumptions. In the present study, we employed two algorithms. The first, GSEA, looks for enrichment of genes in a category at the top or bottom of a ranked list based on a modified Kolmogorov-Smirnov statistic, and has been developed specifically for the detection of biological differences which may be modest relative to technical noise. The second, OntoExpress, identifies categories of genes which are over or under represented in a list of significant genes relative to their representation on the entire chip. Some differences are expected due to the differences between the algorithms [34], but a truly robust finding should be detectable using either method.
62 categories were downregulated in schizophrenia endothelial samples using GSEA and 33 using OntoExpress. The majority of results were comparable regardless of the algorithm used (Table 8), with a small number of exceptions: endothelial-specific and developmental categories were only found significant with GSEA and not OntoExpress, and categories relating to transcriptional and translational processes were identified with OntoExpress but not GSEA. No categories were significantly upregulated, which reflects the bias towards downregulated genes seen in the data (not shown).
Table 8. Results of functional profiling of the endothelial cell data.
| GSEA | OntoExpress | |||||
| GO ID | Name | Size | FDR q value | GO ID | Name | FDR q value |
| RESPONSE TO STIMULUS | ||||||
| GO0007606 | sensory perception of chemical stimulus | 105 | 0.060 | GO:0007600 | sensory perception | 0.097 |
| GO0007608 | sensory perception of smell | 86 | 0.114 | |||
| GO0009581 | detection of external stimulus | 32 | 0.147 | |||
| IMMUNE SYSTEM | ||||||
| GO0051240 | positive regulation of organismal physiological process | 52 | 0.121 | GO:0006952 | defense response | 0.028 |
| GO0050867 | positive regulation of cell activation | 25 | 0.132 | GO:0006955 | immune response | 0.038 |
| GO0046651 | lymphocyte proliferation | 25 | 0.145 | GO:0030154 | cell differentiation | 0.121 |
| GO0050778 | positive regulation of immune response | 48 | 0.148 | |||
| GO0046649 | lymphocyte activation | 82 | 0.149 | |||
| GO0042113 | B cell activation | 32 | 0.150 | |||
| GO0016066 | cellular defense response (sensu Vertebrata) | 27 | 0.156 | |||
| GO0050776 | regulation of immune response | 73 | 0.161 | |||
| GO0050865 | regulation of cell activation | 40 | 0.164 | |||
| GO0045321 | immune cell activation | 96 | 0.193 | |||
| GO0030098 | lymphocyte differentiation | 34 | 0.199 | |||
| GO0051249 | regulation of lymphocyte activation | 38 | 0.201 | |||
| GO0001775 | cell activation | 97 | 0.202 | |||
| GO0006959 | humoral immune response | 137 | 0.140 | |||
| GO0016064 | humoral defense mechanism (sensu Vertebrata) | 100 | 0.212 | |||
| GO:0019735 | antimicrobial humoral response (sensu Vertebrata) | 75 | 0.228 | |||
| GO0019730 | cytokine production | 78 | 0.249 | |||
| GO0009613 | response to pest, pathogen or parasite | 479 | 0.124 | GO:0006954 | inflammatory response | 0.089 |
| GO0009611 | response to wounding | 362 | 0.129 | |||
| GO0006954 | inflammatory response | 198 | 0.132 | |||
| GO0009605 | response to external stimulus | 460 | 0.146 | |||
| GO0019882 | antigen presentation | 39 | 0.139 | |||
| GO0030333 | antigen processing | 32 | 0.150 | |||
| GO0045087 | innate immune response | 55 | 0.106 | |||
| GO0042742 | defense response to bacteria | 53 | 0.117 | |||
| GO0009615 | response to virus | 66 | 0.156 | GO:0009615 | response to virus | 0.034 |
| GO0006968 | cellular defense response | 93 | 0.174 | GO:0006968 | cellular defense response | 0.065 |
| FERTILIZATION | ||||||
