Abstract
Heart disease remains the leading cause of mortality throughout the world. Mammals have an extremely limited capacity to repair lost or damaged heart tissue, thus encouraging biologists to seek out models for heart regeneration. Zebrafish exhibit a robust regenerative capacity in a variety of tissues including the fin, spinal cord, retina, and heart, making it the sole regenerative vertebrate organism currently amenable to genetic manipulation. Future studies will utilize functional approaches to tease apart zebrafish heart regeneration in hopes of unlocking our own regenerative potential.
Introduction
Tissue regeneration has fascinated biologists for centuries. Despite this interest, the cellular and molecular events driving the recovery of lost tissues are poorly understood. At the root of many of these events lie potent progenitor or stem cells capable of reconstituting cell populations of the newly regenerated tissue. This potential, along with the isolation of these progenitor cells from a wide variety of tissues, has impelled the field of regeneration with hopes of future uses in medicine.
Although robust replacement of structural cells has been discovered in mammalian tissues, such as the liver, blood, and skin, most tissues do not share this remarkable ability[1–4]. Perhaps most prominently, the mammalian heart is incapable of significant regeneration following an injury such as an acute myocardial infarction[5, 6]. Loss of oxygenation to ventricular muscle, usually due to occlusion of a coronary artery, will result in necrosis of that tissue. Too often, these injuries result in immediate death. Those fortunate to survive a myocardial infarction replace lost muscle with a scar, and typically are susceptible to compensatory pathology and/or future infarctions. Unlike the mammalian heart, the injured zebrafish heart normally undergoes minimal scarring[7]. Instead, a transient fibrin clot is replaced with new contractile muscle (Fig. 1). This review will focus on recent progress in the field of cardiac regeneration with an emphasis on the zebrafish model system.
Figure 1. Regeneration of the zebrafish heart.
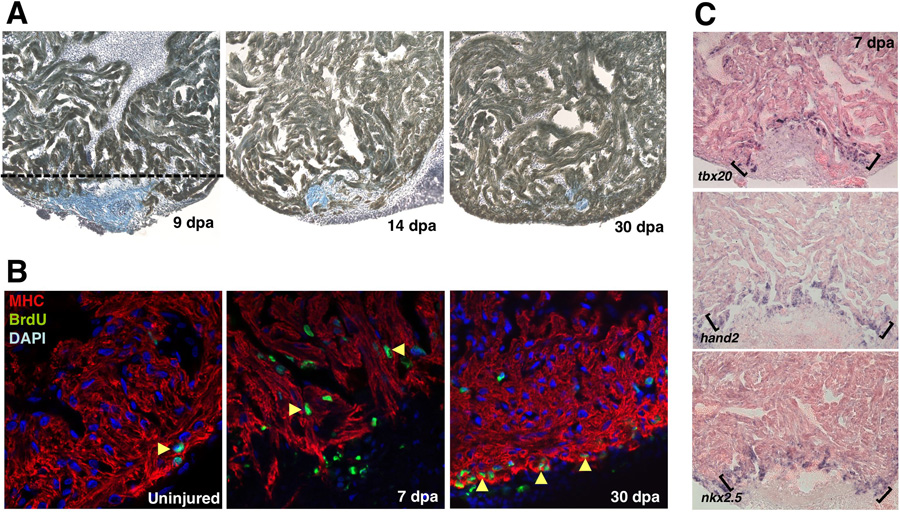
(a) Images of a regenerating zebrafish ventricle following 20% ventricular resection. Tissues were stained for myosin heavy chain to identify cardiac muscle (brown) and aniline blue to identify the fibrin clot (blue). By 7dpa, the wound is sealed by fibrin and is replaced by cardiac muscle by 30 dpa. (b) Proliferation, based on BrdU incorporation, is activated in cardiomyocytes by 7 dpa. The ventricular wall is restored by proliferation at the leading edge of the regenerating tissue. (c) Expression of embryonic heart field markers, hand2, nkx2.5, and tbx20 at the apical edge of the regenerate (brackets). Reproduced with permission from Refs. [7, 38].
