Abstract
Much work has been performed since the development of the lab-on-a-chip concept that has brought microfabricated systems to the forefront of bioanalytical research. The success of using these microchips for performing complicated biological assays faster and cheaper than conventional methods has facilitated their emerging popularity among researchers. A recently exploited advantage of microfabricated technology has led to the creation of single wafers with multiple channel manifolds for high-throughput experiments. Efforts towards parallel microchip development have yielded fascinating new devices for chemical separations showing the potential for replacing conventional multiplexing techniques. This review will focus on recent work towards multiplexed separations on microdevices and complementary detection instrumentation.
Keywords: microfluidics, multiplexed detection, parallel separations, review
1 Introduction
The development towards lab-on-a-chip devices has been greatly advanced since the first introduction of microfluidics for chemical analysis [1, 2], with much progress being made in the area of chemical separations [3–6]. The attractiveness of microfluidics-based separations to analytical chemists owes to its ability to routinely perform rapid and sensitive experiments on a small footprint with minimal use of sample and reagents. Additionally, microfluidic technology facilitates the fabrication of devices with integrated functions [7–9], an advantage which has led to a multitude of microdevices for pertinent biochemical assays. The success of microchips in basic research has led to the birth of specialized commercial devices for multiple bioanalytical applications [10–12]. It is clear that recent advancements made with these tools have placed the use of microchips on the frontier of research in cell biology, genetics, pharmacology, and other biomedical fields.
Many advances with microfluidic devices highlighted above were demonstrated on “single-sample” chips capable of performing only one analysis at a time. An additional advantage of microfluidics is the ease of fabricating several analysis manifolds on a single device, thus potentially increasing throughput while keeping the cost of fabrication essentially the same. Research using such parallel separations will help to realize the continued advancement of microdevices used for real-world applications.
Advances in high-throughput techniques have already shown importance to many fields with conventional multiplexing instrumentation for drug discovery [13], protein characterization [14], and DNA analysis [15]. Clearly, the time and cost savings that these high-throughput methods offer can be increased by utilizing the short analysis times and small reagent volumes associated with micro-scale systems. However, several challenges in developing parallel analysis microchips must be overcome. These challenges include: (i) organizing multiple channel networks on a single device with a small footprint, (ii) coupling multiplexed chips to peripheral equipment (i.e. power supplies, pumping devices), (iii) developing multi-channel detection methods, and (iv) achieving reproducible analyses across parallel channels.
Over the past decade, we have seen the addressing of these concerns with numerous reports of parallel separations on microchips. This paper will serve as a brief review of recent technological advances towards realizing high-throughput microfluidic devices using parallel separations. Though microchip channel organization and operation are critical to performance, design details are commonly influenced by chosen parallel detection methods. As detection is a crucial part of performing parallel separations on microchips, a discussion of various methods of multiplexed detection, consisting mainly of fluorescence techniques, will precede review of recently developed microchips for parallel analysis of biomolecules, affinity assays, and DNA analysis.
2 Multiplexed Detection Methods
One important aspect considered in the development of a microfluidic system with multiple separation channels is how detection will be performed because most detectors are designed for single point detection. Although there have been a multitude of reports using single separation microchips and detectors for measuring low quantities of analyte [16, 17], the development of a multi-channel instrument capable of highly sensitive detection of numerous channels is challenging. Several research groups have developed specialized detectors for use with their own custom-designed microchips that use specific technology providing either high sensitivity, high sampling rates, or ease of use. In broad terms, three basic methods of detecting simultaneous separations on multiple channels have been utilized: i) moving a single point detector across the chip at a frequency high enough to adequately detect separated bands in each channel (e.g. scanning LIF), ii) having a single detector that can continuously monitor all separations while maintaining channel-to-channel resolution (e.g. fluorescence imaging), and iii) assigning independent detectors to each channel on the device (e.g. parallel electrochemical detection).
2.1 Scanning LIF
Fluorescence detection is commonly used with microchips owing to its high sensitivity and ease of use with micrometer-dimensioned channels [18]. Lasers are commonly used for excitation on these devices; however, coupling laser excitation to multiple detection points has been a challenge requiring unique solutions. Rather than using multiple stationary laser spots for excitation, requiring many complicated optical alignments to be made for proper excitation and detection, the majority of research has focused on scanning a single laser spot across several channels. Numerous methods of scanning LIF detection have been demonstrated, ranging from relatively simple stage translation devices for use with a stationary laser to detectors based on a mobile objective lens.
2.1.1 Translation Stage
The basis of translation stage-driven multiplexed LIF detection is moving a microchip over a fixed detection point so that the multiple channels on a device can be sampled. The advantage in using this approach is that detection can be performed on parallel aligned channels without moving any components of the optical train. The first reported scanner of this type, albeit not used for detection with microchips, was reported as an approach to high-throughput DNA sequencing using capillary array electrophoresis (CAE) [19]. This same stage translation detection scheme was later demonstrated for use with parallel separation chips [20]. In this report, the translation stage detector was used for monitoring multiple DNA separations on a chip with 14 parallel channels (12 channels for separations and two for optical alignment). Parallel epi-fluorescence detection was performed using a stationary 20x objective lens while scanning the microchip a distance of 1.2 mm at a speed of 1.0 cm s−1, allowing an overall sampling rate of 3.3 Hz.