| GO0007338 | fertilization (sensu Metazoa) | 35 | 0.158 | GO:0007283 | spermatogenesis | 0.037 |
| GO0009566 | fertilization | 36 | 0.161 | |||
| MOTILITY | ||||||
| GO0051270 | regulation of cell motility | 28 | 0.144 | GO:0006928 | cell motility | 0.097 |
| GO0040012 | regulation of locomotion | 28 | 0.149 | GO:0030155 | regulation of cell adhesion | 0.007 |
| GO0030334 | regulation of cell migration | 28 | 0.153 | |||
| GO:0016477 | cell migration | 95 | 0.226 | |||
| GO0007610 | behavior | 175 | 0.137 | GO:0010033 | response to organic substance | 0.001 |
| GO0042330 | taxis | 107 | 0.144 | |||
| GO0007626 | locomotory behavior | 112 | 0.149 | |||
| GO0006935 | chemotaxis | 107 | 0.155 | |||
| GO:0042221 | response to chemical stimulus | 310 | 0.223 | |||
| ION TRANSPORT | ||||||
| GO0030005 | di-, tri-valent inorganic cation homeostasis | 89 | 0.154 | GO:0006811 | ion transport | 0.093 |
| GO0006875 | metal ion homeostasis | 96 | 0.157 | GO:0006810 | transport | 0.168 |
| GO0030003 | cation homeostasis | 102 | 0.199 | |||
| GO0006817 | phosphate transport | 76 | 0.196 | |||
| NEGATIVE REGULATION OF CELL PROLIFERATION | ||||||
| GO0008285 | negative regulation of cell proliferation | 145 | 0.147 | GO:0016049 | cell growth | 0.036 |
| GO:0006915 | apoptosis | 0.066 | ||||
| DNA METABOLISM | ||||||
| GO0051052 | regulation of DNA metabolism | 31 | 0.161 | GO:0006259 | DNA metabolism | 0.003 |
| GO:0006281 | DNA repair | 0.043 | ||||
| PROTEIN MODIFICATION | ||||||
| GO0045860 | positive regulation of protein kinase activity | 53 | 0.146 | GO:0006464 | protein modification | 0.079 |
| GO0045859 | regulation of protein kinase activity | 123 | 0.161 | |||
| GO0043549 | regulation of kinase activity | 123 | 0.165 | |||
| GO0051338 | regulation of transferase activity | 123 | 0.157 | |||
| GO0000079 | regulation of cyclin-dependent protein kinase activity | 38 | 0.167 | |||
| GO0051347 | positive regulation of transferase activity | 53 | 0.150 | |||
| ENDOTHELIAL-SPECIFIC | ||||||
| GO0001525 | angiogenesis | 61 | 0.246 | |||
| GO0008217 | blood pressure regulation | 29 | 0.175 | |||
| DEVELOPMENT | ||||||
| GO0007398 | ectoderm development | 85 | 0.119 | |||
| GO0008544 | epidermis development | 77 | 0.144 | |||
| GO0009888 | tissue development | 171 | 0.177 | |||
| GO0007519 | striated muscle development | 38 | 0.148 | |||
| GO0007517 | muscle development | 98 | 0.198 | |||
| OTHER | ||||||
| GO0000270 | peptidoglycan metabolism | 26 | 0.129 | |||
| PROTEIN ASSEMBLY/TRANSPORT | ||||||
| GO:0006461 | protein complex assembly | 0.094 | ||||
| GO:0006508 | proteolysis and peptidolysis | 0.100 | ||||
| GO:0015031 | protein transport | 0.077 | ||||
| GO:0006886 | intracellular protein transport | 0.196 | ||||
| SIGNAL TRANSDUCTION | ||||||
| GO:0007186 | G-protein coupled receptor protein signaling pathway | 0.079 | ||||
| GO:0007165 | signal transduction | 0.006 | ||||
| TRANSCRIPTION/TRANSLATION | ||||||
| GO:0045449 | regulation of transcription | 0.032 | ||||
| GO:0006445 | regulation of translation | 0.043 | ||||
| GO:0008380 | RNA splicing | 0.063 | ||||
| GO:0006396 | RNA processing | 0.080 | ||||
| GO:0006260 | DNA replication | 0.195 | ||||
| GO:0006355 | regulation of transcription, DNA-dependent | 0.197 | ||||
| GO:0006350 | transcription | 0.185 | ||||
Gene function categories downregulated in endothelial cells from schizophrenia patients using GSEA (left side) and OntoExpress (right side). Related gene sets as defined by GSEA leading edge analysis are grouped together in column 2, and categories are further grouped by theme. Equivalent categories (identical or closely related in the Gene Ontology) identified by OntoExpress, are indicated in columns 5 and 6.
