Abstract
Background
Chromosome 13q deletion is associated with varying phenotypes, which seem to depend on the location of the deleted segment. Although various attempts have been made to link the 13q deletion intervals to distinct phenotypes, there is still no acknowledged consensus correlation between the monosomy of distinct 13q regions and specific clinical features.
Methods
14 Italian patients carrying partial de novo 13q deletions were studied. Molecular–cytogenetic characterisation was carried out by means of array‐comparative genomic hybridisation (array‐CGH) or fluorescent in situ hybridisation (FISH).
Results
Our 14 patients showed mental retardation ranging from profound–severe to moderate–mild: eight had central nervous system (CNS) anomalies, including neural tube defects (NTDs), six had eye abnormalities, nine had facial dysmorphisms and 10 had hand or feet anomalies. The size of the deleted regions varied from 4.2 to 75.7 Mb.
Conclusion
This study is the first systematic molecular characterisation of de novo 13q deletions, and offers a karyotype–phenotype correlation based on detailed clinical studies and molecular determinations of the deleted regions. Analyses confirm that patients lacking the 13q32 band are the most seriously affected, and critical intervals have been preliminarily assigned for CNS malformations. Dose‐sensitive genes proximal to q33.2 may be involved in NTDs. The minimal deletion interval associated with the Dandy–Walker malformation (DWM) was narrowed to the 13q32.2–33.2 region, in which the ZIC2 and ZIC5 genes proposed as underlying various CNS malformations are mapped.
The 13q‐syndrome is caused by structural and functional monosomy of the 13q chromosomal regions. Carriers of 13q partial deletions may have widely varying phenotypes, but the most common clinical features include moderate–severe mental and growth retardation, craniofacial dysmorphisms, hand and foot anomalies, and brain, heart and kidney defects1,2,3,4 (http://www.ecaruca.net). Several attempts have been made over the years to correlate 13q deletion intervals with distinct phenotypes. Niebuhr5 suggested that distal deletions are closely associated with severe phenotypes, whereas proximal deletions tend to cause fewer major anomalies, with the exception of retinoblastoma. Brown et al3,6 defined 13q32 as the critical band for the most severe phenotypes, showing that monosomy of a 1.2‐Mb region in q32 is related to the development of severe mental and growth retardation, as well as major malformations including brain abnormalities. The ZIC2 (zinc finger protein of cerebellum 2) gene maps to this critical region and has been considered a plausible candidate for brain anomalies. ZIC2 mutations have been associated with holoprosencephaly (HPE),7,8 thus leading to the hypothesis that ZIC2 hemizygosity may contribute to the severe brain malformations of patients with del(13q). Luo et al9 have suggested that one or more of the genes mapping to 13q33–34 may be responsible for the expression of neural tube defects (NTDs) as a result of haploinsufficiency. McCormack et al10 and Alanay et al11 have suggested that the 13q22–33 region is critical for the development of the Dandy–Walker malformation (DWM), which implies the existence of at least one other dose‐sensitive gene (in addition to ZIC2) that has a role in cerebellar development.
There is still no consensus on possible correlations between the monosomy of distinct 13q regions and specific clinical features. Here, we describe 14 cases of de novo 13q partial deletions (seven terminal and seven interstitial) and their characterisation by means of conventional cytogenetics, array‐comparative genomic hybridisation (array‐CGH) or fluorescence in situ hybridisation (FISH), in an attempt to address the karyotype–phenotype correlations.
Methods
Table 1 summarises the clinical characteristics of each patient. Additional clinical details are provided as supplementary information (see http://jmg.bmjjournals.com/supplemental). Case 10 has been previously described.12
Table 1 Cytogenetic and clinical findings of patients with 13q deletions.
