Summary
The sequences encoding the QUAD1 RNAs were initially identified as four repeats in Escherichia coli. These repeats, herein renamed SIB, are conserved in closely related bacteria, though the number of repeats varies. All five Sib RNAs in E. coli MG1655 are expressed, and no phenotype was observed for a five sib deletion strain. However, a phenotype reminiscent of plasmid addiction was observed for overexpression of the Sib RNAs, and further examination of the SIB repeat sequences revealed conserved open reading frames encoding highly hydrophobic 18–19 amino acid proteins (Ibs) opposite each sib gene. The Ibs proteins were found to be toxic when overexpressed and this toxicity could be prevented by co-expression of the corresponding Sib RNA. Two other RNAs encoded divergently in the yfhL-acpS intergenic region were similarly found to encode a small hydrophobic protein (ShoB) and an antisense RNA regulator (OhsC). Overexpression of both IbsC and ShoB led to immediate changes in membrane potential suggesting both proteins affect the cell envelope. Whole genome expression analysis showed that overexpression of IbsC and ShoB, as well as the small hydrophobic LdrD and TisB proteins, has both overlapping and unique consequences for the cell.
Keywords: toxin-antitoxin, small RNA, microarray
Introduction
The identification of small, non-coding RNAs (sRNAs) in all organisms has increased dramatically in recent years [reviewed in (Altuvia, 2007; Livny and Waldor, 2007)]. Computational as well as experimental evidence suggests that at least 70 sRNAs are transcribed from the Escherichia coli MG1655 genome [reviewed in (Storz and Gottesman, 2006)]. A few of the characterized sRNAs bind proteins and modify their activities. The other characterized sRNAs function by base pairing with target mRNAs. Base pairing leads to changes in the stability and/or translation of the target. The majority of the base pairing sRNAs in E. coli act on targets not encoded in the same genetic region and only have limited complementarity with their targets. However, two sRNAs, RdlD and SymR, control the expression of proteins encoded on the opposite strand and thus are perfectly complementary to the target mRNA (Kawano et al., 2002; Kawano et al., 2007). In addition, one sRNA, IstR, is encoded in the same intergenic region as its tisB mRNA target and shares a 23-nucleotide stretch of complementarity with this target (Vogel et al., 2004). Interestingly RdlD, SymR and IstR all control the translation of proteins (LdrD, SymE and TisB, respectively) whose overexpression is toxic to the cell.
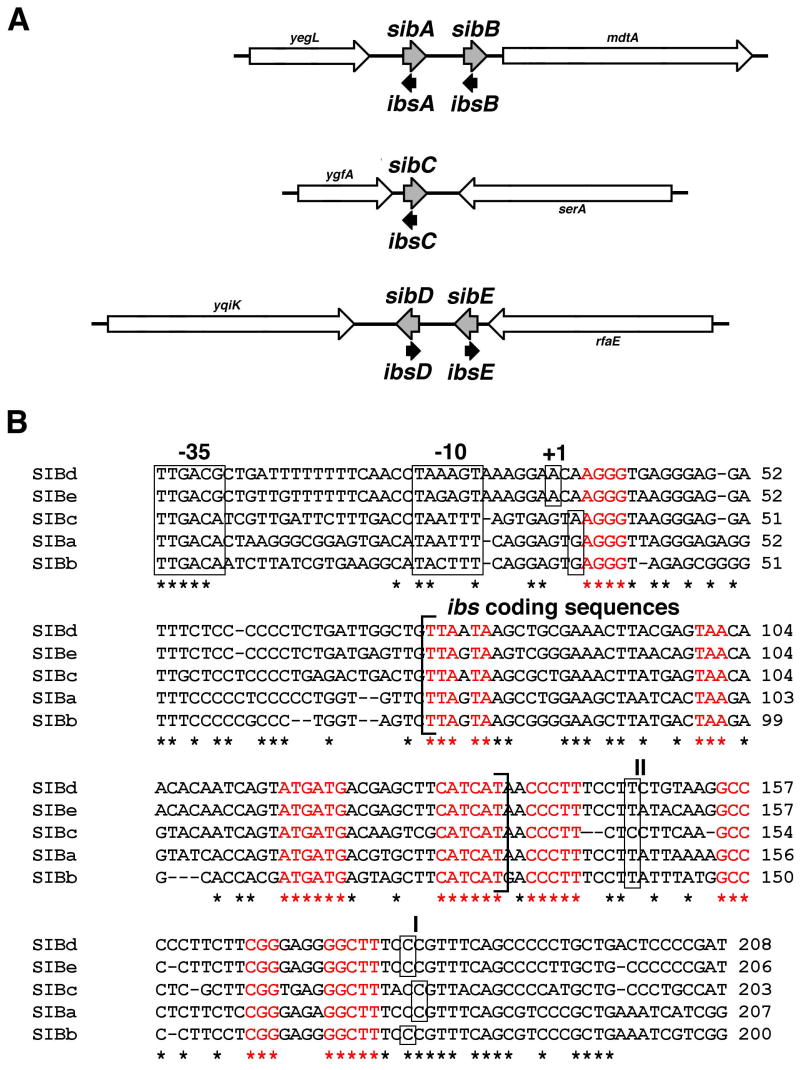
Most genes encoding sRNAs in E. coli are unique sequences; however, the sequences encoding the QUAD1 RNAs were initially identified as four repeat elements in the intergenic regions of E. coli (Rudd, 1999). These sequences were predicted to encode sRNA molecules as they contained potential −35 and −10 promoter elements, but no ribosome binding site or open reading frame (ORF). Subsequent studies demonstrated transcription of these genes in MG1655 and identified homologous sequences in Shigella and Salmonella species (Argaman et al., 2001; Hershberg et al., 2003; Rivas et al., 2001; Wassarman et al., 2001). Two of the QUAD repeats, QUAD1a and QUAD1b, are located in tandem in the intergenic region between yegL-mdtA. The other two QUAD1 sequences, QUAD1c and QUAD1d, are located in the ygfA-serA and yqiK-rfaE intergenic regions, respectively, distant from each other and the QUAD1ab region.
We were interested in determining the function of the sRNAs encoded by the QUAD1a, QUAD1b, QUAD1c and QUAD1d repeats as well as a fifth repeat identified in this study. As the number of these homologous sequences can vary in other E. coli strains, we propose to rename the repeats SIBs for short, intergenic, abundant sequences. We confirmed that all five Sib RNAs are expressed in MG1655. Furthermore, although the deletion of all sib genes was not detrimental under any of the conditions tested, we found multicopy plasmids carrying the SIB sequences could not be introduced into the corresponding deletion strain unless the plasmid-encoded Sib RNA was expressed at high levels. Upon re-examination of the sib gene sequences we discovered the presence of a small ORF encoded opposite each Sib RNA. These genes, termed “ibs” for induction brings stasis, encode very hydrophobic 18–19 amino acid proteins. The Ibs are highly conserved in all species that contain the sib genes. Overexpression of the Ibs proteins is lethal to the cell, but this can be prevented by expression of the corresponding antisense Sib RNA.
We also examined another pair of RNAs, previously referred to as RyfB and RyfC, which are encoded divergently in the same intergenic region but share a 19 nucleotide region of complementarity (Kawano et al., 2005). We found the RyfB RNA also encodes a short hydrophobic ORF (26 amino acids in length) that is lethal upon overproduction and thus we renamed this gene shoB. The RyfC RNA regulates levels of ShoB in the cell, and is now denoted OhsC for oppression of hydrophobic ORF by sRNA.
High levels of both IbsC and ShoB lead to changes in membrane potential suggesting that both act at the membrane, consistent with the hydrophobic nature of the proteins. In addition, whole genome expression analysis upon overexpression of IbsC and ShoB as well as LdrD and TisB, two other short hydrophobic proteins that are toxic upon overexpression, revealed that these small proteins all induce a common set of genes. However, additional genes are strongly induced or repressed by subsets of these proteins indicating that the small, hydrophobic proteins do not act in an identical fashion.
Results
The numbers of SIB repeats vary between species
The QUAD1 repeats were initially identified as four highly homologous sequences located in three separate intergenic regions of the E. coli chromosome (Rudd, 1999). Expression of the four QUAD1 RNAs was reported in separate studies (Argaman et al., 2001; Rivas et al., 2001; Wassarman et al., 2001). Since most sRNAs of E. coli are unique sequences, we sought to determine the function of these duplicated sRNA genes. First, we reexamined the number of QUAD1 repeats encoded by E. coli. A revision of the MG1655 sequence (Hayashi et al., 2006) revealed a 375-nucleotide region had been omitted in the yqiK-rfaE intergenic region, where the QUAD1d sequence is located. A fifth sequence homologous to the QUADs was identified in this region (Fig. 1). As there are five of these repeat sequences in MG1655, the original name QUAD1 is a misnomer. We propose to rename these sequences SIBs for short intergenic abundant sequences with the individual repeats designated as SIBa, SIBb, SIBc, SIBd, and SIBe following the original QUAD1a, QUAD1b, QUAD1c and QUAD1d nomenclature.
Fig. 1. Sib gene homology and genetic organization.