A more recent study also shows the use of stage translation for fluorescence detection of parallel CE separations [21]. In this report, the advantage of having a non-moving optical train has been exploited to facilitate optical gating for sample introduction in parallel channels. Optical gating, previously shown to be compatible with microchips in a study performed by this same group [22], uses high intensity laser light to continuously photobleach sample flowing though a separation channel. An analysis is made by temporarily blocking the laser and letting a fluorescent sample plug separate by CE. Experiments were performed by scanning a five-channel CE microchip across two laser spots (gating and probe beams) which were offset by 400 μm to allow simultaneous detection on one channel and gating on another. A voice coil actuator, which can provide accurate placement from micrometer to centimeter distances, was used to drive the translation stage. It was found that moving the stage at a frequency of greater than 10 Hz produced significant vibrations that increased experimental noise, illustrating a drawback of using this detector for monitoring fast separations.
2.1.2 Scanning Mirror
As translation stage detectors cannot offer the needed speed for detection of fast parallel separations, a drive towards the development of faster scanning detectors led to coupling mirror-based laser scanning with microchip separations. Unlike translation stages, scanning mirror systems raster a laser spot across channels on a stationary chip. The use of galvo-mirrors for scanning laser spots across a short distance has been previously demonstrated by many commercial scanning confocal imaging systems [23]. Some of the scanning mirrors used in these instruments can scan at kHz frequencies, holding potential for improvement over the approximate 10 Hz limit of translation stage detection. However, mirror-driven parallel detection on microchips with kHz rates has yet to be demonstrated.
The earliest mirror-driven laser scanning detectors were built for monitoring simultaneous DNA separations [24, 25]. One study gives a brief description of the laser scanning device, stating that a 5-μm laser spot was scanned across a 48 channel microchip (a distance of 7.4 mm) at rate of 40 Hz [24]. Fluorescence was collected through a stationary objective and detected with a PMT to generate 16-bit images from which parallel electropherograms were extracted. The reported LOD using this detector was 1 molecule Cy3 dye per 100 μm2.
Details of another homemade mirror-based scanning detector were described in a later study [26]. Juxtaposing the previous galvo-mirror detector that was described as scanning at a constant rate across a microchip, the mirror used in this study behaved similarly to a stepping motor allowing fast transitions between channels and longer integration times for a single detection point. This laser scanning technique is advantageous as it increases the duty cycle of the detector yielding higher sensitivity detection. A specific duty cycle of 71% was reported for this detector when scanning over an eight-channel microchip (six channels for separations, two for alignment) at a rate of 7.1 Hz for each channel. An LOD of 30 pM fluorescein was reported using this detector.
2.1.3 Acousto-optical Deflection
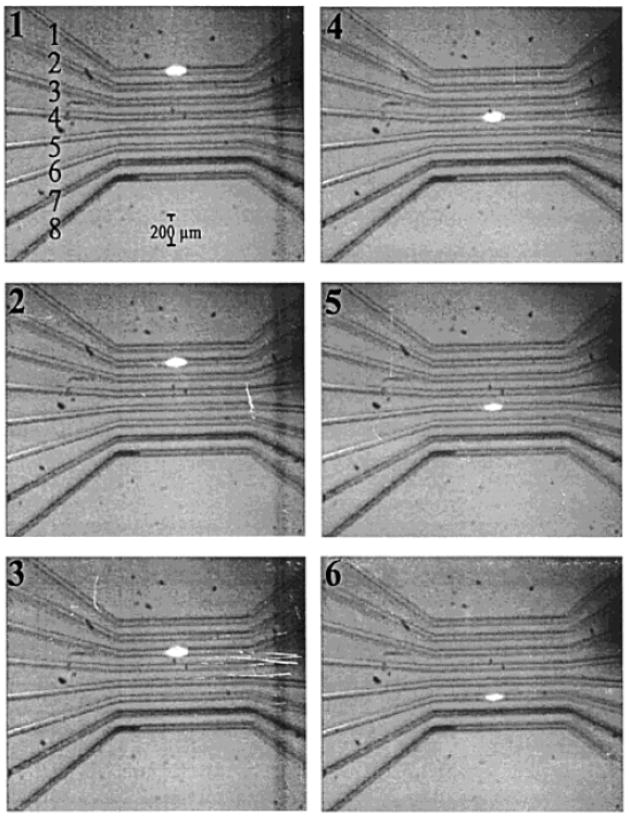
Though mirror scanning detectors can achieve high sampling rates and increased duty cycles, they are still mechanically dependent on moving parts. The use of any mechanically controlled scanner is going to inhibit achieving ultrafast scanning rates due to issues with mechanical noise and the distortion of signal [27]. The Landers group has done much work [27–29] towards eliminating the need for mechanically controlled instrumentation in parallel microchip detectors by employing laser scanning technology based on acousto-optical deflection (AOD). The principles of AOD are centered on laser light diffracting though an optically transparent medium at an angle that is dependent upon the frequency of acoustic waves traveling though the device. The angle of light diffraction can be changed by manipulating the acoustic wave frequency via a piezoelectric transducer. The advantage of using such a device for laser scanning includes the elimination of mechanical noise while achieving fast scan rates (kHz range). The developed detector has been used with a variety of microchips with an on-chip scanning distance of up to 2.4 cm, longer than the highest reported mirror-driven chip scanning distance of 10 mm. Fig. 1 shows a series of CCD images illustrating the scanning of a laser spot across an eight-channel chip. For these studies, fluorescence emission from sample bands was collected though a non-moving objective directed onto a PMT. An additional advantage in using this method of laser scanning is the ability to randomly address microchannels with channel-to-channel transition times down to 11.3 μs. One disadvantage, however, is that the diffraction of light through this device is wavelength dependent, making descanning and epi-fluorescence detection difficult to perform [23].