| Patients | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deleted segment | q13.3‐ qter | q14.11– 21.31 | q14.3– 32.1 | q21.1– 31.3 | q21.33– 31.2 | q22.1/.2– 31.3 | q22.2‐ qter | q22.3– 33.2 | q31.1– 33.1 | q31.1‐ qter | q31.1‐ qter | q32.2‐ qter | q33.3‐ qter | q34‐ qter |
| Sex | F | F | M | M | F | M | F | F | M | F | F | F | F | F |
| Age at karyotyping | 16 w | 11 y | At birth | 21 w | 28 y | At birth | 17 w | 1 y | At birth | At birth | 4 y | 16 y | 5 y | At birth |
| Measures at birth at age (w): | — | NK | 38 | 21 | 38 | NK | — | 40 | NK | 36 | 37 | 40 | 41 | 40 |
| Length (p) | NK | <5 | NK | NK | NK | NK | <3 | <3 | 50 | <3 | NK | |||
| Weight (p) | <3 | NK | 10 | NK | <25 | NK | <3 | <3 | 25 | <3 | <3 | |||
| OFC (p) | NK | >97 | NK | NK | NK | NK | <3 | <3 | 3 | <3 | NK | |||
| Apgar index | — | NK | NK | — | NK | NK | — | NK | NK | 4 | 10, 10 | 2, 3, 6 | 7, 8 | NK |
| Postnatal measures at age | — | 11 y | 4 m | — | 28 y | 4 y | — | 15 y | — | 2 y and 6 m | 5 y | 16 y | 5 y | 22 y |
| Height (p) | NK | <3 | <3 | NK | NK | <3 | <3 | 3 | >97 | <3 | ||||
| Weight (p) | NK | <3 | <3 | NK | NK | <3 | 3 | 3 | >90 | <3 | ||||
| OFC (p) | >97 | <3 | <3 | NK | >10 | <3 | <3 | <3 | <3 | <3 | ||||
| Walked unassisted | — | 30 m | — | — | 3 y | NK | — | – | — | – | – | – | 14 m | 13 m |
| Age at first words | 18 m | 2 y | – | – | NK | – | – | + | 5–6 y | |||||
| Age at first phrases | 18 m | + | – | – | NK | – | – | + | NK | |||||
| Bladder/bowel control | NK | + | NK | – | NK | – | – | + | NR | |||||
| Mental retardation | — | Mild | — | — | Moderate | Severe | — | Severe | — | NK | Profound | Profound | Moderate | Moderate |
| Brain anomalies | ||||||||||||||
| Corpus callosum agenesis | – | – | – | – | – | NK | – | – | NK | + | – | + | – | – |
| DWM | – | – | – | – | – | NK | – | + | NK | – | + | + | – | – |
| Cerebellar hypoplasia | + | – | – | – | – | NK | – | – | NK | – | + | – | – | – |
| Cortical dysplasia | – | – | – | + | – | NK | – | – | NK | – | – | – | – | – |
| NTDs | ||||||||||||||
| Encephalocele | – | – | – | – | – | – | + | – | + | – | – | – | – | – |
| Spina bifida | – | – | – | – | – | – | + | – | – | – | – | – | – | – |
| Hypoplasia of optic nerve | — | + | – | – | – | NK | — | – | – | NK | + | – | – | – |
| Microphthalmos | + | – | – | – | – | – | — | – | – | + | + | + | – | – |
| Iris/choroid retina‐coloboma | — | – | – | – | – | – | — | + | – | + | + | + | – | – |
| Blindness | — | – | – | — | – | – | — | – | – | + | + | + | – | – |
| Hypotonia | — | – | + | — | – | – | — | + | – | + | + | + | + | + |
| Hypertelorism | NK | – | + | – | + | + | NK | + | NK | + | + | + | + | + |
| Epicanthus | NK | + | – | – | – | – | NK | – | NK | + | + | – | + | – |
| Broad prominent nasal bridge | NK | + | + | – | + | – | NK | + | NK | – | + | + | + | – |
| Low‐set ears | NK | – | – | – | + | + | NK | – | + | + | + | – | – | – |
| Malformed ears | NK | + | – | – | – | – | NK | – | – | – | + | – | – | – |
| High and narrow palate | NK | + | – | – | – | – | NK | + | NK | + | – | + | + | – |
| Micrognathia | NK | – | – | – | – | – | NK | – | NK | + | + | + | + | – |
| Congenital heart defects | – | – | + | – | – | + | — | – | – | + | + | – | + | – |
| Ambiguous genitalia | – | – | – | – | – | + | – | – | – | + | – | – | – | – |
| Hand anomalies | – | – | + | – | + | – | — | + | + | + | + | + | + | – |
| Foot anomalies | + | – | + | – | + | – | — | + | + | + | + | + | + | + |
| Skeletal abnormalities | + | + | – | – | – | – | – | – | NK | + | + | + | + | + |
DWM, Dandy–Walker malformation; F, female; M, male; m, months; NK, not known; NR, normal range; OFC, occipitofrontal head circumference; p, percentile; w, weeks; y, years; —, undetectable.