A. Genetic organization of the sib genes. The Ibs ORFs are indicated by black arrows.
B. ClustalW alignment (with minimal manual alignment) of the five SIB repeats from E. coli MG1655. Red sequences are those conserved in all SIB repeats found in enteric bacteria. The predicted −35 and −10 promoter elements for the Sib RNAs are shown. The +1 indicates transcriptional start site as determined by 5′ RACE. The two distinct 3′ ends mapped by 3′ RACE are also indicated by I and II. The Ibs ORFs are indicated by brackets.
Upon searching for homologous sequences in other E. coli strains, we noted that the number of SIB repeats varies between strains (Table 1). Two sequenced EHEC strains, E. coli 0157:H7 EDL933 and E. coli 0157:H7 VT-2 Sakai, contain seven SIB repeats, with one additional repeat in the yegL-mdtA intergenic region and the other additional repeat in the ygfA-serA region. On the other hand, the genomes of E. coli CFT073 and other UPEC/APEC strains are predicted to have only four SIB repeats, with a single repeat in the yqiK-rfaE region. It is worth noting though that the “missing” sib genes of UPEC/APEC strains occur in the yqiK-rfaE intergenic region, the same region where the original sequencing error occurred in MG1655. The genomes of Shigella, Salmonella and Citrobacter also contain SIB repeats of varying numbers (Table 1). Repeats in the same intergenic region are more homologous to each other than with the repeats in other regions indicating that they are likely to be more recent duplications.
Table 1.
Number of sib genes in various enteric strains
| Strain | Number of genes | Genes presenta |
|---|---|---|
| E. coli K-12 MG1655 | 5 | A, B, C, D, E |
| E. coli K-12 W3110 | ||
| E. coli 0157:H7 EDL933 | 7 | A, B, B2, C, C2, D, E |
| E. coli 0157:H7 VT-2 Sakai | ||
| E. coli CFT073 (UPEC)b | 4 | A, B, C, D |
| E. coli 01:K1 (APEC) | ||
| Shigella boydii Sb227 | 7 | A, B, B2, C, C2, D, E |
| Shigella sonnei Ss046 | ||
| Shigella flexneri 2a 2457Tc | 5 | C, C2, D, D2, E |
| Shigella dysenteriae Sd197 | 3 | C, D, E |
| Salmonella typhimurium LT2d | 3 | A, C, D |
| Citrobacter koseri ATCC BAA-895 | 1 | A |
The A, B, C, D, E designation comes from the original QUAD nomenclature with A and B flanked by yegL and mdtA, C flanked by ygfA and serA and D and E flanked by yqiK and rfaE. “2” refers to a duplicated gene.
Two additional UPEC strains, 06:K15:H31 (str.536) and UT189 contain the same four genes.
Two additional S. flexneri strains, 2a str. 301 and 5 str. 840f1 contain the same five genes.
Salmonella enterica serovar Typhi CT18, S. enterica serovar Typhi T2, S. enterica Paratypi, and S. enterica Choleraesuis contain the same three genes.
An alignment of the SIB repeats shows that specific nucleotides are absolutely conserved among the enteric species (Fig. 1). Using the M-fold program to predict the secondary structures of the encoded RNAs, many of the conserved nucleotides are predicted to be in stem structures that are conserved across all strains (Fig. S1). Thus, there is substantial sequence and potentially structural conservation for all Sib RNAs.
All five sib genes in E. coli MG1655 are expressed
Northern analysis was carried out to determine whether E. coli MG1655 expressed all five predicted Sib RNAs. To verify that the five oligonucleotide probes were specific to a given sib gene, we also probed total RNA isolated from mutant strains carrying single deletions (see below) of each of the sib genes. As shown in Fig. 2, transcripts specific to each of the five sib genes were detected during growth in rich as well as minimal medium though overall the levels in minimal medium tend to be higher. Two transcripts were observed per sib gene. The patterns of expression and the ratio of the shorter and longer transcripts varied to some extent; in general, the longer transcript tended to dominate for RNA isolated from stationary-phase cells. In the case of the SibD and SibE RNAs, the longer transcript was expressed more highly in most conditions. Deletion of the one or more sib genes did not appear to alter the expression pattern of the other genes (data not shown). Fusions of the sibC and sibE promoter to the lacZ reporter were also constructed. We observed high levels of β-galactosidase activity under all conditions tested, though again the levels were somewhat higher in minimal compared to rich medium (data not shown).
Fig. 2. Expression of the sib genes.
Total RNA (5 μg) isolated from MG1655 cells grown to OD600 ≈ 0.4 (E), OD600 ≈ 1.8 (L) and OD600 ≈ 5.0 (overnight, S) in LB medium and from MG1655 cells grown to OD600 ≈ 0.4 (E), OD600 ≈ 1.8 (L) and OD600 ≈ 2.5 (overnight, S) in M9 media supplemented with 0.2% glucose were loaded in each lane. The fourth lane in each corresponds to the specific sib deletion strain. No additional bands were observed even upon overexposure.
Based on the Northern analysis, each sib gene produces two distinct transcripts approximately 150 and 110 nucleotides in length. To map the ends of the two observed transcripts, we carried out 5′ and 3′ RACE analysis. These assays showed that there is a single transcriptional start site and two distinct 3′ ends for each RNA. The lengths range from 136 to 145 nucleotides for the longer transcripts and 104 to 112 nucleotides for the shorter transcripts (Fig. 1; Tables S1 and S2). There is no obvious terminator corresponding to either of the 3′ ends, though the end corresponding to the longer transcript is adjacent to a long predicted stem-loop. The 3′ end corresponding to the shorter transcript is in a region predicted to be single stranded so we suggest this product may be a result of processing.
Effects of sib gene deletions
We generated a series of strains carrying single sib gene deletions and various combinations of deletions, such as ΔsibAB, ΔsibDE and ΔsibABCDE, using phage recombination (Court et al., 2003). Growth of these deletion strains was monitored in both rich and minimal medium supplemented with various carbon sources and at different temperatures. Under these conditions, there were no discernable differences between the wild type control and any of the sib deletion strains (data not shown).
Effects of Sib RNA overproduction
To examine the effects of Sib RNA overproduction, the individual genes were placed under the control of the PBAD promoter of plasmid pAZ3 (Kawano et al., 2007). The wild type strain was readily transformed with each overexpression construct, and no significant growth differences were observed between strains carrying the empty vector or the plasmids with inserts, with or without the addition of the inducing agent arabinose. This was not the case when the constructs were transformed into their respective sib gene deletion strains. For example, successful transformations of a plasmid bearing PBAD-sibE into the ΔsibE strain occurred only when agar plates were supplemented with 0.1% and 0.2% arabinose (Table 2 and data not shown). No colonies were obtained when cells were plated on 0.02% and 0.002% arabinose even though these concentrations could induce expression of SibE from the plasmid (data not shown). In contrast, PBAD-sibA, PBAD-sibB, PBAD-sibC and PBAD-sibD plasmid transformants of the same ΔsibE strain could be obtained with and without the inducing agent (Table 2 and data not shown). This was found to be the case for every sib deletion strain except ΔsibA; transformants of ΔsibA, as well as ΔsibAB, with the PBAD-sibA plasmid could be obtained without the addition of arabinose (Table 2 and data not shown).
Table 2.
Efficiency of transformation into different sib deletion strainsa
| vector | arabinose | MG1655 | ΔsibA | ΔsibC | ΔsibE |
|---|---|---|---|---|---|
| pAZ3 | − | 1.6 × 108 | 5.0 × 107 | 2.5 × 108 | 1.6 × 108 |
| + | 1.8 × 108 | 5.2 × 107 | 2.9 × 108 | 1.9 × 108 | |
|
| |||||
| pAZ3-sibA | − | 7.8 × 106 | 3.6 × 107 | 2.6 × 106 | 6.0 × 105 |
| + | 8.8 × 106 | 2.6 × 107 | 4.1 × 106 | 5.0 × 105 | |
|
| |||||
| pAZ3-sibC | − | 1.3 × 106 | 3.8 × 106 | no colonies | 6.0 × 105 |
| + | 1.3 × 106 | 2.1 × 106 | 1.0 × 107 | 1.5 × 106 | |
|
| |||||
| pAZ3-sibE | − | 1.1 × 106 | 4.0 × 106 | 2.4 × 106 | no colonies |
| + | 2.7 × 106 | 2.2 × 106 | 3.8 × 106 | 2.0 × 106 | |
Cells were transformed with 1 ng of DNA. Results similar to those observed for ΔsibC and ΔsibE were obtained for ΔsibB and ΔsibD.
To test whether these observations were plasmid specific, sibE was placed under control of the PLlacO-1 promoter of plasmid pBR-plac (Guillier and Gottesman, 2006). As was the case with the PBAD plasmids, successful transformation into ΔsibE required high (0.1, 0.5 or 1 mM) concentrations of IPTG even though lower (less than 0.1 mM) concentrations of IPTG could induce expression of the SibE RNA (data not shown). This finding suggests that plasmid features such as the antibiotic resistance genes are not responsible for the inability to transform a specific Sib overexpression plasmid into the corresponding deletion strain.