Mammalian and Amphibian Heart Regeneration
As discussed above, mammalian species have little or no ability to replace lost cardiac muscle. This poor regenerative capacity is due in part to the failure for adult cardiomyocytes to undergo proliferation[8]. It is possible that the proliferation of adult cardiomyocytes may be therapeutically stimulated. In support of this, recent reports have shown that although adult mammalian cardiomyocytes show very little or no proliferation when cultured, FGF1 treatment concomitant with p38 MAP kinase inhibition can stimulate their proliferation in culture[9, 10]. In other studies, the discovery of putative progenitor cells within the hearts of adult mammals has led to the suggestion that the heart has the potential for homeostatic or regenerative renewal[11–15]. Yet, it is unclear why such progenitor cell populations are incapable of responding to injury with replacement by new cardiac muscle. Future therapeutic advancements may aim to “switch” on the appropriate programs within these progenitor cells or their environment and stimulate such replacement.
Urodele amphibians, due to their robust capacity to regenerate a wide variety of tissues, including spinal cord, brain, limb, retina, and lens, have historically been a key model system driving regeneration research[16–18]. Unlike adult mammalian cardiomyocytes, newt cardiomyocytes will readily undergo proliferation in tissue culture, thus representing one possible key difference that reflects regenerative capacity[19, 20]. However, although newt heart regeneration does occur to some extent after partial resection of ventricular myocardium, there is only a modest level of tissue replacement that accompanies scarring[21–23]. One suspected conduit for amphibian regenerative success is dedifferentiation, the process by which cells within injured tissue lose the expression of key genes necessary for function of that particular cell type (e.g. contractile genes for cardiac muscle). This functional reduction is thought to reprogram the cells, facilitating proliferation or differentiation to other cell types (transdifferentiation) in support of tissue regeneration. Newt lens, tail, and limb have been shown to undergo dedifferentiation during regeneration[24–26]. Recent data has shown that newt cardiomyocytes have an ability to lose contractile gene expression rapidly following a mechanical injury to the heart[27]. Therefore, regeneration of the urodele heart may be accomplished, at least partially, through dedifferentiation events.
Teleost Heart Regeneration
A combination of forward genetic screens, large clutch size, and external development has made the zebrafish a popular model system for ontogenetic development[28]. In particular, our understanding of heart development has benefited greatly from zebrafish mutants that specifically disrupt cardiovascular form and function[29–31]. Genetic approaches also make zebrafish a favored model system for studying tissue regeneration[32]. Indeed, it remains the only laboratory model system that is both amenable to genetic manipulation and capable of carrying out a robust regenerative response after the loss of complex tissue[7, 33–37].
After acute myocardial infarction in mammalian hearts, fibrin deposition occurs at the injury site. Then, fibrin is replaced by scar tissue, thought to be permanent[5, 6]. In zebrafish, after surgical removal of up to 20% of the ventricle by iridectomy scissors, an initial fibrin clot forms at the wound site. Unlike in mammal infarcts, the fibrin clot is subsequently replaced with new cardiac muscle in a process that takes 1–2 months (Fig. 1)[7].
Zebrafish heart regeneration proceeds through injury-induced proliferation of cardiomyocytes. For instance, BrdU labeling studies revealed labeled cardiomyocytes along the leading edge of the regenerating heart[7]. However, it had not been known until recently whether these BrdU-positive cardiomyocytes result from proliferation of differentiated cardiomyocytes, proliferation of progenitor cells forming new heart tissue, or dedifferentiation, proliferation, and redifferentiation of spared cardiomyocytes. Recent work has tried to address these questions through in vivo developmental timing assays that employed double transgenic zebrafish strains carrying reporter constructs for the cmlc2 (cardiac myosin light chain 2) promoter[38]. In these experiments, transgenes for EGFP and nuclear-DsRed2, both simultaneously driven by separate cmlc2 promoters, reported the contractile state of cardiac cells. Because GFP folds and fluoresces more rapidly than DsRed2, any cardiac progenitors that begin to turn on the cmlc2 promoter, and thus change their contractile status, will be transiently marked as GFP-positive/DsRed2-negative in nascent myocardium[39–42]. Indeed, this was the case in the double-transgenic embryo, as GFP fluorescence is observed up to a day prior to DsRed2 fluorescence within developing cardiomyocytes. Within five days of ventricular resection, a front of GFP-positive/DsRed2-negative cardiomyocytes was found at the apical edge[38]. This suggests that regenerating myocardium matures from undifferentiated, Cmlc2-negative, progenitor cells at the leading edge. In support of this mechanism, this leading edge was found to express a variety of molecular markers for the embryonic heart field, such as hand2, tbx20, and nkx2.5, suggesting that these cells acquire cardiac fate reminiscent of cardiac progenitors that provide the first cardiomyocytes of the developing embryo (Fig. 1).