The ages of developmental milestones are given when available: if a milestone was reached at an unknown age, a plus sign is used; a minus sign indicates that the milestone was not achieved.
In the case of the other features, a plus or minus sign indicates presence or absence. For hands and feet, plus signs denote minor anomalies, such as single palmar crease and syndactyly of the second and third toes.
To define the deletion breakpoints, the patients were investigated by means of FISH, or array‐CGH and FISH.
FISH analysis
To establish the telomeric or interstitial nature of the deletions, metaphases of all the patients were analysed by means of FISH using specific 13q subtelomeric probes (Vysis, Downers Grove, Illinois, USA; and Oncor, Gaithersburg, Maryland, USA) according to the manufacturers' protocols. The maintenance of such sequences in one or both of the 13 homologues indicates a terminal or interstitial deletion. Case 13 was also investigated by means of specific 13/21, 14/22 and 15 centromeric probes (Oncor).
RPCI‐11 BAC clones, selected according to the UCSC Genome Browser (http://genome.ucsc.edu/, release April 2004) and provided by Dr M Rocchi, University of Bari, Italy, were used to characterise the breakpoints. BAC clone cultures, probe labelling, slide preparation and hybridisation were carried out using standard protocols.
Array‐CGH analysis
Genomic DNA was extracted from peripheral blood samples (cases 3, 8, 11), amniocytes (cases 1, 7), fibroblasts (cases 9, 14) or lymphoblast cultures (cases 2, 10) using the GenElute Blood Genomic DNA kit (Sigma‐Aldrich, St Louis, Missouri, USA) according to the supplier's instructions. Patients 1, 2, 3, 7, 8 and 10 were investigated by means of a Spectral Genomics 1 Mb chip (www.spectralgenomics.com) together with SpectralWare software that gives a graphical view of the analysis of the acquired images. The array‐CGH procedures were performed as previously described.13 The array‐CGH analysis of patients 9, 11 and 14 was carried out using the Agilent Human Genome CGH Microarray Kit 44B platform, a high‐resolution 60‐mer oligonucleotide‐based microarray, allowing a genomewide survey and molecular profiling of genomic aberrations with a resolution of about 75 kb. Labelling and hybridisation were carried out as previously described.14
Results
Seven of the de novo 13q deletions were interstitial (patients 2–6, 8 and 9) and seven terminal (cases 1, 7 and 10–14). Table 1 summarises the clinical findings and the size of the 13q‐deleted segment for each patient. The patients are arranged from 1 to 14 according to the breakpoint localisation (from centromere to telomere) on 13q. Additional clinical details, including whole facial dysmorphisms, congenital heart defects, and hand, foot and skeletal abnormalities, are given as supplementary information. Three of the 14 patients (1, 4 and 7) were diagnosed prenatally, and the ultrasound results of all of them are available: the three pregnancies were voluntarily interrupted, but only patient 4 underwent fetal autopsy. Our patient series consists of nine XX females, three XY males and two XY patients with ambiguous genitalia; patient 11 is characterised by a minor genital abnormality. The ages of the post‐natal patients range from 4 months to 28 years. Growth parameters were poor in nine patients, with head growth concordantly reduced in six; postnatal overgrowth was recorded in one patient (case 13). Mental retardation ranged from severe–profound (four patients) to mild–moderate (four patients); among the mild–moderate patients, the age at which developmental milestones were reached varied (table 1); three of the 11 postnatal patients were not assessed for cognitive performance. Eight of the 14 patients show CNS anomalies (including NTDs). Six