Synthesis of toxic Ibs proteins
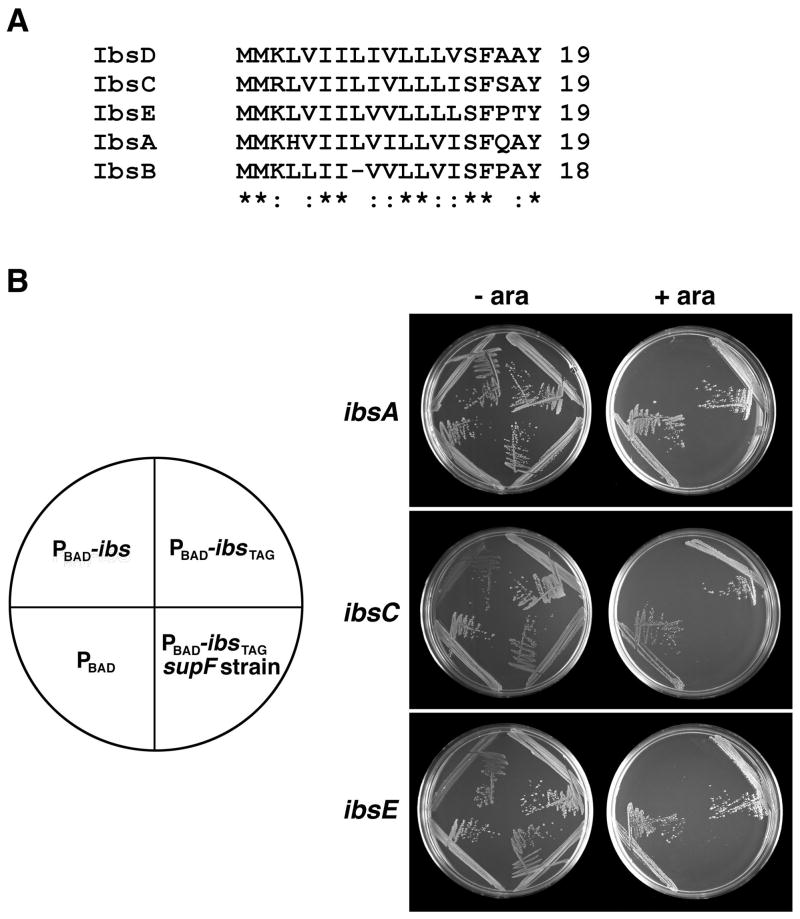
The need for high levels of the Sib sRNAs for plasmid transformation into the deletion strains was reminiscent of what has been reported for some plasmid addiction modules. In these cases, cells carrying plasmids encoding a stable toxin and unstable antitoxin are killed when the plasmid is lost and the antitoxin can no longer be synthesized [reviewed in (Gerdes et al., 2005; Hayes, 2003)]. For the well-characterized Hok-Sok plasmid addiction module of plasmid R1, the toxin is the 52-amino acid Hok protein and the antitoxin is the Sok antisense RNA [reviewed in (Gerdes and Wagner, 2007)]. The requirement for high levels of sib gene expression from plasmids in the deletion strains led us to hypothesize that the Sib RNAs might regulate the expression of a toxic protein. Upon closer examination of the sib genes, we noted a conserved small ORF encoded opposite each sib gene (Fig. 1). Whole genome expression analysis using tiled arrays for E. coli showed that indeed transcripts were expressed at low levels from the strand opposite the sib genes (data not shown). The antisense genes were predicted to encode proteins of 18–19 amino acids containing many hydrophobic residues (Fig. 3A). The small hydrophobic proteins are also conserved in Shigella, Salmonella and Citrobacter, and when the protein sequences are used in tblastn searches homologs can even be found in Haemophilus and Mannheimia.
Fig. 3. Overexpression of the Ibs proteins is toxic.
A. ClustalW alignment of the Ibs sequences from E. coli MG1655.
B. Growth of E. coli harboring either empty vector (pAZ3) pAZ3-ibsA, pAZ3-ibsATAG, pAZ3-ibsC, pAZ3-ibsCTAG, pAZ3-ibsE or pAZ3-ibsETAG grown in the presence or absence of 0.2% arabinose. The orientation of the strains is shown at left.
To determine whether the small ORFs might be toxic and thus be responsible for the plasmid transformation defect observed with the Δsib strains, we cloned the ORFs for the SIBa, SIBc and SIBe repeats together with approximately 70 nucleotides upstream of the predicted start codons behind the arabinose-inducible promoter of pAZ3. In liquid cultures with low concentrations of arabinose, we observed a cessation of growth with slow recovery for strains carrying these plasmids (data not shown). We thus propose the name ibs (induction brings stasis) for the genes, with ibsA encoded opposite sibA etc. At higher concentrations of the inducing agent, there was an irreversible stop in growth with a significant drop in the ability of the strains to form colonies (Fig. S2 and data not shown). In addition, strains carrying these plasmids were unable to growth on agar plates supplemented with 0.2 % arabinose (Fig. 3B). To confirm that overexpression of the Ibs proteins was responsible for the lack of growth, the fourth codon of each clone was mutated to TAG. Overexpression of these mutant constructs did not impair growth in a wild type strain; however, a supF suppressor strain was still susceptible to the toxicity of the mutant proteins (Fig. 3B). In contrast to the case with the PBAD-sibA clone, the PBAD-ibsA clone gave a phenotype that was similar to the other clones.
We propose that the toxicity of the PBAD-sibB, PBAD-sibC, PBAD-sibD, and PBAD-sibE plasmids in the respective deletion strains is due to transcription of the ibsB, ibsC, ibsD and ibsE genes encoded on the respective plasmids. Without expression of the corresponding sib gene, the cells were unable to grow. In the wild type strains, expression of the chromosomal copy of sib genes was sufficient to repress the ibs mRNA expressed from the plasmid.
Synthesis of the toxic ShoB protein
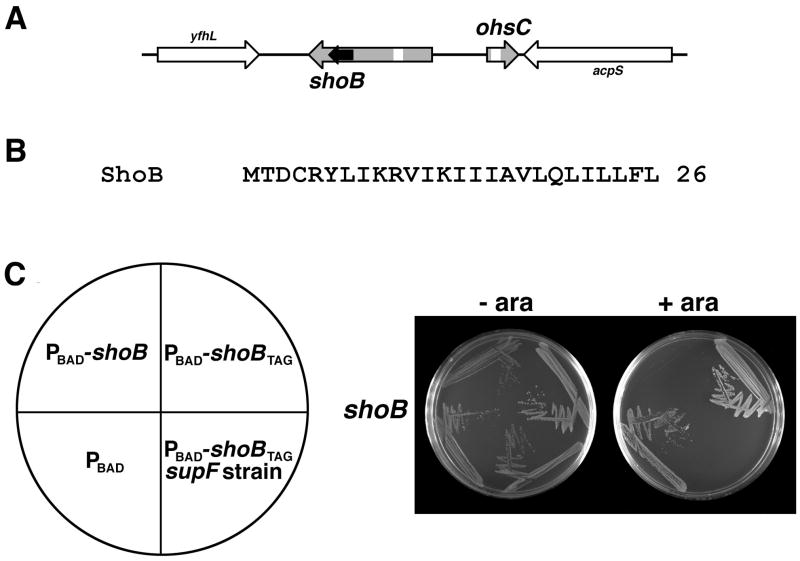
The realization that the sib-ibs regions encoded both a small toxic protein and the corresponding antitoxin sRNA, prompted us to examine other intergenic regions encoding two RNAs. One such region is the yfhL-acpS interval encoding the 280–320 nucleotide RyfB and 60–63 nucleotide RyfC RNAs (Kawano et al., 2005). These two RNAs are encoded divergently on opposite strands but share 19 nucleotides of complementarity suggesting possible regulation by base pairing (Fig. 4A). Upon examination of the ryfB sequence, we predicted that it could encode a hydrophobic protein of 26 amino acids (Fig. 4B). As was carried out for the IbsA, IbsC and IbsE proteins, the region corresponding to the entire RyfB RNA was cloned in pAZ3. A strain harboring this plasmid was unable to grow with arabinose (Fig. 4C and Fig. S2), indicating that, like IbsA, IbsC and IbsE, high levels of the 26-amino acid protein are toxic. Upon mutation of the sixth codon of the ORF to a stop codon, the strain could survive overexpression. Again, a supF suppressor strain restored the toxicity of the mutant clone under conditions of arabinose induction (Fig. 4C). As ryfB encodes a short hydrophobic ORF, we have renamed this gene shoB and ryfC is denoted ohsC (oppression of hydrophobic ORF by sRNA). Unlike the more broadly conserved ibs-sib loci, the shoB and ohsC genes appear to be confined to E. coli and Shigella.
Fig. 4. Overexpression of the ShoB protein is toxic.
A. Genetic orientation of shoB (formerly ryfB) and ohsC (formerly ryfC). The ShoB ORF is indicated by the black arrow and the 19 nucleotide regions of complementarity between shoB and ohsC are indicated by the white boxes.
B. Sequence of ShoB from E. coli MG1655.
C. E. coli harboring either pAZ3, pAZ3-shoB, pAZ3-shoBTAG grown in the presence or absence of 0.2% arabinose. The orientation of the strains is shown at left.