It is not known from what source(s) these apparent progenitor cells are derived. One possibility is that dedifferentiation of mature cardiac cells supply the regenerate with new progenitors. Although this possibility was examined by developmental timing assays similarly as above and was not witnessed, it cannot be ruled out entirely[38]. Lineage analyses will be necessary to determine possible contributions of dedifferentiation. For example, experimentally pulsing the expression of a permanent genetic marker within mature cardiomyocytes just before injury will help determine whether new muscle cells are derived from existing cardiomyocytes during regeneration.
The Role of the Epicardium During Zebrafish Heart Regeneration
Recent results have shed light on the role of the outer non-muscle layer of the heart, the epicardium, in cardiac regeneration. During embryogenesis, the epicardium migrates out as a sheet from the proepicardium, a cluster or mesoderm-derived cells near the liver primordium and the septum transversum, to envelop the developing myocardial tube[43]. Following this encasement, some epicardial-derived cells undergo an epithelial-to-mesenchymal transition (EMT) into the subepicardial space and invade the myocardium to contibute endothelial and smooth muscle tissue of the coronary vasculature[44, 45].
Interestingly, adult zebrafish epicardial cells retain this plastic behavior. Perhaps the most striking aspect of the zebrafish epicardium during regeneration is that it undergoes an extremely rapid and dynamic response to injury[38]. Its developmental activation can be assayed by raldh2 and tbx18 expression, two genes known to be expressed within the embryonic epicardium (Fig. 2)[46, 47]. As early as six hours post-ventricular resection, raldh2 expression initiates within the epicardium of the outflow tract and atrium and then follows within the ventricle by 24 hours. In addition, tbx18 expression is observed within the atrial and ventricular epicardium by 1–2 days. Later on, cells positive for tbx18 and/or raldh2 appear at the wound site. This organ-wide response is intriguing for multiple reasons. First, the response is extremely rapid. The molecular details that connect the injury to this early response are unknown but are of interest, as their understanding will shed light on how heart regeneration is initiated. Second, the response is seen first distant from the wound site, within the atrium. The significance of this distal activation and how this signal is transferred over these distances are unknown, but are of interest to the field.
Figure 2. Participation of the epicardium during regeneration.