Figure 1.
Six CCD images showing the positioning of a laser spot scanned across eight microfluidic channels using AOD. This type of laser scanning was used to monitor parallel separations on a microchip with a PMT detector. [27]
2.1.4 Rotary Confocal
Attempts to maximize the number of microfluidic channels fabricated on a single device have led to the exploration of new channel organization methods. One design that has been proven useful for the arrangement of hundreds of channels on a single device is the radial alignment of separation channels in which fluidic reservoirs are situated close to the outside edge of a circular chip and separation channels converge to a common waste reservoir in the center. The Mathies group, responsible for the introduction of this parallel network design, has also developed a detector suitable for use with radially aligned channels [30]. Detection with this device, continually improved upon to allow use with higher channel densities from 96 to 384 [31, 32], is based on rotating an objective lens around an off-center axis for laser-excited confocal fluorescence in a circular path. For this device, the larger mechanically-based movements of the optical train have allowed dramatic increases in the number of channels on which detection can be performed. The detector uses a hollow stepping motor shaft with a rhombic prism and objective lens placed atop to allow displacement of laser light traveling up a vertical axis (orthogonal to the microchip surface) by 1 cm. The prism and objective are then rotated resulting in a scanning path of a 2 cm diameter circle. Fluorescence is collected though the same objective and directed towards a four-color confocal detector. It has been reported that sampling frequencies as high as 20 Hz can be used with a spatial resolution of ~12.6 μm, however, lower rotational speed is typically reported for experiments with this detector. LODs as low as ~1 pM (S/N = 2) have been reported using this device.
2.2 Fluorescence Imaging Detectors
CCDs and other imaging detectors have been used in conjunction with the scanning laser instruments featured above; however, mostly for the purpose of detector characterization and alignment rather than for detection in actual separation experiments. In cases when highly sensitive detection is not required, it may be advantageous to use an imaging detector due to the 100% duty cycle capabilities, commercial availability, lack of moving parts, and relative ease of use. Indeed, parallel fluorescence detection of multiple separations on a microchip has been performed on standard commercially available fluorescence microscopes using arc lamp excitation and CCD camera detection [33, 34]. Commercially available CCD cameras attain frame collection rates up to hundreds of frames per second making them useful for detection of fast separations. Additionally, the use of commercial imaging detectors requires little optical alignment. Disadvantages of an imaging detector include the risk of channel crosstalk (controlled by proximity of channels in the detection region and image resolution) and a defined imaging area that may be difficult to manipulate if a specific image magnification is required.
Several groups have demonstrated laser-induced excitation with CCD detection [35–39]. In these studies, laser light was shaped into a line using a cylindrical lens and directed across all sample channels on a chip, exciting sample plugs passing through it. Laser light was directed at an angle (typically around 45°) towards the chip surface while fluorescence was collected through perpendicular optics that focused an image of the detection region onto a CCD chip. Parallel separation data were then produced by plotting fluorescence intensities versus time from several regions of the images that corresponded to microfluidic channels. This detection scheme was used to monitor up to 10 parallel channels with a sampling frequency of 30 Hz [36]. An interesting modified version of this detector was demonstrated by a system that positioned a laser directed across parallel channels though the side of the microchip (perpendicular and in the same plane as channels) [40]. Imaging detection from 48 samples across a 140 mm width was completed with a CCD.
2.3 Electrochemical Detection
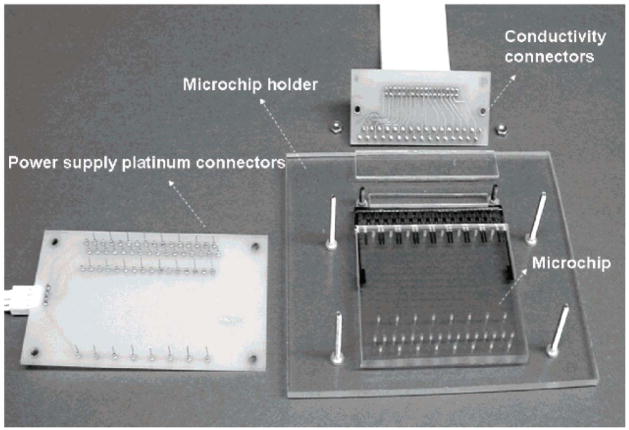
Electrochemical detection is another method for monitoring separations that has been used for parallel analysis on microchips owing to the ease of microfabricating detection electrodes on the same scale as microchannels. Although detection limits are not as low as have been achieved with fluorescence detection, electrochemical monitoring does offer reasonable LODs while requiring relatively simple instrumental setups. Electrochemical detection for parallel microchips has recently been demonstrated [41]. In this study, parallel electrochemical detection was performed with a contact conductivity array used to monitor simultaneous separations performed in 16 channels on a microchip. In contrast to other electrochemical techniques such as amperometry, conductivity detection does not require the analyte to be electroactive [42], suggesting that this device can be used with a wide variety of native samples. Detection at each channel was controlled by independent pairs of microfabricated gold electrodes (60 μm wide with a 5 μm space) and electrical circuits (16 circuits in total). Parallel separations were monitored using a bipolar pulse voltage waveform with an amplitude of ±0.6 V and frequency of 6 kHz. An image of the system is shown in Fig. 2. LODs as low as 1.6 μM were reported from using this detector with separations of peptides.