SibC and OhsC RNA repression of ibsC and shoB expression
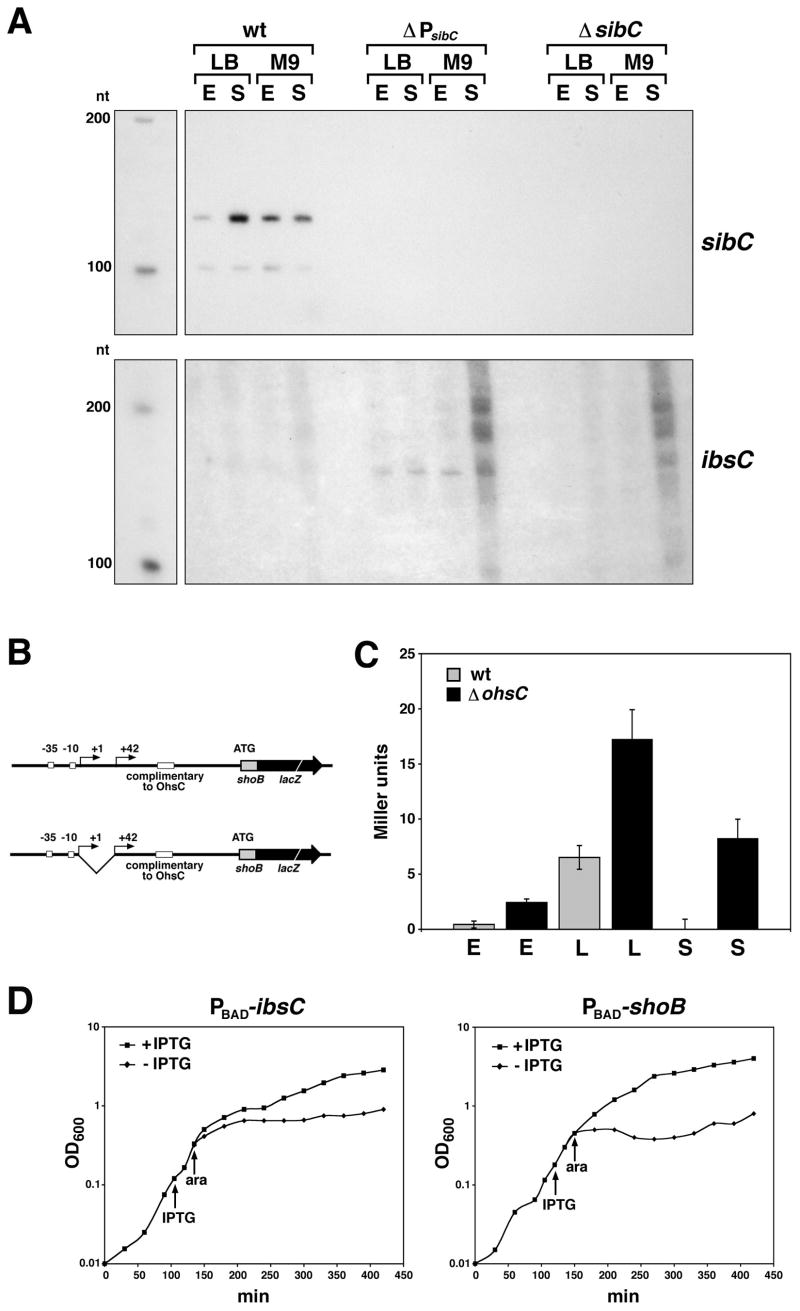
The complete complementarity between the sib and ibs transcripts and the 19 nucleotides of complementarity between the ohsC and shoB transcripts led us to propose that the Sibs and OhsC RNAs repress expression of the potentially toxic Ibs and ShoB proteins by base pairing with their respective mRNAs. We examined the effects of the SibC RNA on ibsC expression by deleting the sibC promoter and monitoring the levels of the ibsC mRNA (Fig. 5A). SibC clearly has a negative effect on ibsC transcript levels. In fact, by Northern analysis, we were only able to detect the ibsC mRNA in a sibC promoter deletion strain. The levels of the ibsC transcript were similar under the four growth conditions tested discounting the higher background for the sample isolated from cells grown to stationary phase in minimal medium. We also deleted the ohsC gene and examined shoB mRNA levels. In contrast to what we observed for the ibsC mRNA, we did not detect a difference in shoB mRNA levels in the presence or absence of the OhsC RNA (Fig. S3).
Fig. 5. Repression of IbsC and ShoB synthesis by the SibC and OhsC RNAs.
A. Northern analysis of ibsC mRNA levels. Total RNA was isolated from MG1655, ΔPsibC and ΔsibC/ibsC cells grown in LB or M9 with 0.2% glucose to OD600 ≈ 0.4 (E) or overnight (S, OD600 ≈ 5.0 in LB and OD600 ≈ 2.5 in M9 glucose).
B. Schematic of shoB-lacZ fusions constructed.
C. β-galactosidase levels of the 5′-UTR shoB-lacZ fusion with the short 5′ untranslated region in MG1655 (gray) and a ohsC deletion strain (black) grown to OD600 ≈ 0.4 (E), OD600 ≈ 1.8 (L) and overnight (S, OD600 ≈ 5.0) in LB medium. The average (in Miller units) of three independent cultures done in triplicate is given.
D. Overexpression of the SibC and OhsC RNAs can prevent IbsC and ShoB toxicity. DJ624 (a mal::lacI derivative of MG1655) was transformed with pBR-plac-sibC together with pEF21-ibsC or pBR-plac-ohsC together with pEF21-shoB (shown schematically in Fig. S4). The strains were grown in LB to OD600 ≈ 0.1, the cultures were split and IPTG (1 mM final concentration) was added to half of each culture. After 30 min, arabinose (0.002% final concentration) was added to all samples. OD600 was measured over time.
To further test OhsC RNA regulation of shoB expression, we constructed several translational fusions of the shoB promoter and 5′ untranslated sequence with the lacZ reporter gene (Fig. 5B). Multiple 5′ ends have been mapped for the ShoB transcript (Kawano et al., 2005). When the shoB promoter together with the longest 5′ sequence was fused to lacZ, no β-galactosidase activity was detected (data not shown), possibly due to inhibitory secondary structures formed by the long 5′ untranslated region. We also constructed a fusion in which the sequence between the first and second mapped 5′ ends was deleted (nucleotides 2698358 to 2698399) leaving the region of complementarity (nucleotides 2698294 to 2698313). The corresponding fusion gave low, but measurable, levels of β-galactosidase activity. Under all growth conditions tested, the levels of β-galactosidase activity were more than 2-fold higher in strains deleted for ohsC compared to the wild type strain (Fig. 5C), consistent with the hypothesis that the OhsC RNA negatively regulates translation of the ShoB message.
SibC and OhsC RNA repression of IbsC and ShoB toxicity
Our data suggested that the Sib and OhsC RNAs repress Ibs and ShoB protein synthesis, respectively, thereby limiting the toxicity of the small hydrophobic proteins. This was tested directly by cloning the ibsC and shoB genes behind the PBAD promoter of a derivative of pBAD33 (Guzman et al., 1995) and cloning the corresponding SibC and OhsC RNA genes behind the IPTG-inducible Plac promoter on the compatible plasmid pBR-plac. The ibsC stop codon mutation described above was also introduced into the sibC clone to eliminate potential expression of the IbsC protein. Cells carrying both the small protein-expression plasmid and the corresponding sRNA-expression plasmid were grown to early exponential phase (OD600 ≈ 0.1). The culture was split and IPTG was added to half of each culture to induce expression of the SibC and OhsC RNAs. After 30 min, arabinose was added to all cultures to induced expression of the IbsC and ShoB proteins. As shown in Fig. 5D, induction of IbsC and ShoB led to cell stasis of the cultures to which no IPTG was added, while the cultures treated with IPTG continued to grow. Thus the SibC and OhsC RNAs, respectively, prevent the toxic effects of IbsC and ShoB overproduction. The results also show that the SibC and OhsC RNAs do not need to be encoded in cis in order to repress IbsC and ShoB synthesis.
Reduction in membrane potential upon IbsC and ShoB overexpression
The features of the ibs and shoB genes are very similar to the hok gene of plasmid R1 (Gerdes and Wagner, 2007); all encode small hydrophobic proteins that are toxic upon overexpression and whose synthesis is repressed by sRNAs that have extensive complementarity with the corresponding mRNA. The Hok protein has been suggested to form pores in the membrane given that high levels of the protein dissipate the proton motive force (Gerdes et al., 1986). To examine whether high levels of the IbsC and ShoB proteins are having similar detrimental effects on the membrane, we tested the ability of cells to take up the dye DiBAC4(3) [bis-(1,3-dibarbituric acid)-trimethine oxanol]. This dye enters cells, leading to increased fluorescence, upon membrane depolarization (Wickens et al., 2000). Cells taken 0, 5 and 20 min after induction of IbsC or ShoB by the addition of arabinose were incubated with DiBAC4(3) for 20 min and then analyzed by flow cytometry (Fig. 6). No changes were observed for the plasmid control pAZ3. In contrast, induction of IbsC or ShoB had rapid and dramatic effects on the membrane integrity; after 5 min of induction, 50% and 98% of cells, respectively, had depolarized membrane and after 20 min of induction 97% and 99% of all cells were depolarized.