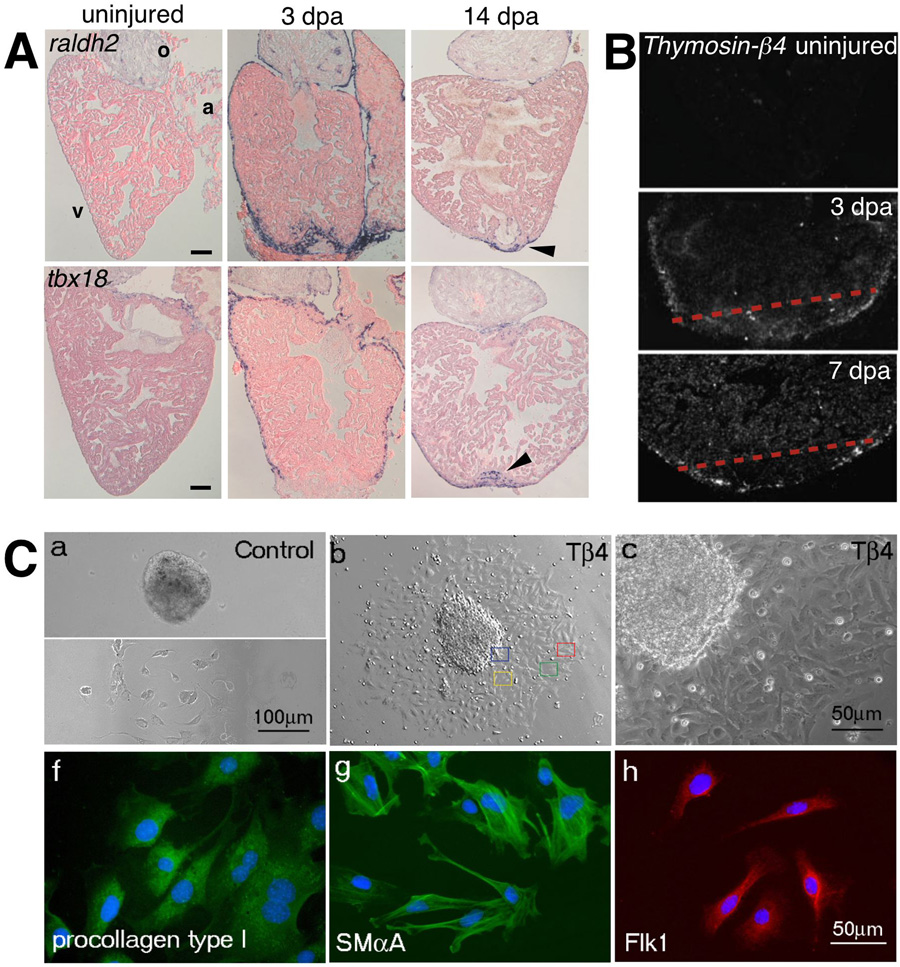
(a) Expression of the embryonic epicardial markers, raldh2 (top) and tbx18 (bottom) are induced in the adult epicardium after ventricular resection. Expression of both genes becomes localized to the wound by 14 dpa. The outflow tract (o), atrium (a), and ventricle (v) are labeled accordingly. (b) Radioactive in situ hybridization for Thymosin- 4 in sham operated control and amputated hearts 3 and 7 dpa. Expression of Thymosin- 4 is upregulated in regions surrounding the wound and the compact myocardium following injury. (c) Adult mouse heart explants cultured in vitro show increased migration of epicardial cells (confirmed by staining for epicardin – data not shown here) following treatment with Thymosin- 4. These cells then begin to express markers for smooth muscle cells (SM A – smooth muscle alpha-actin), fibroblasts (procollagen type I), and endothelial cells (Flk-1). Reproduced with permission from Refs. [38, 50, 52].
After ventricular resection induces developmental gene expression and proliferation within the epicardial layer, activated epicardial cells soon surround the wound with a portion of them penetrating several cell layers deep into the wound and regenerating muscle. Concomitantly, the new myocardium is substantially vascularized[38]. Thus it is assumed that the epicardial cells have similar roles as in the embryonic heart; that is, as a progenitor tissue that contributes smooth muscle and/or endothelial cells during neovascularization.
In the regenerating zebrafish heart, Fgf receptors 2 and 4 are expressed in epicardial or epicardial-derived cells at or near the injury site, which was shown to express at least one Fgf ligand. Furthermore, signaling by Fibroblast growth factors (Fgfs) is necessary for epicardial cell activity during regeneration, as ectopic expression of a dominant negative transgene that inhibits signaling through Fgf receptors disrupts this invasion of epicardial-derived cells, arresting regeneration (Fig. 3). This stimulatory role for Fgf signaling in adult zebrafish epicardial cells in vivo appears to mirror its effects on cultured epicardial cells in in vitro EMT assays[48]. Together, these data indicate a specific role for Fgf signaling in directing the EMT of epicardial-derived cells that ultimately vascularize the regenerate. Interestingly, treatment of injured rodent hearts by Fgf supplementation along with p38 MAP kinase inhibition stimulated neovascularization, decreasing infarct size and the level of scarring (Fig. 3)[9, 10]. These results suggest that both mammalian and non-mammalian vertebrates are responsive to Fgfs after injury as a means to increase neovascularization, but only selected species like zebrafish naturally utilize Fgfs to support myocardial regeneration. It is likely that additional factors will emerge that affect the production of coronary vessels in both zebrafish and mammalian models.