have eye abnormalities, including three (patients 10–12) who are blind owing to microphthalmos, coloboma and hypoplasia of the optic nerve. Nine patients show facial dysmorphisms, the most frequent of which are hypertelorism (8/9 cases) and a broad and prominent nasal bridge (7/9); other minor anomalies are less frequent, such as a high and narrow palate (5/9), low‐set ears (4/9) and epicanthus (4/9). Eight patients have both hand and foot anomalies, and two have either hand or foot anomalies. Absent or hypoplastic thumbs and brachyphalangy of the middle phalanx of the little finger have been reported as the main digital malformations in patients with 13q deletions,15 and patients 10 and 11 show absent or hypoplastic thumbs; patients 3, 11 and 13 show fifth finger clinodactyly, with other phalangeal abnormalities identified in patients 3 and 9. Two patients (5 and 12) have brachymetacarpia or brachymetatarsia. Five patients show congenital heart defects. Two patients (8 and 10) have gastrointestinal malformations (severe gastro‐oesophageal reflux and stenotic anus) and one (case 11) has a minor renal anomaly consisting of hypoplastic kidneys with normal renal function.
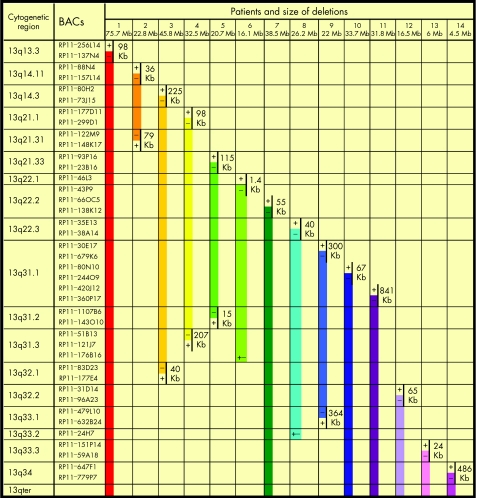
Figure 1 shows the BAC clones used for the analyses, their cytogenetic localisation, the distances between the clones defining the breakpoints and the extension of the deleted regions. Additional cytogenetic data are provided as supplementary information. The size of the deletions varies from 4.2 to 75.7 Mb.
Figure 1 FISH results obtained using BAC clones targeting the 13q arm. For each patient, the size of the deleted regions (coloured bars) and the distances between the BACs defining the breakpoints are indicated. +, presence of the hybridisation signal on the deleted chromosome; −, absence of the hybridisation signal on the deleted chromosome; +/−, diminished intensity of the hybridisation signal on the deleted chromosome.
Discussion
Our study is the first comprehensive molecular characterisation of de novo 13q deletions by means of array‐CGH and FISH, and allows us to address karyotype–phenotype correlations depending on the extent of the deletions. In agreement with previous reports,6 the overall phenotype severity in our series varies widely, with some patients being only slightly affected and carrying only dysmorphic features and minor anomalies, and others being seriously compromised.
Three patient groups have been distinguished on the basis of previous attempts to correlate defined 13q‐deletion intervals with specific phenotypic signs3: patients carrying proximal deletions not extending into the q32 band and characterised by variable dysmorphic features, mild or moderate mental retardation and growth retardation (group 1); patients with deletions including at least part of q32 and the most serious phenotype, with severe mental and growth retardation and one or more major malformations most often involving the brain, eyes, distal limbs, genitourinary and gastrointestinal traits (group 2); and patients with more distal deletions involving bands q33–34, who usually show growth and severe mental retardation, but without gross malformations (group 3). In our cohort, four patients (2, 4, 5 and 6) fit into group 1, as they carry proximal deletions not extending to q32; eight patients (1, 3, 7–12) show the deletion of the entire 13q32 band (with the exception of patient 3 who lacks only the q32.1 sub‐band) and are assigned to group 2; and two patients (13 and 14) seem to belong to group 3 as they carry more distal deletions exclusively involving the q33–q34 bands. Table 2 lists the clinical features indicated as being typical of patients with 13q32 deletions,3,6 and their presence or absence in our three patient groups.