Fig. 6. Changes in membrane polarization upon IbsC and ShoB overproduction.
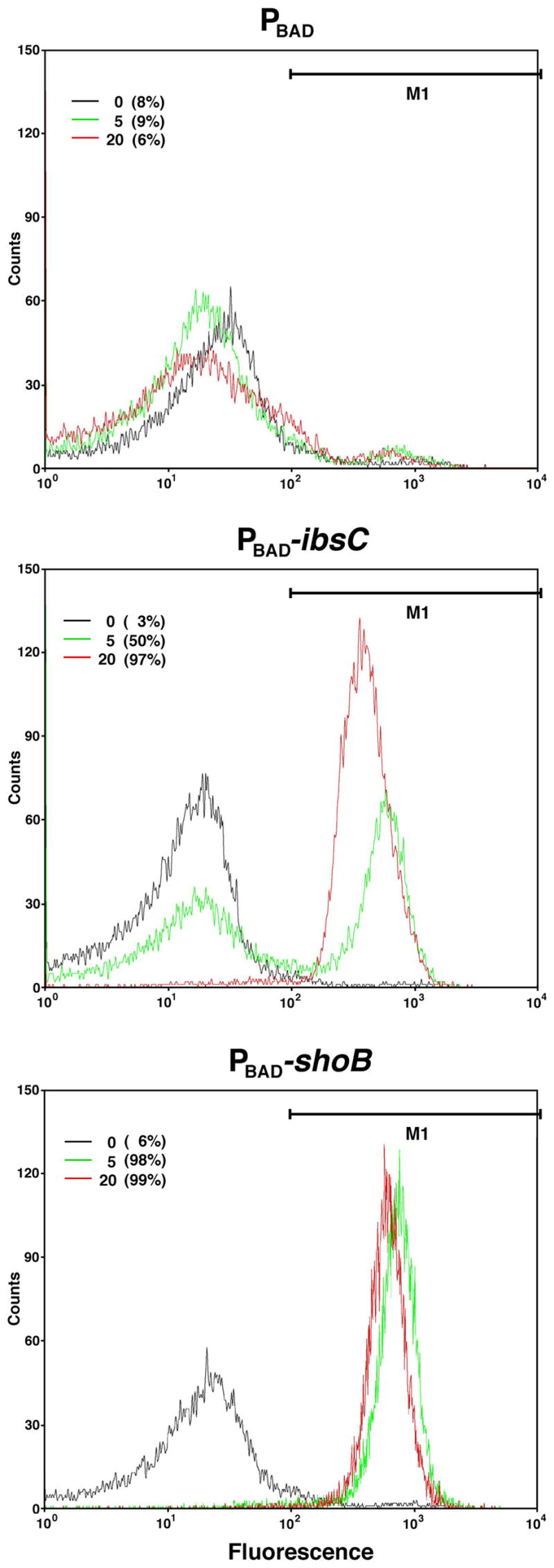
Control cells and cells overexpressing IbsC and ShoB were examined by flow cytometry after staining with DiBAC4(3), an indicator of membrane depolarization. MG1655 harboring pAZ3, pAZ3-ibsC or pAZ3-shoB was grown to exponential phase (OD600 ≈ 0.3). At 0, 5, and 20 min following the addition of 0.2% arabinose, cells were stained with DiBAC4(3) for 20 min and 15,000 cells were counted. The percentage of cells with a depolarized membrane was calculated from the region denoted M1. The graphs are representative of three repetitions.
Global effects of IbsC, ShoB, LdrD and TisB overexpression
Remnants of the five hok-sok modules have been detected on the E. coli K-12 genome, though all appear to have degenerated with mutations and transposon insertions (Pedersen and Gerdes, 1999). However, the four ldr and the unique tisB genes (Kawano et al., 2002; Vogel et al., 2004) all also encode small hydrophobic proteins whose synthesis is regulated by sRNAs. We confirmed that as for the 19-amino acid IbsC protein and 26-amino acid ShoB proteins, ovexpression of the 35-amino acid LdrD protein and 29-amino acid TisB protein leads to the inhibition of cell growth and reduction in colony forming units (Fig. S2). We wondered whether the shared properties of the proteins meant that overexpression causes identical changes in the cells. To address this question and also to begin explore the biological functions of these proteins, we carried out whole genome expression analysis after inducing IbsC, ShoB, LdrD or TisB expression for 20 min (the complete data set for three independent experiments is given in Table S3).
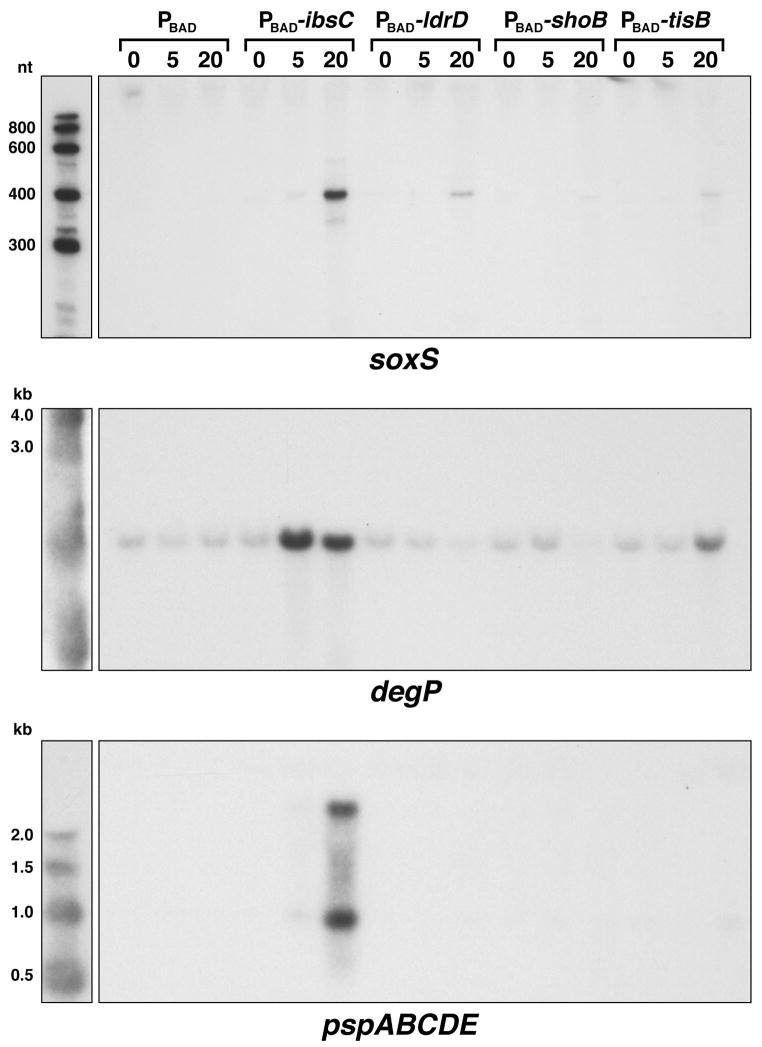
Overall, elevated levels of IbsC lead to the largest changes in gene expression; 65 genes were induced more than three-fold and 43 genes were repressed more than three-fold in three independent experiments. The numbers of genes induced and repressed by ShoB, LdrD and TisB were lower and the fold induction varied more between experiments, especially for ShoB. We do not know the cause of the variation; possibly the levels of ShoB synthesis differed between experiments due to additional levels of regulation. Nonetheless, a number of conclusions can be drawn by examining the expression of operons for which genes are induced ≥ 10-fold (Table 3) or repressed ≤ 5-fold (Table 4) by overexpression of at least one of the proteins. First, a common group of genes is induced by overexpression of all four proteins. Many of these genes encode stress-response or membrane proteins. Most striking among these is the soxS mRNA, which encodes a regulator of the superoxide stress response. Northern analysis confirmed that this mRNA indeed is strongly induced by IbsC overexpression as well as to a more limited extent by high levels of ShoB, LdrD and TisB (Fig. 7). Other genes induced by all four proteins encode tryptophanase and proteins involved in maltose transport. These genes are induced by many different stress responses (Jones and Rudd, unpublished), and several are regulated by CRP. Some strongly-responsive members of the heat shock regulon are also induced by overexpression of all of the small hydrophobic proteins.
Table 3.