Figure 3. Fgf signaling is necessary for zebrafish heart regeneration and cardioprotection in rat hearts. Myocardial infarction was induced and treated in vivo.
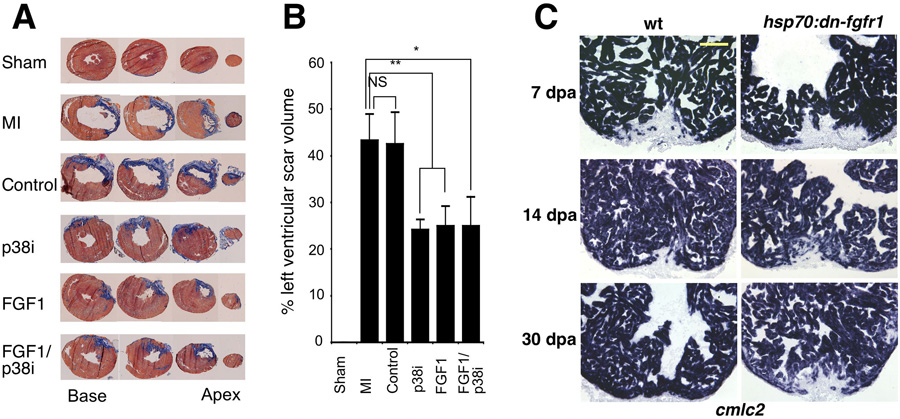
(a) Trichrome stains on heart sections to indicate scar tissue (blue). Sections shown were taken from base to apex. FGF1, p38 MAP kinase inhibition (p38i), and combined treatment significantly reduced the level of scar tissue formed after two weeks. This result is depicted graphically in panel (b). (c) cmlc2 expression at 7, 14, and 30 dpa in control and hsp70:dn-fgfr1 zebrafish. Treated animals failed to complete regeneration leaving a large wound by 30 dpa. Reproduced with permission from Refs. [10, 38].
Other recent studies support the idea that zebrafish have naturally optimized regenerative machinery that can function in both non-mammalian and mammalian injured hearts. The G-actin sequestering protein, Thymosin- 4, induces outgrowths from mammalian epicardial explants in vitro (Fig. 2)[49, 50]. Treatment of epicardial explants with Thymosin- 4 induces the differentiation of fibroblasts, endothelial, and smooth muscle cells as assessed by gene expression and cellular morphology (Fig. 2). In addition, in vivo Thymosin- 4 treatment can partially restore cardiac survival and function following coronary ligation in the mouse heart[51]. Notably, during zebrafish heart regeneration, Thymosin- 4 expression is induced in the wound and compact myocardium, indicating that fish naturally release this epicardial stimulant upon injury (Fig. 2)[52].
Genetic Approaches to Heart Regeneration in Zebrafish
Genetic approaches in zebrafish have revealed a large number of mutants affecting embryonic development. Most of these mutants exhibit embryonic or larval lethality, thus making it impossible to assess roles for specific genes in adult tissue regeneration, unless such mutations have effects in heterozygous animals. Investigators have circumvented this issue in a small number of studies by searching for conditional (temperature-sensitive) or hypomorphic alleles of genes that may be important for regeneration. Unfortunately, the idea of a direct screen for heart regeneration mutants comes with many challenges. Transgenic fish allowing inducible ectopic expression of a wild-type gene or a dominant-negative construct have been used recently and give similar benefits as conditional mutants[7, 38, 52, 53]. Future studies are also likely to take advantage of lineage tracing tools that have been utilized in mice for progenitor cell studies, to define progenitor/progeny relationships during heart regeneration[54–57].