Discussion
In the present study we show molecular alterations in the vasculature of schizophrenia patients. Based on these data alone it is not possible to infer whether alterations in RNA expression reflect a functional impairment in the blood brain barrier. Nonetheless, the results of gene expression profiling indicate a downregulation of genes involved in ion transport, cell proliferation and adhesion, which are consistent with such an impairment. Furthermore, downregulation of genes related to immune system function was identified, including GO:0006954 “inflammatory response” which was identified using both OntoExpress and GSEA. Hanson and Gottesman have proposed inflammation of the cerebral microvasculature as the source of blood-brain barrier dysfunction in schizophrenia, with systemic effects [9]. However the data from this study, and other studies from our laboratory on T cell function in schizophrenia [35], point more towards a hypo-inflammatory state in schizophrenic patients. This is consistent with a growing body of evidence in the field, including the negative association between schizophrenia and rheumatoid arthritis [36]–[38], lower antibody reactions to vaccination [39], and decreased skin sensitivity to the type IV antigen test [40]. However, schizophrenia has been positively linked to other auto-immune disorders [41]. At the molecular level, increased levels of acute phase proteins have been reported in schizophrenia pointing towards a pro-inflammatory state [42], [43], and conflicting data exists on the role of inflammatory cytokines in the disorder [44]. A potential explanation for these apparently opposing results is overall dysregulation of inflammation, leading to an inappropriate response (either too much or too little inflammation) depending on the stimulus and site. Such a dysfunction in the blood brain barrier is consistent with the broader implications of Hanson & Gottesman's hypothesis, and could result in slower response and lower resistance to brain injury/insult, affecting the regulation of supply of substances to the brain. Furthermore, abnormal inflammatory processes may have downstream effects on angiogenesis [45], and thus may further impact vascular abnormalities. However, conclusions cannot be firmly drawn without further in vivo study of the microvasculature in schizophrenia patients.
The blood brain barrier is composed of endothelial cells, pericytes, astrocyte end-feet and neuronal processes. A method for isolating pure vascular endothelium has been developed [46]; however, as this method is specific for endothelial cells and involves numerous steps, its use would preclude comparison of the resulting cell population with neurons and other cell types. Furthermore, cerebral microvascular function arises from the interactions between its various components, therefore global data for the intact microvessel is of greater interest. In this context, our result of downregulation of genes in the GO category “spermatogenesis” is of interest as numerous functional links exist between the blood-brain barrier and the blood-testis barrier, including shared properties of astrocytes and oligodendrocytes and the Sertoli and Leydig cells of the testis [47], [48]. We speculate that this result may indicate altered gene expression in non-endothelial components of the blood-brain barrier. Interestingly, a recent study has found a decreased number of oligodendrocytes per unit length of capillary in post mortem prefrontal cortex from schizophrenia patients [49] which provides a potential explanation for some of the results seen here. Further investigation of these cell types is clearly warranted.