Genes most highly induced upon IbsC, ShoB, LdrD and TisB overexpressiona
| Gene | Descriptionb | Regulationc | Average fold induction | |||
|---|---|---|---|---|---|---|
| IbsC | LdrD | TisB | ShoB | |||
| soxS3 | Transcriptional regulator | SoxR, SoxS | 67.3 | 13.7 | 8.7 | 8.1 |
| tnaC (tnaL) | Tryptophanase | TorR, Crp | 26.7 | 19.6 | 4.9 | 23.5 |
| tnaA | 34.7 | 26.1 | 4.3 | 20.0 | ||
| tnaB | 4.0 | 6.2 | 1.6 | 3.8 | ||
| malE | Maltose transport | MalT, Crp | 5.8 | 11.3 | 3.6 | 16.3 |
| malF | 2.7 | 4.7 | 1.3 | 9.1 | ||
| malG | 2.9 | 6.0 | 1.6 | 12.7 | ||
| malK | Maltose transport | MalT, Crp | 18.6 | 32.7 | 2.6 | 48.6 |
| lamB | 5.3 | 13.3 | 2.1 | 19.2 | ||
| malM | 3.3 | 9.1 | 1.7 | 20.0 | ||
| ibpB | Heat shock | σ32 | 39.0 | 24.2 | 4.6 | 36.4 |
| ibpA | Heat shock | 24.4 | 20.2 | 4.1 | 34.2 | |
| htpG | Heat shock chaperone | σ32 | 11.7 | 8.2 | 2.0 | 16.6 |
| clpB | Protease; chaperone | σ32 | 11.5 | 14.3 | 2.5 | 24.5 |
| yceP/bssS | Regulator | 12.9 | 3.3 | 2.9 | 6.8 | |
| cpxP (b3913) | Regulator; chaperone | CpxR | 17.3 | 3.2 | 8.0 | 5.7 |
| degP (htrA) | Protease | CpxR, H-NS | 21.0 | 1.3 | 2.7 | 1.7 |
| spy | Periplasmic | CpxR, BaeSR | 20.4 | 1.8 | 2.9 | 1.3 |
| ydeH | Unknown | CpxR | 49.2 | 1.1 | 6.1 | 1.3 |
| yebE | Unknown | CpxR | 44.8 | 1.6 | 7.3 | 2.9 |
| yncJ (b1436) | Unknown; signal peptide | 63.1 | 1.6 | 7.3 | 3.1 | |
| yhjX | Putative transporter | 21.0 | 44.1 | 2.6 | 165.6 | |
| pspA | Regulator; phage shock protein | PspF, IHF | 30.7 | 1.1 | 1.5 | 1.8 |
| pspB | Regulator; phage shock protein | 32.2 | 1.6 | 1.6 | 1.5 | |
| pspC | Regulator; phage shock protein | 56.9 | 1.7 | 2.0 | 2.1 | |
| pspD | Inner membrane | 33.1 | 1.2 | 1.4 | 1.2 | |
| pspE | Sulfurtransferase | 9.5 | 1.2 | 1.2 | 1.5 | |
| pspG (yjbO) | Inner membrane | PspF, IHF | 37.9 | 1.3 | 0.7 | 0.9 |
| ytfK | Unknown | PhoB | 22.0 | 1.9 | 3.8 | 3.0 |
| rbsD | Ribose transport | RbsR, Crp | 2.6 | 5.0 | 1.8 | 8.2 |
| rbsA | Ribose transport | 1.2 | 2.4 | 1.0 | 5.4 | |
| rbsC | Ribose transport | 1.8 | 4.7 | 1.6 | 21.3 | |
| rbsB | Ribose transport | 2.3 | 2.7 | 1.4 | 6.6 | |
| rbsK | Ribose transport | 0.8 | 2.6 | 1.2 | 9.0 | |
| rbsR | Ribose transport | 1.1 | 1.4 | 0.8 | 2.8 | |
| manX | Mannose permease | NagC, DgsA, Crp | 4.2 | 2.1 | 1.1 | 5.0 |
| manY | Mannose permease | 3.4 | 2.6 | 1.1 | 5.2 | |
| manZ | Mannose permease | 6.5 | 5.7 | 1.0 | 16.7 | |
Genes and operons which are induced ≥10-fold in three independent experiments are listed with the average fold induction for the three experiments. Boxes are shaded for those genes and operons induced ≥ 2-fold in all experiments.
Descriptions of gene function are taken from EcoGene (http://ecogene.org/).
Information about regulation is taken from RegulonDB (http://regulondb.ccg.unam.mx/).
Table 4.
Genes most highly repressed upon IbsC, ShoB, LdrD and TisB overexpressiona
| Gene | Descriptionb | Regulationc | Average fold repression | |||
|---|---|---|---|---|---|---|
| IbsC | LdrD | TisB | ShoB | |||
| dusB/yhdG | tRNA dihydrouridine synthase B | CRP, IHF, Fis | 6.5 | 0.9 | 1.1 | 4.3 |
| fis | Transcriptional activator | 7.7 | 0.9 | 1.1 | 3.8 | |
| yeeF | Putative transporter | 17.8 | 0.8 | 1.2 | 4.7 | |
| apt | Adenine phosphoribosyl transferase | 6.1 | 0.9 | 1.2 | 2.3 | |
| efeU (ycdN) | Inactive ferrous iron permease | CpxR | 21.2 | 1.2 | 2.1 | 1.2 |
| efeO (ycdO) | Inactive ferrous iron transporter | 14.3 | 1.0 | 1.8 | 1.2 | |
| efeB (ycdB) | Inactive ferrous iron transporter | 2.6 | 1.8 | 0.9 | 1.2 | |
| ompF | Porin | OmpR, EnvYd | 6.1 | 1.4 | 0.9 | 0.5 |
| yagU | Unknown | 26.7 | 1.7 | 2.5 | 1.2 | |
Genes and operons which are repressed ≥5-fold in three independent experiments are listed with the average fold repression for the three experiments. Boxes are shaded for those genes and operons repressed ≥ 2-fold in all experiments.
Descriptions of gene function are taken from EcoGene (http://ecogene.org/).
Information about regulation is taken from RegulonDB (http://regulondb.ccg.unam.mx/).
Also regulated by Crp, IHF, Lrp, CpxR, RstA, Fur as well as σE (Rhodius et al., 2006).
Fig. 7. Transcripts induced by IbsC, LdrD, ShoB and TisB overproduction.
Northern analysis of soxS, degP and the pspABCDE mRNA levels after IbsC, LdrD, ShoB and TisB overproduction. Total RNA was isolated from MG1655 harboring pAZ3, pAZ3-ibsC, pAZ3-ldrD, pAZ3-shoB or pAZ3-tisB at 0, 5, and 20 min following the addition of 0.2% arabinose to exponentially growing cultures (OD600 ≈ 0.3).
Other sets of genes were only induced or repressed by overexpression of subsets of the proteins. For example, overexpression of IbsC and TisB induced many members of the Cpx regulon, such as degP and spy, which are part of the cell envelope stress response. We also examined degP (Fig. 7) and spy expression (data not shown) by Northern analysis. While the degP and spy mRNA levels are clearly induced by IbsC and TisB overexpression, the levels appear to be repressed by LdrD and ShoB overexpression (although this is not reflected in the array data). Among the genes repressed by high levels of both IbsC and ShoB is the dusB-fis operon. Some sets of genes were affected by overexpression of only one protein. The pspABCDE operon as well as the pspG gene, which all encode phage shock proteins, were only induced by elevated IbsC levels. This specific induction was again confirmed by Northern analysis (Fig. 7) where we detected strong induction of the expected 700 and 2100 nucleotide bands reported for the pspABCDE operon (Brissette et al., 1991).
Together the whole genome expression analysis shows that while overexpression of IbsC, ShoB, LdrD and TisB leads to the induction of a common set of genes, the small proteins also have unique effects. It is interesting to note that many of the genes whose expression is induced encode membrane proteins or are members of the heat shock or envelope stress responses. We suggest that the induction of these genes is a downstream consequence of the changes--to the membrane or other components of the cell--caused by the small, hydrophobic proteins. We do not think the toxic effects of the proteins are exerted through the induced genes, since, for example, overexpression of IbsC is still toxic in a ΔpspABCDE ΔpspG deletion strain (data not shown). However, the finding that overexpression of some proteins induces unique genes, indicates the proteins are not all acting in an identical manner.
Elevated pspABCDE expression in absence of SibC RNA repression of ibsC
While overexpression of IbsC, ShoB, LdrD and TisB from multicopy plasmids clearly has dramatic consequences for the cell, we wondered whether expression of the proteins from the chromosome could also affect gene expression. To examine the effects of decreased SibC RNA levels and the concominant increase in ibsC mRNA levels, we probed the total RNA samples in Fig. 5A for pspABCDE expression. As shown in Fig. 8, the levels of the pspABCDE transcript were clearly induced in the sibC promoter deletion strain in the LB-stationary phase culture and even more highly in the M9-exponential culture. No induction was observed for either the wild type strain or the ΔsibC deletion strain in which the ibsC gene is also deleted. Thus even low levels of the IbsC protein, which do not have detrimental effects on growth, lead to changes in gene expression.
Fig. 8. Transcripts induced by derepressed levels of IbsC.