Table 2 Main clinical features of patients with 13q deletions grouped according to the inclusion or exclusion of the 13q32 band.
| Features | Group 1 patients: deletions proximal to q32 | Group 2 patients: deletions including q32 | Group 3 patients: deletions distal to q32 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 5 | 6 | 1 | 3 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| OFC | ↑ | ↑ | ↓ | NK | — | ↓ | — | → | NK | ↓ | ↓ | ↓ | ↓ | ↓ |
| Growth | NK | ↓ | ↓ | NK | ↓ | ↓ | — | ↓ (pre‐n) | NK | ↓ | ↓ | ↓ | ↑ (post‐n) | ↓ |
| Mental retardation | Mild | — | Moderate | Severe | — | — | — | Severe | — | — | Profound | Profound | Moderate | Moderate |
| Brain anomalies | – | + | – | NK | + | – | – | + | NK | + | + | + | – | – |
| NTDs | – | – | – | – | – | – | + | – | + | – | – | – | – | – |
| Other major malformations | – | – | – | +* | +† | – | +‡ | – | – | +§ | +¶ | +** | – | – |
OFC, occipitofrontal head circumference; NTDs, neural tube defects; NK, not known; pre‐n, prenatal; post‐n, postnatal. ↓, reduced; →, normal; ↑, increased; +/–, presence/absence of the features; —, undetectable.
*Ambiguous genitalia.
†Microphthalmos.
‡Diaphragmatic hernia.
§Eye abnormalities with blindness; absent or hypoplastic thumbs; atrial and ventricular septal defects; patent ductus arteriosus; ambiguous genitalia; stenotic anus.
¶Hypoplastic thumbs; cleft lip and palate; eye abnormalities with blindness.
**Eye abnormalities with blindness.
Patients of groups 1 and 3 do not perfectly fit the suggested phenotypic features,3 especially with regard to mental retardation: in group 1, this was scored mild and moderate in patients 2 and 5, but severe in patient 6; and, in group 3, patients 13 and 14 show only moderate mental retardation, but they lack brain anomalies and other major malformations, according to Brown et al.3 However, our findings confirm that group 2 patients (lacking the 13q32 band) are the most seriously affected: all of the patients for whom data are available show severe and profound mental retardation, growth delay, microcephaly and brain anomalies or NTDs (except for patient 3, who lacks only the q32.1 sub‐band; table 2). The microphthalmia and coloboma previously reported in patients with the 13q32 deletion6 are confined to our group 2 patients (table 1 patients 1, 8, 10–12). Patients 8 and 10–12 share coloboma (table 1) and a common deleted region ranging from 13q32.2 to 13q33.2 (fig 1). In addition to coloboma, patients 10–12 show microphthalmia and blindness (table 1), and a common deleted interval extending from 13q33.2 (the distal bkp of the “coloboma” region) to 13q33.3 (the deletion breakpoint of patient 13 who does not display any eye abnormality; fig 1). The EFNB2 (ephrin‐B2) gene maps within the 13q33.2–13q33.3 interval. This gene belongs to a family of ligands with specificity for the eph receptors, which, in animal models, participate in several aspects of visual system development.16 It might be hypothesised that haploinsufficiency in this gene is responsible for the more severe eye anomalies found in patients 10–12 as compared with patient 8. Hand and foot anomalies in our sample are not restricted to group 2 (table 1), but patients 10 and 11 show the absent or hypoplastic thumbs considered to be the most representative of these features.3,15,17 Given that patients 10–12 present with the most severe eye abnormalities, but diverge from the absent or hypoplastic thumbs phenotype, assignment to 13q33.2–13q33.3 of gene or genes involved in absent or hypoplastic thumbs cannot be definitely inferred.