Laser capture microdissection is a technically challenging method due to the small amounts of tissue and the variability that may be introduced at various stages of the analytical process, compounded by the effects of working with post mortem human tissue, and the present work involved extensive optimization and validation of the methods prior to study commencement. In the present study, although cell-type specific changes could be detected in neurons, no schizophrenia-related alterations could be reliably detected in neurons collected from the same brain region as the endothelial cells (dorsolateral prefrontal cortex). Although we (data not shown) and others [50]–[52] have found that RNA amplification gives reproducible results, it does result in loss and/or truncation of transcripts, leading to a smaller number of high-quality arrays than would otherwise be expected, and furthermore the final mRNA population analysed will not contain the full range of information found in the original sample [50], [53], [54]. Thus in this context the neuronal result cannot be considered a true negative. Although no directly comparable study has been carried out, there is much evidence for neuronal alterations in schizophrenia at the molecular and structural level [55], including data derived using similar methods in the same [56], [57] and other [2] brain regions. However, as the endothelial data were collected in an identical manner, the results do provide compelling preliminary evidence for molecular alterations in the microvasculature of schizophrenia patients. More targeted and functional studies of the blood-brain barrier, including investigation of its subcomponents, are now required, and further investigation is necessary to determine whether, if proven, blood-brain barrier dysfunction can directly explain the impairment in glucose utilization in the brains of schizophrenic patients. The potential effects of antipsychotic medication on the blood brain barrier should also be assessed. Furthermore, investigation of other cell types such as astrocytes, and further investigation of peripheral tissues, is key to elucidating the role of metabolic abnormalities in the pathophysiology of schizophrenia.
Supporting Information
A document containing demographics variables for the cell type analysis, and additional details of the microarray data processing
(0.05 MB DOC)
RNA 5′ -3′ signal bias. (a) RNA digestion plot showing signal from probes decreases with distance of target sequence from 3′ end of transcript. Note the inter-chip variability in the gradient of the curves. (b) The degree of RNA 5′-3′ signal bias within a chip, as measured by the slope of RNA digestion curve, displays strong positive correlation with the number of probe-sets on the chip which are flagged as present.
(7.00 MB TIF)
Detection and removal of RNA 5 -3signal bias from Affymetrix GeneChip data on the expression profiles of endothelial cells (E) and neurons (N). (a) PCA of RMA expression data reveals that the major component of the variation in the data (PC1) is not related to differential expression between endothelial cells and neurons. (b) PC1 of the RMA expression data shows strong correlation with the RNA 5 -3signal bias within each chip. The slope of a chip's RNA digestion curve was used as the measure of 5 -3signal bias. (c) Following a transformation to remove 5 -3signal bias (see text for details), the major source of variation in the data set is now differential gene expression between the two cell types, which are now clearly separable on PC1. (d) PC1 of transformed data is not correlated with the 5 -3signal bias within a chip.
(8.23 MB TIF)
Number of probesets on the Affymetrix GeneChip detecting differential ex- pression between endothelial cells and neurons at a range of false discovery rates (FDR), before and after a correction was applied for 5′-3′ signal bias. For details of systematic bias and correction, see text.
(6.77 MB TIF)
Acknowledgments