It will be important to examine new cardiac injury models in zebrafish. The current models produce a mechanical injury[7]. While this injury might indeed stimulate the strongest possible regenerative response, it will also be interesting to better mimic the infarct models utilized in mammals. A recent study used the reducing power of Nitro-reductase (NTR), expressed by a tissue-specific promoter, to convert non-toxic metronidazole (Mtz) into a cytotoxic substance in transgenic zebrafish[58]. This approach facilitated cardiac lesions in embryonic zebrafish under temporal control determined by Mtz addition, and may have additional uses in studies of adult heart regeneration[58]. For instance, one could create patches of lesioned myocardium, or specifically ablate different cardiac cell types to address their roles in regeneration. Potentially, the NTR/Mtz system or other genetic toxins might be adapted to expedite a genetic screen for heart regeneration mutants.
Conclusions
Within the past few years, we have witnessed a growing interest in regeneration biology. Amphibians, mammals, and teleosts have different capacities to repair lost cardiac tissue, an it is from these differences we can learn best how to minimize scarring and maximize regeneration aftery injury. Zebrafish represent a highly useful system because of the combination of available genetic tools and a robust regenerative capacity. Through continued studies of zebrafish heart regeneration, we stand to learn how cardiac progenitor cells are successfully utilized for regeneration, and how the epicardium is optimally activated and directed to support this regeneration. Additional molecular studies in the future will increase the resolution of regenerative pathways and bring the idea of cardiac regeneration in humans closer to reality.
Acknowledgements
We thank Felix Engel, Ellen Lien, and Paul Riley for original figure panels, and the National Heart, Lung, and Blood Institute, the American Heart Association, the Whitehead Foundation, and the Pew Charitable Trusts for funding our research on heart regeneration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5(10):836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 2.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu Rev Med. 2005;56:509–538. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 3.Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128(3):445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma AD, et al. The role of stem cells in physiology, pathophysiology, and therapy of the liver. Stem Cell Rev. 2006;2(1):51–58. doi: 10.1007/s12015-006-0009-8. [DOI] [PubMed] [Google Scholar]
- 5.Azevedo CF, Cheng S, Lima JA. Cardiac imaging to identify patients at risk for developing heart failure after myocardial infarction. Curr Heart Fail Rep. 2005;2(4):183–188. doi: 10.1007/BF02696648. [DOI] [PubMed] [Google Scholar]
- 6.Rumyantsev PP. Interrelations of the proliferation and differentiation processes during cardiact myogenesis and regeneration. Int Rev Cytol. 1977;51:186–273. [PubMed] [Google Scholar]
- 7.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 8.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90(10):1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 9.Engel FB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19(10):1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel FB, et al. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A. 2006;103(42):15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans SM, Mummery C, Doevendans PA. Progenitor cells for cardiac repair. Semin Cell Dev Biol. 2007;18(1):153–160. doi: 10.1016/j.semcdb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114(6):763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 13.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433(7026):647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh H, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100(21):12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moretti A, et al. Cardiovascular development: towards biomedical applicability : Biology of Isl1 (+) cardiac progenitor cells in development and disease. Cell Mol Life Sci. 2007;64(6):674–682. doi: 10.1007/s00018-007-6520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mochii M, Taniguchi Y, Shikata I. Tail regeneration in the Xenopus tadpole. Dev Growth Differ. 2007;49(2):155–161. doi: 10.1111/j.1440-169X.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 17.Endo T, et al. Brain regeneration in anuran amphibians. Dev Growth Differ. 2007;49(2):121–129. doi: 10.1111/j.1440-169X.2007.00914.x. [DOI] [PubMed] [Google Scholar]
- 18.Mescher AL, Neff AW. Limb regeneration in amphibians: immunological considerations. ScientificWorldJournal. 2006;6:1–11. doi: 10.1100/tsw.2006.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat Embryol (Berl) 2002;205(3):235–244. doi: 10.1007/s00429-002-0249-6. [DOI] [PubMed] [Google Scholar]
- 20.Oberpriller JO, et al. Stimulation of proliferative events in the adult amphibian cardiac myocyte. Ann N Y Acad Sci. 1995;752:30–46. doi: 10.1111/j.1749-6632.1995.tb17404.x. [DOI] [PubMed] [Google Scholar]