Figure 2.
Illustration of a workstation consisting of a microfluidic chip, chip holder, and necessary electrical connection devices for parallel CE separations and electrochemical detection. The device, fabricated from polycarbonate, allowed 16 parallel separations of amino acids, peptides, proteins, and DNA fragments. [41]
3 Parallel Microchip Applications
The multi-channel detectors reviewed have been used with a wide variety of parallel microchips, with most applications involving parallel chemical separations. Specific designs of parallel microchips have ranged from simple arrays of CE channels to more complicated systems including devices for parallel single cell lysing and separation [43] and on-chip PCR coupled to CE analysis [44]. This section will highlight advances towards microchips for parallel separations of biomolecules, affinity assays, and DNA analysis.
3.1 Separations of Biomolecules
One of the earliest examples of parallel amino acid separations on a microchip was performed on the previously described optical gating translation stage multiplexed detector [21, 45]. The nature of optical gating allowed chip designs to consist of parallel-aligned straight channels with no integrated injectors. In these studies, parallel separations of NBD-labeled amino acids (arginine, phenylalanine, glycine, and glutamic acid) were performed on a five-channel microchip with a 750 V cm−1 field strength and 5 mm separation distance. Reported detection limits for these separations were 1 mM, 500 μM, 500 μM, and 3.3 mM for arginine, phenylalanine, glycine, and glutamic acid, respectively. Additionally, parallel separation of DNA fragments was performed by this group using a similar chip design and detector [45].
Another microchip for parallel CE separations of biomolecules has been reported more recently [46]. This chip, while performing separations on only four parallel channels, required a more complicated design to allow for electrokinetic injections to be made using four single-tee injectors. Injections and separations on a single device were controlled by a high voltage power supply consisting of six high voltage modules and 12 high voltage relays. This study reported separations of FITC-labled glycine, arginine, phenylalanine, and lysine in a separation field of 400 V cm−1 over a distance of 40 mm. The same chip was used to separate myoglobulin, ovalbumin, BSA, and conalbumin using similar experimental procedures (same separation field and distance). In an additional study by this group, a similar parallel chip and detector were used for multi-channel chiral separations [38].
Further increasing the number of parallel separations of biomolecules performed on a single device, a 16-channel polycarbonate chip has been used to separate amino acids, peptides, proteins, and DNA fragments in a single experiment [41]. The 16 channels on the device were arranged into eight pairs of independent channels, allowing simultaneous separations of multiple sample types to be performed with relatively different experimental conditions. The chip performance was evaluated by performing CZE analysis of amino acids (alanine, valine, glutamine, tryptophan), peptides (leucine enkephalin, methionine enkephalin, oxytocin), and proteins (chymotrypsinogen A, cytochrome C, BSA) and CEC separations of an oligonucleotide ladder. Separations of all analytes were completed in less than 4 minutes in a field of 90 V cm−1 yielding detection limits as low as 7.1 μM, 1.6 μM, 3.3 μM, and 0.09 μg/μL for amino acids, peptides, proteins, and oligonucleotides, respectively.
Although it has been shown that radial organization of channels allows hundreds of parallel separations to be conducted on a single chip [31], the parallel organization of channels on a device can still be advantageous due to its compatibility with several detector types, ease of sample loading [41], and simple design that facilitates integrated electrodes for CE control. The latter advantage was demonstrated through a device that used a unique 12-channel parallel design for separations of DNA fragments [34]. The chip used I-shaped microchannels fabricated in PDMS (tapered at the injection point to act as a passive stop valve) to perform injections of sample and CE separations in a channel with only two fluidic reservoirs. The elimination of reservoirs used for sample injections facilitated the fabrication of parallel microchannels with integrated electrodes by allowing the close proximity of parallel networks. Functionality of the device was tested by performing parallel separations of 10 fragments of a 100 – 1000 bp DNA ladder.
3.2 Affinity Assays
The versatile organization of channel layouts offered by microfluidics makes the technique attractive for use with biochemical and affinity assays. For this reason, microchips have been frequently used in studies of protein and enzyme interactions [47]. Though microchips offer an increase in analysis speed for a single sample, the overall speed of multiple assay completion on a single sample device is still lower compared to the multiplexing abilities of standard larger-scale methods such as plate-readers or gels with multiple lanes. The creation of microdevices for parallel affinity assays increases the overall throughput of homogenous assays in microchip format, making rate of completion comparable with standard assay techniques.