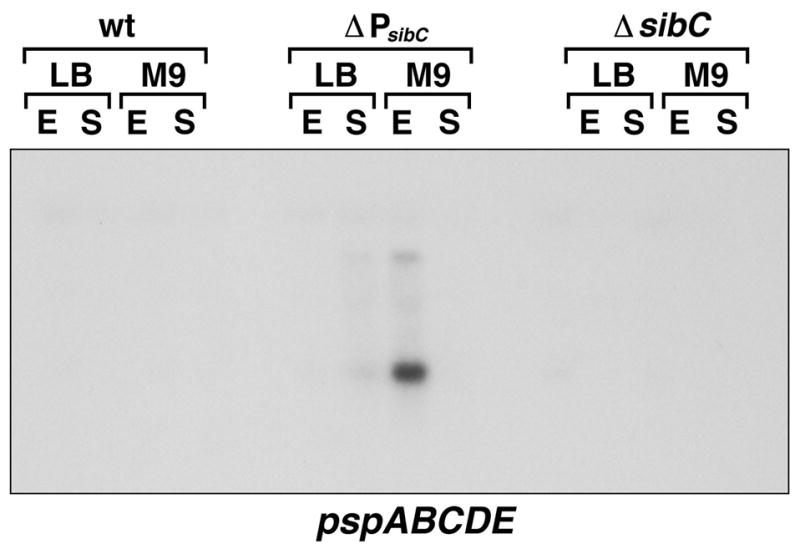
Northern analysis of pspABCDE mRNA levels in the same MG1655, ΔPsibC and ΔsibC-ibsC RNA samples probed in Fig. 5A.
Discussion
In this work, we characterized the Sib RNAs of E. coli MG1655. The sequences encoding these sRNAs were initially referred to as QUAD1a, 1b, 1c, and 1d (Rudd, 1999). However, as the number of repeats encoding these sRNAs varies even within E. coli strains, we propose to replace the QUAD1 designation with SIB (short, intergenic, abundant sequences). Although we did not observe any growth defects associated with the deletion of the sib genes, individually or in combination, we found multicopy plasmids encoding each sib gene could not be maintained in a given sib deletion strain unless the corresponding Sib RNA was expressed at high levels. Upon closer examination of the sib gene regions, we noted the presence of an ORF denoted ibs (induction brings stasis) encoding a small hydrophobic protein on the opposite strand of each sib gene. We demonstrated that overexpression of Ibs proteins is toxic, and this toxicity is prevented by the complementary Sib RNAs. We also characterized two short transcripts from the yfhL-acpS intergenic region. The longer transcript encodes a 26 amino acid protein, denoted ShoB (short hydrophobic ORF), that is toxic upon overproduction, and the shorter transcript acts as a sRNA, renamed OhsC (oppression of hydrophobic ORF by sRNA), to represses ShoB synthesis by pairing with the shoB mRNA.
The organization of the ibs-sib genes mirrors that of the ldr-rdl genes (Kawano et al., 2002) and the organization of the shoB-ohsC genes is similar to that of tisB-istR (Vogel et al., 2004). In all four of these cases, the mRNA encodes a short hydrophobic protein that is toxic when overexpressed, and the sRNA acts to repress synthesis by base pairing with the mRNA. These features make these loci members of the Type I family of toxin-antitoxin where the toxin is a small, hydrophobic protein and the antitoxin is an antisense RNA [reviewed in (Gerdes and Wagner, 2007)] and more than doubles the number of Type I toxin-antitoxin systems identified on the E. coli chromosome.
Regulation by the Sib and OhsC RNAs
We suggest that the Sib and OhsC RNAs repress the synthesis of the Ibs and ShoB protein upon base pairing with corresponding mRNAs by interfering with translation as has been found for IstR-1 RNA repression of TisB synthesis (Darfeuille et al., 2007). The sRNAs may also impact the stability of the mRNAs, since we only detected the ibsC mRNA in strains carrying a deletion of the sibC promoter or the ribonuclease III gene (data not shown). Possibly the shorter Sib transcript is generated by cleavage that occurs upon base pairing with the ibs mRNAs. The Sib and OhsC sRNAs have the potential for extensive base pairing with their targets, but the extent of the interaction and how it occurs is not known. The RNA chaperone Hfq, which is required for the regulation by sRNAs that have only limited complementarity with their targets, does not impact the levels of the SibC and ibsC RNAs or expression of the shoB-lacZ fusion (data not shown).
Several observations about the Sib and OhsC RNAs have consequences for their roles as antisense regulators. The Sib RNAs appear to be expressed constitutively since we detected the sib transcripts in E. coli MG1655 under all conditions tested, though the levels are generally highest in minimal media. This constitutive expression is similar to what has been observed for the SymR antisense RNA regulator, which represses toxic SymE protein synthesis (Kawano et al., 2007). Possibly, SymR and the Sib RNAs simply serve as dampers to constitutively maintain low levels of the corresponding toxic proteins. It is also possible that only a fraction of the sRNA molecules is functional for base pairing with the toxin mRNA and that this fraction differs under specific conditions. The Sib RNAs as well as the ShoB and SymR RNAs are predicted to have extensive secondary structures. The structures are likely to impact the stabilities of the sRNAs or their abilities to base pair with the ibs, shoB and symE mRNAs. In addition, the sib, ohsC and symR genes may be subject to transcriptional regulation that we have not yet uncovered.
It is probable that the synthesis of the Ibs and ShoB proteins is subject to regulation beyond repression by the Sib and OhsC RNAs. We suggest the reason why the SibA overexpression plasmid did not require induction in the ΔsibA strain, although overexpression of IbsA itself is toxic, is that the IbsA protein is expressed at lower levels due to additional regulation. Further levels of regulation may also explain the observation that no expression was detected for a lacZ translational fusion to the shoB promoter and full length 5′ untranslated region, despite the fact that the shoB mRNA could readily be detected by Northern analysis (Kawano et al., 2005). In addition, only limited expression was observed when the fusion was made with a shortened 5′ untranslated region. The 5′ portion of the shoB mRNA is predicted to fold into an extensive secondary structure which may preclude ribosome binding, as has been observed for the tisB mRNA (Darfeuille et al., 2007). Similar to tisB and symE, which are both repressed by LexA and are induced by DNA damage (Kawano et al., 2007; Vogel et al., 2004), transcription of the ibs and shoB genes may also be induced by specific growth conditions. Finally, the Ibs and ShoB proteins might be subject to degradation by specific proteases as has been observed for SymE (Kawano et al., 2007).
Toxicity of small hydrophobic proteins
The synthesis of the E. coli Ibs proteins, Ldr, ShoB and TisB proteins presumably is tightly regulated because high levels of these hydrophobic proteins are toxic to the cell. It is not clear why these proteins in particular are so toxic since we have found that high levels of other small hydrophobic proteins identified by Hemm et al. (submitted) do not have similarly detrimental effects (Fig. S2 and data not shown). The exact mechanism of Ibs, Ldr, ShoB and TisB toxicity also is unknown. Given their hydrophobic nature, we suggest that they insert into membranes and/or interact with other proteins. Consistent with this hypothesis, we found that induction of high levels of IbsC and ShoB led to a rapid loss of membrane potential.
Overexpression of IbsC, LdrD, ShoB and TisB all lead to the induction of a core set of genes, indicating some commonalities in the effects of these proteins. The observation that many of the induced genes encode proteins that are localized to the membrane is also consistent with the possibility that all four proteins affect the membrane. Perhaps more interesting, we found that overexpression of subsets of the proteins leads to the induction and repression of unique sets of genes. Only high levels of IbsC induced the phage shock proteins (pspABCDE, pspG). Induction of this regulon can occur in response to multiple environmental stresses, all of which are believed to dissipate proton motive force [reviewed in (Darwin, 2005)], and the PspA protein was shown to interact with phospholipids and serve to block movement of protons across damaged membranes (Kobayashi et al., 2007). Perhaps the overproduced IbsC proteins are forming pores in the membrane, leading to induction of the psp regulon. However, the observation that only IbsC overexpression leads to the induction of the psp genes may also mean that IbsC interacts with specific proteins that are not targeted by LdrD, ShoB or TisB.
The biological roles of the Ibs, Ldr, ShoB and TisB proteins expressed from the chromosome are unknown. In the case of TisB, which is expressed in response to DNA damage, it has been proposed that its expression may not normally kill the cell, but lead to growth arrest, which would allow for DNA repair prior to cell division (Vogel et al., 2004). Although we did not observe any growth defects for a strain with de-repressed IbsC expression, we did find that expression of the pspABCDE operon was induced indicating that low levels of IbsC does have consequences for the cell. Future experiments aimed at characterizing the changes in gene expression upon de-repression of the chromosomally-encoded ibs, ldr, shoB and tisB genes should give insights into why these genes are maintained by the cell.
Possible role of multiple genes
The ibs-sib genes, similar to ldr-rdl genes, are present as multiple copies on the chromosome. What is the role of the many copies? That fact that we observe constitutive expression of the Sib RNAs suggests that the repeats are not maintained for differential regulation of the sib genes, though it is possible that the ibs genes are differentially expressed. We did observe exclusive specificity; the lethal effects of the Sib overexpression plasmids were only seen with the strains carrying deletions of the same sib gene suggesting that the Sib RNAs do not have overlapping roles in ibs regulation. However, given the high degree of homology among the Ibs proteins, it seems unlikely that the proteins have different functions. In addition, the variation in the number of ibs-sib genes across even the different species of E. coli suggests that different numbers of these genes provide a selective advantage. Ongoing studies to further investigate the regulation by the antisense RNAs, the causes of toxicity upon overexpression of the small proteins, phenotypes associated with endogenous levels of the genes, as well as the genome-wide distribution of these Type I toxin-antitoxin modules should begin to provide insight into cellular roles of this large number of small, hydrophobic proteins.