Patients 1 and 2 carry deletions including the RB1 locus (13q14.2). No signs of retinoblastoma were evident at prenatal ultrasound examination in patient 1, whereas patient 2 did not develop the tumour until age 11 years. In contrast with most patients with germline RB1 mutations, a relevant proportion of those carrying a cytogenetically visible deletion involving the 13q14 band show unilateral disease and some develop no tumours at all.18,19 This is the case in our postnatal patient 2.
Congenital heart defects are heterogeneous and do not seem to be associated with 13q32 or any other specific deleted segments; and renal and gastrointestinal malformations are infrequent, even in group 2.
Bartchs et al20 suggested that the 13q32.2–q34 region plays an important role in genital development. Two of our group 2 patients show ambiguous genitalia (patient 10) and a minor genital abnormality (patient 11; table 1), thus confirming the possible involvement of the q32 band in genital development. However, patient 6 (carrying a 13q22.1 or q22.2–q31.3 deletion) shows ambiguous genitalia, thus suggesting that other proximal genes may be involved in genital anomalies due to haploinsufficiency.
The critical 13q32 band responsible for the severe phenotypes of group 2 patients has subsequently been restricted to a 1.2‐Mb region between markers D13S136 and D13S147,6 containing the ZIC2 gene at q32.3.7,8 The observation that heterozygotic ZIC2 mutations are associated with HPE in patients without chromosomal abnormalities led to the hypothesis that ZIC2 haploinsufficiency may partially underlie the brain malformations seen in patients with the 13q deletion and, in line with this hypothesis, the only one of our group 2 patients without CNS anomalies (patient 3) retains the q32.3 sub‐band and, consequently, the ZIC2 gene. Luo et al9 have proposed a critical region at 13q33.2‐qter, distal to and not overlapping Brown's critical band specifically involved in NTDs, and suggested that one or more genes mapping to this band and distal to ZIC2 may cause NTDs as a result of haploinsufficiency. Two of our patients show NTDs (tables 1 and 2): patient 7 with a 13q22‐qter deletion, and patient 9 with an interstitial 13q31.1–q33.1 deletion and the maintenance of the entire proposed critical region. The distal breakpoint at q33.1 indeed maps 1.5 Mb proximally to the microsatellite markers D13S274‐D13S1311, fixing the q33.2 breakpoint of the terminal deletion indicated by Luo et al.9 On the basis of these observations, we presume that other dose‐sensitive genes proximal to q33.2 may be involved in NTDs if they are haploinsufficient, and this favours the hypothesis of the presence of more than one locus for NTDs in the 13q32‐qter region.6 The role of ZIC2 mutations as a common cause of human NTDs was excluded by Brown et al,21 but it should be noted that ZIC5 (another member of the ZIC gene family) maps 16 kb proximally to ZIC2. Deficiency of the Zic5 mouse orthologue is associated with NTDs,22 which makes the hypothesis that ZIC5 deletion may be responsible for human NTDs attractive even if no human ZIC5 mutations have yet been described.23
Another locus within 13q22–33 (different from ZIC2) has been proposed as a cause of DWM, a specific CNS abnormality associated with distal 13q deletion.10,11 Our patient 8 has DWM, and patients 11 and 12 show DWMs respectively associated with cerebellar hypoplasia and agenesis of the corpus callosum. As the breakpoints of their deletions (fig 1) are localised distally to the centromeric boundary of the previously reported DWM critical region,10 we narrowed the minimal DWM‐associated deletion interval to 13q32.2–33.2, and a recent report of a 13q31.2/32.1‐qter deletion in a fetus with DWM and other malformations supports our finding.24
The first locus involved in human DWM has been localised to an interval encompassing the ZIC1 and ZIC4 genes in the 3q2 region and, on the basis of the observation that heterozygotic deletions of both mouse orthologues Zic1 and Zic4 cause a DWM‐like phenotype, it has been hypothesised that a heterozygotic loss of both ZIC1 and ZIC4 genes may cause DWM in patients with 3q deletion.25 ZIC1 and ZIC4 genes map to chromosome 3 in a configuration that is paired with that of ZIC2 and ZIC5 on chromosome 13. Like the four previously reported patients with DWM,10,11,24 our patients with DWM lack the 13q32.3 band encompassing the ZIC2 and ZIC5 genes. It is therefore possible to assume that the loss of both ZIC2 and ZIC5 may cause DWM in patients with the 13q deletion, like the loss of ZIC1 and ZIC4 in patients with the 3q deletion. Grinberg et al23 have suggested that the loss of ZIC2 may lead to HPE, and the loss of both ZIC2 and ZIC5 may lead to DWM with HPE. However, all our patients with CNS anomalies lack both genes and have different CNS features, thus making it difficult to support this assumption. Clinical studies on patients with 13q deletions including only one of the two genes may be useful for establishing their possible role in determining CNS‐specific phenotypes, although further studies on human mutations and their phenotype effects are necessary to clarify the possible role of ZIC5 in CNS anomalies.