3.2.1 Immunoassays
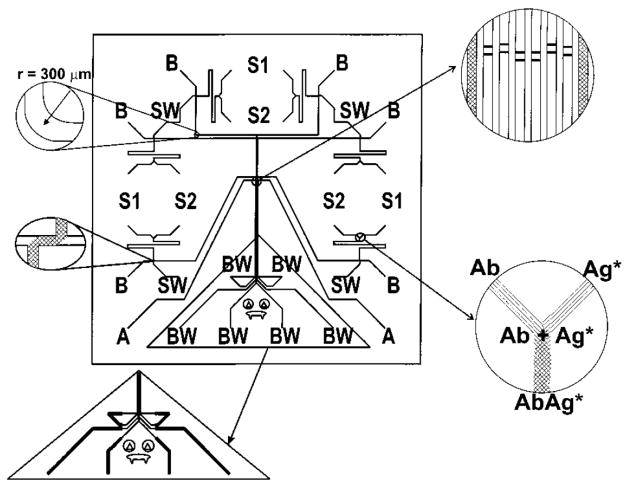
The first microfluidic device for the parallel completion of CE-based immunoassays was characterized with direct assays for ovalbumin and anti-estradiol [26]. The device, illustrated in Fig. 3, consisted of eight separate channel manifolds, two for detector alignment and six capable of supporting online mixing of immunoassay reagents and CE-LIF analysis. Mixing of reagents, reaction, and separation of products was completed within 60 s for all experiments and within 30 s in optimized conditions, demonstrating the potential of high-speed parallel immunoassays on a chip. Detection was performed with a scanning LIF instrument built in-house. The chip was used to demonstrate both simultaneous immunoassays of a single sample and simultaneous calibration and analysis in a single experiment, allowing a complete calibrated assay to be performed within 30 s. Detection limits reported for an anti-estradiol assay were 4.3 nM in any single channel and 6.4 nM for the cross-channel calibration mode.
Figure 3.
Illustration of a microchip design for six simultaneous CE-based immunoassays. Each channel manifold is capable of online mixing of immunoassay reagents (from reservoirs S1 and S2) followed by injection onto capillary electrophoresis channels for separation and detection. [26]
A dramatic increase in the number of simultaneous CE-based immunoassays performed on a microchip was demonstrated with a 48-channel device [48]. While offering an increased rate of immunoassay product separation and detection, this device was comprised of only electrophoretic injection and separation channels and lacked the ability for on-chip mixing of reagents and calibration. Sample preparation was performed completely off-chip and consisted of a 45 minute incubated reaction. The chip was used for a TNT assay with a reported 1 ng mL−1 LOD and dynamic range of 1 – 300 ng mL−1.
A recently developed chip has demonstrated the use of parallel immunoassays for the monitoring of cellular secretions from multiple independent biological samples housed on the device [49]. With this device, insulin secretion was continuously sampled from four separate pancreatic islets and quantified every 6.25 s using online mixing and reacting of immunoassay reagents coupled to parallel CE-LIF analysis. Over 700 assays were completed in a 20 min experiment, noting a significant decrease in immunoassay reagent costs compared to typical insulin ELISA kits. Parallel fluorescence detection was performed using a commercial scanning confocal microscope operated in line scan mode. An LOD for insulin was reported as 7 nM. A 15-sample microchip based on the same assay has also been developed [50].
Additional parallel immunoassays, though not utilizing CE, have been developed for sandwich-based assays [51, 52]. One report examined a polymer CD-shaped chip that utilized centrifugal force for mixing and dispensing immunoassay reagents in precisely defined volumes [51]. The chip, when rotated at sufficient speed, would introduce solutions of biotinylated antibodies against a target molecule into 104 parallel on-chip microcolumns packed with streptavidin-coated particles. Solutions of sample and then fluorescently labeled detection antibody were then introduced in a similar manner after which fluorescence of the packed beds was used to determine sample binding. Assays for α-fetoprotein, interleukin-6, and carcinoembryonic antigen with reported LODs of 0.15, 1.25, and 1.31 pM, respectively, were performed at speeds up to 104 immunoassays in 50 min.
3.2.2 Enzyme Assays
A parallel microchip for enzyme assays was reported [53] that used a similar chip design and translation stage optical gating detector from previous work [21, 45]. To monitor multiple enzymatic reactions, continuous serial injections of an enzyme/substrate mixture were made into parallel microchannels for CE-LIF analyses. The chip was used for monitoring the hydrolysis of fluorescein mono-β-D-galactopyranoside (FMG) by β-galactosidase (β-Gal) with and without phenylethyl β-D-thio-galactoside (PETG), a competitive inhibitor. The separation of enzyme reaction products was performed in a separation field of 500 V cm−1 with 30 s temporal resolution. Results from the parallel studies were used to calculate Km values from Lineweaver-Burk plots. Additionally, the simultaneous monitoring of the hydrolysis of FMG with different inhibitors (no inhibitor, lactose, and PETG) was performed by testing each inhibitor on a different channel. The results from this study illustrate the potential for microfluidics-based high-throughput screening of drug candidates for potential activity.
3.3 DNA Analysis and Integrated Devices
To address the needs of the human genome project, many researchers strived for the development of methods to improve the speed, cost-effectiveness, and throughput of DNA sequencing [54]. The developments of capillary array gel electrophoresis have offered techniques equaling the high-number DNA sample processing capabilities of conventional slab gel electrophoresis, making it commonly used for DNA analysis. Owing to the simple, low cost fabrication of microfluidic devices and potential for high speed separations, it has not been surprising to see rapid development of microchips for parallel DNA separations and sequencing. The capabilities of the first parallel chip for DNA analysis were tested by performing 12 simultaneous separations of pBR322 DNA samples and also by genotyping HLA-H [20]. While this device was not able to provide higher throughput than the current CAE techniques, it demonstrated potential for multiplexed genetic analysis on chips.
The ease of fabricating parallel channels in microfluidic devices has allowed rapid improvements to this 12-channel design to be made over the past decade. There are several reviews available that include a discussion on the development of these devices [32, 54–56]. The Mathies group has pioneered much progress towards high-throughput genetic analysis on microchips by developing 48- [24], 96- [30, 57], and 384-lane [31] devices for DNA separations. The development of these chips was a significant advancement as the throughput of DNA microdevices was improved while the cost of fabrication did not increase substantially. Potential applications of parallel DNA chips have been demonstrated through single nucleotide polymorphism genotyping [58, 59], single-strand conformation polymorphism analysis [60], and short tandem repeat typing [61].