Experimental procedures
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Tables S4 and S5, and the sequences of all oligonucleotides used in the study are listed in Table S6. The mini-λ-Red recombination system was used to generate deletion strains of the sib genes (Court et al., 2003; Datsenko and Wanner, 2000; Yu et al., 2000). In all cases, pKD4 (Datsenko and Wanner, 2000), which encodes kanamycin resistance, was used as the template plasmid in PCR reactions together with oligonucleotide primers containing 20 bases of pKD4 sequence and approximately 40 nucleotides of homology to the chromosomal region. Subsequently, the kanamycin cassette was removed using pCP20 (Cherepanov and Wackernagel, 1995).
Plasmids pAZ3 (Kawano et al., 2007), a plasmid with a pBR origin that is a derivative of pBAD18 with an EcoRI site at +1, and pEF21, a plasmid with a pACYC origin that is a derivative of pBAD33 (Guzman et al., 1995) with the introduction of a PstI site at +1, were used to overproduce RNAs from the PBAD promoter. For cloning into pAZ3, the sibA, sibC, sibD and sibE genes along with approximately 50 base pairs downstream of the mapped 3′ end was amplified from MG1655 genomic DNA, digested with EcoRI and HindIII and cloned into the corresponding sites of pAZ3. The ibsA, ibsC, ibsE and yoaJ genes were cloned into pAZ3 in the same fashion although in these cases the 5′ end were not known. The sibB gene was similarly cloned into pAZ3, except that the fragment was digested with EcoRI and XbaI. For cloning into pEF21, the ibs sequences were amplified from the genomic DNA, digested with PstI and HindII and cloned into the corresponding sites in pEF21. Plasmid pBR-plac (Guillier and Gottesman, 2006) was used to express RNAs from the Plac promoter. Gene sequences were amplified from genomic DNA, digested with AatI and EcoRI and cloned into the corresponding sites of pBR-plac. All experiments utilizing pBR-plac were performed in the strain DJ624, a ΔlacX74 derivative of MG1655. Plasmid DNA was always isolated using the Qiagen Mini Plasmid Kit, and PCR purification was performed either using Qiagen PCR Purification Kit or Qiagen Gel Extraction Kit.
The translational lacZ fusion to shoB assayed in Fig. 5C was generated using PCR (Horton et al., 1989). PCR product A was the result of amplifying from -75 to -1 of shoB message, with additional 15 nucleotides at the 3′ end that overlapped the first 15 nucleotides of PCR product B. PCR product B was from second mapped 5′ end of the shoB mRNA (nucleotide 2698357) to the seventh codon of ShoB. The two products were then spliced together using the external primers. This product was digested with EcoRI and BamHI and cloned into the corresponding sites of pRS552 (Simons et al., 1987). The plasmid was then recombined onto λRS45, which was subsequently used to lysogenize DJ480.
Growth conditions
E. coli K-12 MG1655 was grown in Luria-Bertani (LB with 10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) or M9 minimal media supplemented with 1 mM MgSO4, 0.1 mM CaCl2, 1 μg/ml thiamine and 0.2% glucose at 37°C. Arabinose was added as indicated to final concentrations of 0.002, 0.02, 0.1, 0.2%, and IPTG was added as indicated to final concentrations of 01, 0.5 or 1 mM. Antibiotics were added when needed to the following concentrations: 30 μg/ml kanamycin, 25 μg/ml chloramphenicol and 100 μg/ml ampicillin.
RNA extraction
Cells for RNA extraction were grown in LB or M9 + 0.2% glucose and harvested at OD600 ≈ 0.4, OD600 ≈ 1.8, or from overnight cultures (OD600 ≈ 5.0 in LB; OD600 ≈ 2.2 in M9). Total RNA was isolated by hot acid-phenol as described previously (Kawano et al., 2002).
Northern analysis
For the detection of the Sib, ibs and soxS transcripts, total RNA (5 μg for Sib and ibs and 10 μg for soxS) was separated on a denaturing 8% polyacrylamide-8 M urea gel and transferred to a Zeta-Probe Membrane (Bio-Rad). Oligonucleotide probes, specific for the individual Sib RNAs, were labelled with 32P using T4 polynucleotide kinase (New England Biolab). Hybridization and wash steps were as described previously (Opdyke et al., 2004). For the detection of the degP and pspABCDE mRNAs, total RNA (5 μg) was separate on 1X MOPS-1% agarose gel, transferred to a Zeta-Probe Membrane (Bio-Rad) and hybridized and washed as described previously (Opdyke et al., 2004).
5′ and 3′ RACE
5′ and 3′ RACE analysis was carried out as described (Argaman et al., 2001). Primers used to amplify sib cDNA are found in Table S6. The amplified cDNA fragments were then cloned into vector pCRII Topo (Invitrogen) and sequenced. The results of these analyses are found in Tables S1 and S2.
Transformation efficiency
To determine transformation efficiency, overnight cultures were diluted 1:500 into 50 ml LB and grown to OD600 ≈ 0.5. Cells were harvested and washed twice, once with 50 ml cold water, and once with 2 ml cold water. Cell pellets were resuspended in 200 μl cold water. A total of 40 μl of cells were transformed with 1 ng plasmid by electroporation. Cells were then resuspended in a final volume of 1 ml SOC media and placed at 37°C with shaking for 1 h. Cells were plated onto LB agar plates supplemented as indicated and incubated for 16 h at 37°C.
β-galactosidase assays
The β-galactosidase assays were carried out as described by (Miller, 1972).
Flow cytometry
MG1655 harboring pAZ3, pAZ3-ibsC or pAZ3-shoB were grown to OD600 ≈ 0.4 in LB and then induced with arabinose at a final concentration of 0.2%. At 0, 5 or 20 min, samples were diluted in 500 μl of filtered M9 + 10μg/ml DiBAC4(3) (Biotium) to approximately 5×106 CFU/ml. After a 20 min incubation at room temperature in the dark, cells were analyzed by flow cytometry in a FACSalibur (Beckton Dickinson) with a 488 nm laser. All parameters were collected as logarithmic signals. Samples were run at a low flow rate setting such that the event rate was 500 events per second. Green fluorescence from stained cells was collected in the FL1 channel (530 +/− 15nm). On average, cells with depolarized membrane become 50-fold more fluorescent in presence of DiBAC4(3). The two populations of cells were easily separable in each staining experiment.
Microarray analysis
MG1655 harboring pAZ3, pAZ3-ibsC, pAZ3-shoB, pAZ3-ldrD and pAZ3-tisB were grown to OD600 ≈ 0.3 in LB and was induced with arabinose at a final concentration of 0.2%. Samples were harvested at 20 min post-induction. RNA was prepared as described in (Kawano et al., 2002) with these modifications: following extraction with hot acid phenol:chloroform, the supernatant was extracted twice with phenol:chloroform and once with chloroform, whereupon the supernatant was ethanol precipitated. Preparation of the cDNA and hybridization to Affymetrix E. coli antisense arrays was done as described by (Massé et al., 2005).
Acknowledgments
We thank V. Gallegos, A. Kimchi, Z. Li, J. Saud, G. Tolun and M. Valledor for technical assistance and S. Gottesman and members of the Storz lab for helpful discussions and comments. This research was supported by NIH grant number R01-GM58560 (K.E.R.), the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (E.M.F., M.K., F.F., K.S.M and G.S.), a research fellowship from the Japan Society for the Promotion of Science (M.K) and a Research Associateship from the National Research Council (E.M.F).
References
- Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr Opin Microbiol. 2007;10:257–261. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Brissette JL, Weiner L, Ripmaster TL, Model P. Characterization and sequence of the Escherichia coli stress-induced psp operon. J Mol Biol. 1991;220:35–48. doi: 10.1016/0022-2836(91)90379-k. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Court D, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Darfeuille F, Unoson C, Vogel J, Wagner EGH. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Darwin AJ. The phage-shock-protein response. Mol Microbiol. 2005;57:621–628. doi: 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Bech FW, Jørgensen ST, Løbner-Olesen A, Rasmussen PB, Atlung T, Boe L, Karlstrom O, Molin S, von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986;5:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Wagner EG. RNA antitoxins. Curr Opin Microbiol. 2007;10:117–124. doi: 10.1016/j.mib.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol. 2006;2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–1499. doi: 10.1126/science.1088157. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Kawano M, Reynolds AA, Miranda-Rios J, Storz G. Detection of 5′ and 3′ UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acid Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R, Suzuki T, Yoshida M. Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol Microbiol. 2007;66:100–109. doi: 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sE stress response in related genomes. PLoS Biol. 2006;4:e. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Rudd KE. Novel intergenic repeats of Escherichia coli K-12. Res Microbiol. 1999;150:653–664. doi: 10.1016/s0923-2508(99)00126-6. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Storz G, Gottesman S. Versatile roles of small RNA regulators in bacteria. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 567–594. [Google Scholar]
- Vogel J, Argaman L, Wagner EGH, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens HJ, Pinney RJ, Mason DJ, Gant VA. Flow cytometric investigation of filamentation, membrane patency, and membrane potential in Escherichia coli following ciprofloxacin exposure. Antimicrob Agents Chemother. 2000;44:682–687. doi: 10.1128/aac.44.3.682-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]