Key points
Chromosome 13q deletion is associated with varying phenotypes and there is still no acknowledged consensus correlation between the monosomy of distinct 13q regions and specific clinical features.
We studied 14 Italian patients carrying de novo 13q deletions by means of array‐comparative genomic hybridisation or fluorescent in situ hybridisation.
All our patients showed severe to mild mental retardation: eight had central nervous system (CNS) anomalies, six had eye abnormalities, nine had facial dysmorphisms and 10 had hand and/or feet anomalies. The size of the deleted regions varied from 4.2 to 75.7 Mb.
Our study confirms that patients lacking the 13q32 band are the most seriously affected. Dose‐sensitive genes proximal to q33.2 may be involved in neural tube defects (NTDs). The minimal deletion interval associated with the Dandy–Walker malformation (DWM) was narrowed to the 13q32.2–33.2 region and we proposed ZIC2 and ZIC5 genes as underlying various CNS malformations.
An additional consideration is that the loss of ZIC2 and/or ZIC5 may underlie the various CNS malformations seen in patients with the 13q deletion. Indeed, loss of the same genes leading to different CNS phenotypes can be ascribed to the simultaneous loss of contiguous but different chromosomal regions that may influence their expression. Alternatively, variable expression of such genes may be caused by modifier loci modulating the severity of the traits. Furthermore, it is well known that patients carrying deletions of precisely the same chromosomal regions may show different phenotypic features depending on the genetic background in which they are placed.
For more information visit the website http://jmg.bmj.com/supplemental
Abbreviations
array‐CGH - array comparative genomic hybridisation
BAC - —
CNS - central nervous system
DWM - Dandy–Walker malformation
FISH - fluorescent in situ hybridisation
HPE - holoprosencephaly
NTDs - neural tube defects
ZIC2 - —
Footnotes
Competing interests: None declared.
For more information visit the website http://jmg.bmj.com/supplemental
References
- 1.Allderdice P W, Davis J G, Miller O J, Klinger H P, Warburton D, Miller D A, Allen F H, Jr, Abrams C A, McGilvray E. The 13q‐deletion syndrome. Am J Hum Genet 196921499–512. [PMC free article] [PubMed] [Google Scholar]
- 2.Tranebjaerg L, Brøndum‐Nielsen K, Tommerup N, Warburg M, Mikkelsen M. Interstitial deletion 13q: further delineation of the syndrome by clinical and high resolution chromosome analysis of five patients. Am J Med Genet 198829739–753. [DOI] [PubMed] [Google Scholar]
- 3.Brown S, Gersen S, Anyane‐Yeboa K, Warburton D. Preliminary definition of a “critical region” of chromosome 13 in q32: report of 14 cases with 13q deletions and review of the literature. Am J Med Genet 19934552–59. [DOI] [PubMed] [Google Scholar]
- 4.Van Buggenhout G, Trommelen J, Hamel B, Fryns J P. 13q deletion syndrome in an adult mentally retarded patient. Genet Couns 199910(2)177–181. [PubMed] [Google Scholar]
- 5.Niebuhr E. Partial trisomies and deletions of chromosome 13. In: Yunis JJ, ed. New chromosomal syndromes. New York: Academic Press, 1977273–299.