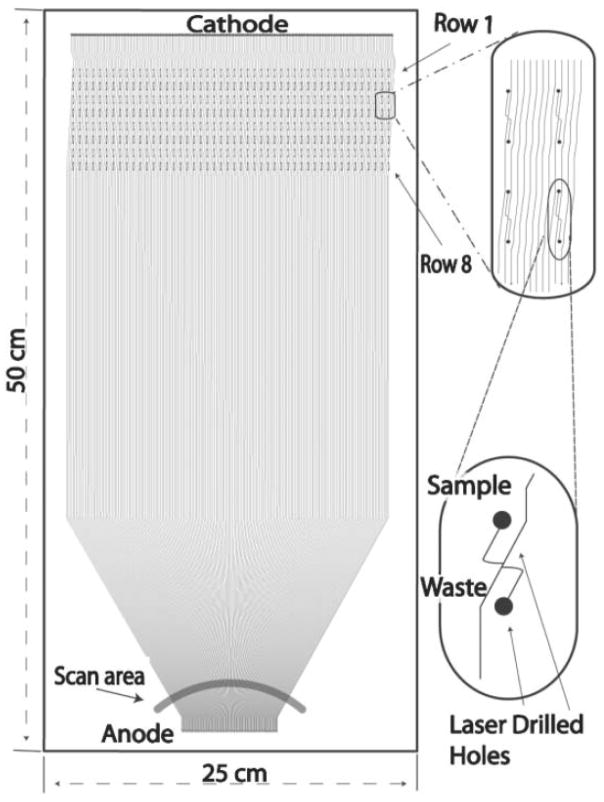
Additionally, a recent paper has reported a system with 768 parallel channels for high throughput DNA sequencing [62]. This system consisted of two 25 cm × 50 cm microfabricated plates, one illustrated in Fig. 4, with 384 separation channels each. Analysis was performed on an automated custom-build instrument for sample loading, separation, detection, and plate regeneration (one plate was prepared for experiments while another was used for analysis). A scanning epi-fluorescence detector rotating at 3 Hz, compatible with the 384 lanes of a single chip, was used for single plate detection. Once the separations were completed on one chip, the laser was then relocated to the adjacent chip for a second analysis.
Figure 4.
One of two microfabricated plates used in a 768-lane system for DNA sequencing. Each plate consists of 384 parallel channels. Separation and detection is performed on one plate while another is regenerated. Details on the automated instrumentation used are available here. [62]
While these devices show unprecedented capabilities for performing hundreds of separations simultaneously, they are comprised of only simple injection and separation channels and have no other integrated function capabilities. Recent advances in integrated parallel DNA analysis on microchips include devices for PCR amplification of samples and CE separations [44, 63]. On these chips, precise temperature of four parallel nanoliter sized PCR chambers was controlled with resistance temperature detectors and heaters integrated into the devices. Flow through these chambers and onto separation channels were controlled by PDMS valves. Though these reports present a down-sizing of the number of parallel analyses compared to previous DNA microchips, there exists potential for an increase in throughput through the addition of microchannel networks and heating elements.
4 Concluding Remarks
The research devoted to developing lab-on-a-chip devices for multiplexed analysis along with complimentary detection instrumentation has yielded a number of exceptional systems that exemplify the potential for their widespread use. Work towards this goal is still needed; however, preliminary success in this field has shown outstanding results. Widespread use of parallel microchips is beginning to be realized through the commercial development of high-throughput microdevices by several companies. Future work in this direction should focus on the integration of sensitive detectors on parallel analysis chips, eliminating the need for specialized instrumentation and facilitating portability and use in clinical, environmental, and research lab locations. The advancements outlined by this review lead the way for a greater transition from microchips being used only in basic research labs to their specialized design and use for real-world situations.