We gratefully acknowledge the donations of the SMRI brain collection, courtesy of Drs. Michael B. Knable, E Fuller Torrey, Maree Webster, Serge Weis and Robert H. Yolken. Also Anita Thorn and Gavin Hardy of GE Healthcare for technical support and Svante Paabo for provision of laboratory resources and helpful comments on the work. We are also grateful to the students of Bildungsinstitut Pscherer gGmbH, Lengenfeld, Germany, for technical assistance, and to Paul Guest, Dave Bailey and Deena Gendoo for comments on the manuscript and bioinformatics support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded as part of the Molecular Evolution of Human Cognition project (EU Sixth Framework Programme grant PKB 140404). The Stanley Medical Research Institute (SMRI) provided centre support. GE Healthcare UK provided CodeLink Human Whole Genome Bioarrays. ML is the recipient of an NIH-Cambridge health scholarship. MR is the recipient of a NARSAD Young Investigator Award. SB holds a NARSAD Essel Independent Investigator Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Glahn DC, Ragland JD, Abramoff A, Barrett J, Laird AR, et al. Beyond hypofrontality: a quantitative meta-analysis of functional neuroimaging studies of working memory in schizophrenia. Hum Brain Mapp. 2005;25:60–69. doi: 10.1002/hbm.20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 3.Iwamoto K, Bundo M, Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum Mol Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- 4.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697, 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 6.Regenold WT, Phatak P, Kling MA, Hauser P. Post-mortem evidence from human brain tissue of disturbed glucose metabolism in mood and psychotic disorders. Mol Psychiatry. 2004;9:731–733. doi: 10.1038/sj.mp.4001517. [DOI] [PubMed] [Google Scholar]
- 7.Holmes E, Tsang TM, Huang JT, Leweke FM, Koethe D, et al. Metabolic profiling of CSF: evidence that early intervention may impact on disease progression and outcome in schizophrenia. PLoS Med. 2006;3:e327. doi: 10.1371/journal.pmed.0030327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang JT, Leweke FM, Oxley D, Wang L, Harris N, et al. Disease biomarkers in cerebrospinal fluid of patients with first-onset psychosis. PLoS Med. 2006;3:e428. doi: 10.1371/journal.pmed.0030428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson DR, Gottesman II. Theories of schizophrenia: a genetic-inflammatory-vascular synthesis. BMC Med Genet. 2005;6:7. doi: 10.1186/1471-2350-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson CJ, Lin A, Cogan S, Cashman F, Warsh JJ. The niacin challenge test: clinical manifestation of altered transmembrane signal transduction in schizophrenia? Biol Psychiatry. 1997;41:507–513. doi: 10.1016/s0006-3223(96)00112-6. [DOI] [PubMed] [Google Scholar]
- 11.Messamore E, Hoffman WF, Janowsky A. The niacin skin flush abnormality in schizophrenia: a quantitative dose-response study. Schizophr Res. 2003;62:251–258. doi: 10.1016/s0920-9964(02)00311-0. [DOI] [PubMed] [Google Scholar]
- 12.Puri BK, Easton T, Das I, Kidane L, Richardson AJ. The niacin skin flush test in schizophrenia: a replication study. Int J Clin Pract. 2001;55:368–370. [PubMed] [Google Scholar]
- 13.Rybakowski J, Weterle R. Niacin test in schizophrenia and affective illness. Biol Psychiatry. 1991;29:834–836. doi: 10.1016/0006-3223(91)90202-w. [DOI] [PubMed] [Google Scholar]
- 14.Shah SH, Vankar GK, Peet M, Ramchand CN. Unmedicated schizophrenic patients have a reduced skin flush in response to topical niacin. Schizophr Res. 2000;43:163–164. [PubMed] [Google Scholar]
- 15.Malaspina D, Harkavy-Friedman J, Corcoran C, Mujica-Parodi L, Printz D, et al. Resting neural activity distinguishes subgroups of schizophrenia patients. Biol Psychiatry. 2004;56:931–937. doi: 10.1016/j.biopsych.2004.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz SK, O'Leary DS, Boles Ponto LL, Arndt S, Magnotta V, et al. Age and regional cerebral blood flow in schizophrenia: age effects in anterior cingulate, frontal, and parietal cortex. J Neuropsychiatry Clin Neurosci. 2002;14:19–24. doi: 10.1176/jnp.14.1.19. [DOI] [PubMed] [Google Scholar]
- 17.Brambilla P, Cerini R, Fabene PF, Andreone N, Rambaldelli G, et al. Assessment of cerebral blood volume in schizophrenia: A magnetic resonance imaging study. J Psychiatr Res. 2007;41:502–510. doi: 10.1016/j.jpsychires.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Cohen BM, Yurgelun-Todd D, English CD, Renshaw PF. Abnormalities of regional distribution of cerebral vasculature in schizophrenia detected by dynamic susceptibility contrast MRI. Am J Psychiatry. 1995;152:1801–1803. doi: 10.1176/ajp.152.12.1801. [DOI] [PubMed] [Google Scholar]