- 21.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187(2):249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 22.Becker RO, Chapin S, Sherry R. Regeneration of the ventricular myocardium in amphibians. Nature. 1974;248(444):145–147. doi: 10.1038/248145a0. [DOI] [PubMed] [Google Scholar]
- 23.Neff AW, Dent AE, Armstrong JB. Heart development and regeneration in urodeles. Int J Dev Biol. 1996;40(4):719–725. [PubMed] [Google Scholar]
- 24.Eguchi G, Abe SI, Watanabe K. Differentiation of lens-like structures from newt iris epithelial cells in vitro. Proc Natl Acad Sci U S A. 1974;71(12):5052–5056. doi: 10.1073/pnas.71.12.5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc Natl Acad Sci U S A. 1993;90(15):7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298(5600):1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- 27.Laube F, et al. Re-programming of newt cardiomyocytes is induced by tissue regeneration. J Cell Sci. 2006;119(Pt 22):4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- 28.Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22(9):473–478. doi: 10.1016/j.tig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13(6):507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- 30.Auman HJ, Yelon D. Vertebrate organogenesis: getting the heart into shape. Curr Biol. 2004;14(4):R152–R153. [PubMed] [Google Scholar]
- 31.Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2(1):39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 32.Keating MT. Genetic approaches to disease and regeneration. Philos Trans R Soc Lond B Biol Sci. 2004;359(1445):795–798. doi: 10.1098/rstb.2004.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44(3):289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 34.Otteson DC, Hitchcock PF. Stem cells in the teleost retina: persistent neurogenesis and injury-induced regeneration. Vision Res. 2003;43(8):927–936. doi: 10.1016/s0042-6989(02)00400-5. [DOI] [PubMed] [Google Scholar]
- 35.Becker T, et al. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377(4):577–595. doi: 10.1002/(sici)1096-9861(19970127)377:4<577::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 36.Akimenko MA, et al. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226(2):190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- 37.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226(2):202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 38.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 39.Verkhusha VV, et al. An enhanced mutant of red fluorescent protein DsRed for double labeling and developmental timer of neural fiber bundle formation. J Biol Chem. 2001;276(32):29621–29624. doi: 10.1074/jbc.C100200200. [DOI] [PubMed] [Google Scholar]
- 40.Verkhusha VV, et al. High stability of Discosoma DsRed as compared to Aequorea EGFP. Biochemistry. 2003;42(26):7879–7884. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- 41.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97(22):11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20(1):83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 43.Reese DE, Mikawa T, Bader DM. Development of the coronary vessel system. Circ Res. 2002;91(9):761–768. doi: 10.1161/01.res.0000038961.53759.3c. [DOI] [PubMed] [Google Scholar]
- 44.Olivey HE, Compton LA, Barnett JV. Coronary vessel development: the epicardium delivers. Trends Cardiovasc Med. 2004;14(6):247–251. doi: 10.1016/j.tcm.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Dettman RW, et al. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol. 1998;193(2):169–181. doi: 10.1006/dbio.1997.8801. [DOI] [PubMed] [Google Scholar]
- 46.Moss JB, et al. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199(1):55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- 47.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100(1):83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 48.Morabito CJ, et al. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 2001;234(1):204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- 49.Bock-Marquette I, et al. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432(7016):466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- 50.Smart N, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445(7124):177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 51.Cavasin MA. Therapeutic potential of thymosin-beta4 and its derivative N-acetyl-seryl-aspartyl-lysyl-proline (Ac-SDKP) in cardiac healing after infarction. Am J Cardiovasc Drugs. 2006;6(5):305–311. doi: 10.2165/00129784-200606050-00003. [DOI] [PubMed] [Google Scholar]
- 52.Lien CL, et al. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4(8):e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A. 2003;100 Suppl 1:11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dor Y, et al. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 56.Meilhac SM, et al. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development. 2003;130(16):3877–3889. doi: 10.1242/dev.00580. [DOI] [PubMed] [Google Scholar]
- 57.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curado S, et al. Conditional targeted cell ablation in zebrafish: A new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]