Abbreviations
- AOD
acousto-optical deflection
- BSA
bovine serum albumen
- ELISA
enzyme-linked immuno-sorbent assay
- NBD
4-chloro-7-ni-trobenz-2-oxa-1, 3-diazole
- PCR
polymerase chain reaction
- PDMS
poly(dimethylsiloxane)
- TNT
2,4,6-trinitrotoluene
References
- 1.Manz A, Graber N, Widmer HM. Sens Actuat B 1. 1990:224–248. [Google Scholar]
- 2.Harrison DJ, Manz A, Fan Z, Ludi H, Widmer HM. Anal Chem. 1992;64:1926–1932. [Google Scholar]
- 3.Effenhauser CS, Bruin GJM, Paulus Electrophoresis. 1997;18:2203–2213. doi: 10.1002/elps.1150181211. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson SC, Hergenroder R, Koutny LB, Ramsey JM. Anal Chem. 1994;66:1114–1118. [Google Scholar]
- 5.Dolnik V, Liu S, Jovanovich SB. Electrophoresis. 2000;21:41–54. doi: 10.1002/(SICI)1522-2683(20000101)21:1<41::AID-ELPS41>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 6.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson SC, Koutny LB, Hergenroder R, Moore AW, Ramsey JM. Anal Chem. 1994;66:3472–3476. [Google Scholar]
- 8.Woolley AT, Hadley D, Landre P, deMello AJ, Mathies RA, Northrup MA. Anal Chem. 1996;68:4081–4086. doi: 10.1021/ac960718q. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson SC, Hergenroder R, Moore AW, Ramsey JM. Anal Chem. 1994;66:4127–4132. [Google Scholar]
- 10.Panaro NJ, Yuen PK, Sakazume T, Fortina P, Kricka LJ, Wilding P. Clin Chem. 2000;46:1851–1853. [PubMed] [Google Scholar]
- 11.Yin H, Killeen K. J Sep Sci. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- 12.Yin H, Killeen K, Brennen R, Sobek D, Werlich M, van de Goor T. Anal Chem. 2005;77:527–533. doi: 10.1021/ac049068d. [DOI] [PubMed] [Google Scholar]
- 13.Wan H, Bergstrom F. J Liq Chromatogr R T. 2007;30:681–700. [Google Scholar]
- 14.Ducret A, Van Oostveen I, Eng JK, Yates JR, Aebersold R. Protein Sci. 1998;7:706–719. doi: 10.1002/pro.5560070320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang HC, Quesada MA, Mathies RA. Anal Chem. 1992;64:967–972. doi: 10.1021/ac00042a021. [DOI] [PubMed] [Google Scholar]
- 16.Dittrich PS, Tachikawa K, Manz A. Anal Chem. 2006;78:3887–3907. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 17.Roman GT, Kennedy RT. J Chromatogr A. 2007;1168:170–188. doi: 10.1016/j.chroma.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Tao L, Kennedy RT. TrAC - Trend Anal Chem. 1998;17:484–491. [Google Scholar]
- 19.Mathies RA, Huang XC. Nature. 1992;359:167–169. [Google Scholar]
- 20.Woolley AT, Sensabaugh GF, Mathies RA. Anal Chem. 1997;69:2181–2186. doi: 10.1021/ac961237+. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Roddy TP, Lapos JA, Ewing AG. Anal Chem. 2002;74:5517–5522. doi: 10.1021/ac025773f. [DOI] [PubMed] [Google Scholar]
- 22.Lapos JA, Ewing AG. Anal Chem. 2000;72:4598–4602. doi: 10.1021/ac000581a. [DOI] [PubMed] [Google Scholar]
- 23.Pawley JB. Handbook of Biological Confocal Microscopy. Plenum Press; New York: 1995. [Google Scholar]
- 24.Simpson PC, Roach DJ, Woolley AT, Thorsen T, Johnston R, Sensabaugh GF, Mathies RA. Proc Natl Acad Sci. 1998;95:2256–2261. doi: 10.1073/pnas.95.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu S, Ren H, Gao Q, Roach DJ, Loder RT, Armstrong TM, Mao Q, Blaga I, Barker DL, Jovanovich SB. Proc Natl Acad Sci. 2000;97:5369–5374. doi: 10.1073/pnas.100113197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng SB, Skinner CD, Taylor J, Attiya S, Lee WE, Picelli G, Harrison DJ. Anal Chem. 2001;73:1472–1479. doi: 10.1021/ac0007938. [DOI] [PubMed] [Google Scholar]
- 27.Huang Z, Munro N, Huhmer AFR, Landers JP. Anal Chem. 1999;71:5309–5314. doi: 10.1021/ac990740u. [DOI] [PubMed] [Google Scholar]
- 28.Sanders JC, Huang Z, Landers JP. Lab Chip. 2001;1:167–172. doi: 10.1039/b107835f. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z, Jin L, Sanders JC, Zheng Y, Dunsmoor C, Tian H, Landers JP. IEEE T Bio-med Eng. 2002;49:859–866. doi: 10.1109/TBME.2002.800767. [DOI] [PubMed] [Google Scholar]
- 30.Shi Y, Simpson PC, Scherer JR, Wexler D, Skibola C, Smith MT, Mathies RA. Anal Chem. 1999;71:5354–5361. doi: 10.1021/ac990518p. [DOI] [PubMed] [Google Scholar]
- 31.Emrich CA, Tian H, Medintz IL, Mathies RA. Anal Chem. 2002;74:5076–5083. doi: 10.1021/ac020236g. [DOI] [PubMed] [Google Scholar]
- 32.Medintz IL, Paegel BM, Blazej RG, Emrich CA, Berti L, Scherer JR, Mathies RA. Electrophoresis. 2001;22:3845–3856. doi: 10.1002/1522-2683(200110)22:18<3845::AID-ELPS3845>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 33.Shackman JG, Munson MS, Ross D. Anal Chem. 2007;79:565–571. doi: 10.1021/ac061759h. [DOI] [PubMed] [Google Scholar]