- 19.Kreczmanski P, Schmidt-Kastner R, Heinsen H, Steinbusch HW, Hof PR, et al. Stereological studies of capillary length density in the frontal cortex of schizophrenics. Acta Neuropathol (Berl) 2005;109:510–518. doi: 10.1007/s00401-005-1003-y. [DOI] [PubMed] [Google Scholar]
- 20.Shusta EV, Boado RJ, Mathern GW, Pardridge WM. Vascular genomics of the human brain. J Cereb Blood Flow Metab. 2002;22:245–252. doi: 10.1097/00004647-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Burnet PW, Eastwood SL, Harrison PJ. Laser-assisted microdissection: methods for the molecular analysis of psychiatric disorders at a cellular resolution. Biol Psychiatry. 2004;55:107–111. doi: 10.1016/s0006-3223(03)00642-5. [DOI] [PubMed] [Google Scholar]
- 22.Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 23. www.palm-microlaser.com. [Google Scholar]
- 24.Jackson ES, Wayland MT, Fitzgerald W, Bahn S. A microarray data analysis framework for postmortem tissues. Methods. 2005;37:247–260. doi: 10.1016/j.ymeth.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry R, Gautier L, Bolstad B, Miller C, Astrand M, et al. Affy: Methods for Affymetrix Oligonucleotide Arrays 2005 [Google Scholar]
- 27.Smyth G. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. GSEA http://www.broad.mit.edu/gsea/ [Google Scholar]
- 31.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 32.Cheadle C, Becker KG, Cho-Chung YS, Nesterova M, Watkins T, et al. A rapid method for microarray cross platform comparisons using gene expression signatures. Mol Cell Probes. 2006 doi: 10.1016/j.mcp.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Lledo PM, Alonso M, Grubb MS. Adult neurogenesis and functional plasticity in neuronal circuits. Nat Rev Neurosci. 2006;7:179–193. doi: 10.1038/nrn1867. [DOI] [PubMed] [Google Scholar]
- 34.Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- 35.Craddock RM, Lockstone HE, Rider DA, Wayland MT, Harris LJ, et al. Altered T-cell function in schizophrenia: a cellular model to investigate molecular disease mechanisms. PLoS ONE. 2007;2:e692. doi: 10.1371/journal.pone.0000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eaton WW, Hayward C, Ram R. Schizophrenia and rheumatoid arthritis: a review. Schizophr Res. 1992;6:181–192. doi: 10.1016/0920-9964(92)90001-l. [DOI] [PubMed] [Google Scholar]
- 37.Gorwood P, Pouchot J, Vinceneux P, Puechal X, Flipo RM, et al. Rheumatoid arthritis and schizophrenia: a negative association at a dimensional level. Schizophr Res. 2004;66:21–29. doi: 10.1016/s0920-9964(03)00017-3. [DOI] [PubMed] [Google Scholar]
- 38.Oken RJ, Schulzer M. At issue: schizophrenia and rheumatoid arthritis: the negative association revisited. Schizophr Bull. 1999;25:625–638. doi: 10.1093/oxfordjournals.schbul.a033407. [DOI] [PubMed] [Google Scholar]
- 39.Russo R, Ciminale M, Ditommaso S, Siliquini R, Zotti C, et al. Hepatitis B vaccination in psychiatric patients. Lancet. 1994;343:356. doi: 10.1016/s0140-6736(94)91193-2. [DOI] [PubMed] [Google Scholar]
- 40.Riedel M, Spellmann I, Schwarz MJ, Strassnig M, Sikorski C, et al. Decreased T cellular immune response in schizophrenic patients. J Psychiatr Res. 2007;41:3–7. doi: 10.1016/j.jpsychires.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Eaton WW, Byrne M, Ewald H, Mors O, Chen CY, et al. Association of schizophrenia and autoimmune diseases: linkage of Danish national registers. Am J Psychiatry. 2006;163:521–528. doi: 10.1176/appi.ajp.163.3.521. [DOI] [PubMed] [Google Scholar]
- 42.Wan C, La Y, Zhu H, Yang Y, Jiang L, et al. Abnormal changes of plasma acute phase proteins in schizophrenia and the relation between schizophrenia and haptoglobin (Hp) gene. Amino Acids. 2007;32:101–108. doi: 10.1007/s00726-005-0292-8. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Wan C, Li H, Zhu H, La Y, et al. Altered levels of acute phase proteins in the plasma of patients with schizophrenia. Anal Chem. 2006;78:3571–3576. doi: 10.1021/ac051916x. [DOI] [PubMed] [Google Scholar]
- 44.Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, et al. Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biol Psychiatry. 2008;63:801–808. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 45.Kirk SL, Karlik SJ. VEGF and vascular changes in chronic neuroinflammation. J Autoimmun. 2003;21:353–363. doi: 10.1016/s0896-8411(03)00139-2. [DOI] [PubMed] [Google Scholar]
- 46.Kinnecom K, Pachter JS. Selective capture of endothelial and perivascular cells from brain microvessels using laser capture microdissection. Brain Res Brain Res Protoc. 2005;16:1–9. doi: 10.1016/j.brainresprot.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Davidoff MS, Middendorff R, Kofuncu E, Muller D, Jezek D, et al. Leydig cells of the human testis possess astrocyte and oligodendrocyte marker molecules. Acta Histochem. 2002;104:39–49. doi: 10.1078/0065-1281-00630. [DOI] [PubMed] [Google Scholar]
- 48.Holash JA, Harik SI, Perry G, Stewart PA. Barrier properties of testis microvessels. Proc Natl Acad Sci U S A. 1993;90:11069–11073. doi: 10.1073/pnas.90.23.11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vostrikov V, Orlovskaya D, Uranova N. Deficit of pericapillary oligodendrocytes in the prefrontal cortex in schizophrenia. World J Biol Psychiatry. 2008;9:34–42. doi: 10.1080/15622970701210247. [DOI] [PubMed] [Google Scholar]
- 50.Baugh LR, Hill AA, Brown EL, Hunter CP. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 2001;29:E29. doi: 10.1093/nar/29.5.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaposi-Novak P, Lee JS, Mikaelyan A, Patel V, Thorgeirsson SS. Oligonucleotide microarray analysis of aminoallyl-labeled cDNA targets from linear RNA amplification. Biotechniques. 2004;37:580, 582–586, 588. doi: 10.2144/04374ST02. [DOI] [PubMed] [Google Scholar]
- 52.Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA. Accurate and reproducible gene expression profiles from laser capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn. 2003;5:9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boelens MC, te Meerman GJ, Gibcus JH, Blokzijl T, Boezen HM, et al. Microarray amplification bias: loss of 30% differentially expressed genes due to long probe - poly(A)-tail distances. BMC Genomics. 2007;8:277. doi: 10.1186/1471-2164-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122(Pt 4):593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- 56.O'Connor JA, Hemby SE. Elevated GRIA1 mRNA expression in Layer II/III and V pyramidal cells of the DLPFC in schizophrenia. Schizophr Res. 2007;97:277–288. doi: 10.1016/j.schres.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, et al. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatry. 2007;12:385–397. doi: 10.1038/sj.mp.4001954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A document containing demographics variables for the cell type analysis, and additional details of the microarray data processing
(0.05 MB DOC)
RNA 5′ -3′ signal bias. (a) RNA digestion plot showing signal from probes decreases with distance of target sequence from 3′ end of transcript. Note the inter-chip variability in the gradient of the curves. (b) The degree of RNA 5′-3′ signal bias within a chip, as measured by the slope of RNA digestion curve, displays strong positive correlation with the number of probe-sets on the chip which are flagged as present.
(7.00 MB TIF)
Detection and removal of RNA 5 -3signal bias from Affymetrix GeneChip data on the expression profiles of endothelial cells (E) and neurons (N). (a) PCA of RMA expression data reveals that the major component of the variation in the data (PC1) is not related to differential expression between endothelial cells and neurons. (b) PC1 of the RMA expression data shows strong correlation with the RNA 5 -3signal bias within each chip. The slope of a chip's RNA digestion curve was used as the measure of 5 -3signal bias. (c) Following a transformation to remove 5 -3signal bias (see text for details), the major source of variation in the data set is now differential gene expression between the two cell types, which are now clearly separable on PC1. (d) PC1 of transformed data is not correlated with the 5 -3signal bias within a chip.
(8.23 MB TIF)
Number of probesets on the Affymetrix GeneChip detecting differential ex- pression between endothelial cells and neurons at a range of false discovery rates (FDR), before and after a correction was applied for 5′-3′ signal bias. For details of systematic bias and correction, see text.
(6.77 MB TIF)