- 34.Inoue A, Ito T, Makino K, Hosokawa K, Maeda M. Anal Chem. 2007;79:2168–2173. doi: 10.1021/ac0616097. [DOI] [PubMed] [Google Scholar]
- 35.Roddy ES, Price M, Ewing AG. Anal Chem. 2003;75:3704–3711. doi: 10.1021/ac034223u. [DOI] [PubMed] [Google Scholar]
- 36.Dang F, Tabata O, Kurosawa M, Ewis AA, Zhang L, Yamaoka Y, Shinohara S, Shinohara Y, Ishikawa M, Baba Y. Anal Chem. 2005;77:2140–2146. doi: 10.1021/ac0485031. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Yin X. Electrophoresis. 2007;28:1281–1288. doi: 10.1002/elps.200600553. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y, Shen Z, Wang H, Dai Z, Lin B. Electrophoresis. 2005;26:4774–4779. doi: 10.1002/elps.200500283. [DOI] [PubMed] [Google Scholar]
- 39.Smith EM, Xu H, Ewing AG. Electrophoresis. 2001;22:363–370. doi: 10.1002/1522-2683(200101)22:2<363::AID-ELPS363>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 40.Simpson JW, Ruiz-Martinez MC, Mulhern GT, Berka J, Latimer DR, Ball JA, Rothberg JM, Went GT. Electrophoresis. 2000;21:135–149. doi: 10.1002/(SICI)1522-2683(20000101)21:1<135::AID-ELPS135>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Shadpour H, Hupert ML, Patterson D, Liu C, Galloway M, Stryjewski W, Goettert J, Soper SA. Anal Chem. 2007;79:870–878. doi: 10.1021/ac0612168. [DOI] [PubMed] [Google Scholar]
- 42.Solinova V, Kasicka V. J Sep Sci. 2006;29:1743–1762. doi: 10.1002/jssc.200600167. [DOI] [PubMed] [Google Scholar]
- 43.Munce NR, Li J, Herman PR, Lilge L. Anal Chem. 2004;76:4983–4989. doi: 10.1021/ac0496906. [DOI] [PubMed] [Google Scholar]
- 44.Liu CN, Toriello NM, Mathies RA. Anal Chem. 2006;78:5474–5479. doi: 10.1021/ac060335k. [DOI] [PubMed] [Google Scholar]
- 45.Xu H, Roddy ES, Roddy TP, Lapos JA, Ewing AG. J Sep Sci. 2004;27:7–12. doi: 10.1002/jssc.200301593. [DOI] [PubMed] [Google Scholar]
- 46.Shen Z, Liu X, Long Z, Liu D, Ye N, Qin J, Dia Z, Lin B. Electrophoresis. 2006;27:1084–1092. doi: 10.1002/elps.200500689. [DOI] [PubMed] [Google Scholar]
- 47.Bilitewski U, Genrich M, Kadow S, Mersal G. Anal Bioanal Chem. 2003;377:556–569. doi: 10.1007/s00216-003-2179-4. [DOI] [PubMed] [Google Scholar]
- 48.Bromberg A, Mathies RA. Electrophoresis. 2004;25:1895–1900. doi: 10.1002/elps.200305818. [DOI] [PubMed] [Google Scholar]
- 49.Dishinger JF, Kennedy RT. Anal Chem. 2007;79:947–954. doi: 10.1021/ac061425s. [DOI] [PubMed] [Google Scholar]
- 50.Dishinger JF, Kennedy RT. 2008 in preparation. [Google Scholar]
- 51.Honda H, Lindberg U, Andersson P, Hoffmann S, Takei H. Clin Chem. 2005;51:1955–1961. doi: 10.1373/clinchem.2005.053348. [DOI] [PubMed] [Google Scholar]
- 52.Herrmann M, Veres T, Tabrizian M. Lab Chip. 2006;6:555–560. doi: 10.1039/b516031f. [DOI] [PubMed] [Google Scholar]
- 53.Xu H, Ewing AG. Electrophoresis. 2005;26:4711–4717. doi: 10.1002/elps.200500620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrilho E. Electrophoresis. 2000;21:55–65. doi: 10.1002/(SICI)1522-2683(20000101)21:1<55::AID-ELPS55>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 55.Kan CW, Fredlake CP, Doherty EAS, Barron AE. Electrophoresis. 2004;25:3564–3588. doi: 10.1002/elps.200406161. [DOI] [PubMed] [Google Scholar]
- 56.Zhang L, Dang F, Baba Y. J Pharmaceut Biomed. 2003;30:1645–1654. doi: 10.1016/s0731-7085(02)00510-1. [DOI] [PubMed] [Google Scholar]
- 57.Paegel BM, Emrich CA, Wedemayer GJ, Scherer J, Mathies RA. Proc Natl Acad Sci. 2002;99:574–579. doi: 10.1073/pnas.012608699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Medintz IL, Wong WW, Sensabaugh G, Mathies RA. Electrophoresis. 2000;21:2352–2358. doi: 10.1002/1522-2683(20000701)21:12<2352::AID-ELPS2352>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 59.Mendintz I, Wong WW, Berti L, Shiow L, Tom J, Scherer J, Sensabaugh G, Mathies RA. Genome Res. 2001;11:413–421. doi: 10.1101/gr.164701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian H, Emrich CA, Scherer JR, Mathies RA, Andersen PS, Larsen LA, Christiansen M. Electrophoresis. 2005;26:1834–1842. doi: 10.1002/elps.200410205. [DOI] [PubMed] [Google Scholar]
- 61.Yeung SHI, Greenspoon SA, McGuckian A, Crouse CA, Emrich CA, Ban J, Mathies RA. J Forensic Sci. 2006;51:740–747. doi: 10.1111/j.1556-4029.2006.00153.x. [DOI] [PubMed] [Google Scholar]
- 62.Aborn JH, El-Difrawy SA, Novotny M, Gismondi EA, Lam R, Matsudaira P, Mckenna BK, O’Neil T, Streechon P, Ehrlich DJ. Lab Chip. 2005;5:669–674. doi: 10.1039/b501104c. [DOI] [PubMed] [Google Scholar]
- 63.Toriello NM, Liu CN, Mathies RA. Anal Chem. 2006;78:7997–8003. doi: 10.1021/ac061058k. [DOI] [PubMed] [Google Scholar]