- 6.Brown S, Russo J, Chitayat D, Warburton D. The 13q‐ syndrome: the molecular definition of a critical deletion region in band 13q32. Am J Hum Genet 199557859–866. [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S A, Warburton D, Brown L Y, Yu C Y, Roeder E R, Stengel‐Rutkwoski S, Hennekam R C, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd‐paired. Nat Genet 199820180–183. [DOI] [PubMed] [Google Scholar]
- 8.Brown L Y, Odent S, David V, Blayau M, Dubourg C, Apacik C, Delegado M A, Hall B D, Reynolds J F, Sommer A, Wieczorek D, Brown S A, Muenke M. Holoprosencephaly due to mutations in ZIC2: alanine tract expansion mutations may be caused by parental somatic recombination. Hum Mol Genet 200110791–796. [DOI] [PubMed] [Google Scholar]
- 9.Luo J, Balkin N, Stewart J F, Sarwark J F, Charrow J, Nye J S. Neural tube defects and the 13q deletion syndrome: evidence for a critical region in 13q33‐34. Am J Med Genet 200091227–230. [DOI] [PubMed] [Google Scholar]
- 10.McCormack W M, Shen J J, Curry S M, Berend S A, Kashork C, Pinar H, Potocki L, Bejjani B A. Partial deletions of the long arm of chromosome 13 associated with holoprosencephaly and the Dandy‐Walker malformation. Am J Med Genet 2002112384–389. [DOI] [PubMed] [Google Scholar]
- 11.Alanay Y, Aktas D, Utine E, Talim B, Onderoglu L, Caglar M, Tuncbilek E. Is Dandy‐Walker malformation associated with “distal 13q syndrome”? Findings in a fetus supporting previous observations. Am J Med Genet 2005136A265–268. [DOI] [PubMed] [Google Scholar]
- 12.Guala A. Dellavecchia C, Mannarino S, Rognone F, Giglio S, Minelli A, Danesino C. Ring chromosome 13 with loss of the region D13S317‐D13S285: phenotypic overlap with XK sindrome, Am J Med Genet 199772319–323. [PubMed] [Google Scholar]
- 13.Ciccone R, Giorda R, Gregato G, Guerrini R, Giglio S, Carrozzo R, Bonaglia M C, Priolo E, Lagana C, Tenconi R, Rocchi M, Pramparo T, Zuffardi O, Rossi E. Reciprocal translocations: a trap for cytogenetists? Hum Genet 2005117(6)571–582. [DOI] [PubMed] [Google Scholar]
- 14. Bonaglia MC , Giorda R , Mani E , Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. Published Online First: 11 November 2005, doi: 10. 1136/jmg. 2005. 038604 [DOI] [PMC free article] [PubMed]
- 15.Grindel S, Sandlin C, Wood V. Hand involvement in 13q deletion syndrome.J Pediatr Orthop 199919(5)620–623. [PubMed] [Google Scholar]
- 16.Williams S E, Mann F, Erskine L, Sakurai T, Wei S, Rossi D J, Gale N W, Holt C E, Mason C A, Henkemeyer M. Ephrin‐B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron 200339919–935. [DOI] [PubMed] [Google Scholar]
- 17.Amor D J, Voullaire L, Bentley K, Savarirayan R, Choo K H. Mosaic monosomy of a neocentric ring chromosome maps brachyphalangy and growth hormone deficiency to 13q31.1‐13q32.3. Am J Med Genet 2005133A151–157. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga E. Retinoblastoma: host resistance and 13q‐ chromosomal deletion. Hum Genet 198056(1)53–58. [DOI] [PubMed] [Google Scholar]
- 19.Baud O, Cormier‐Daire V, Lyonnet S, Desjardins L, Turleau C, Doz F. Dysmorphic phenotype and neurological impairment in 22 retinoblastoma patients with constitutional cytogenetic 13q deletion. Clin Genet 199955478–482. [DOI] [PubMed] [Google Scholar]
- 20.Bartsch O, Kuhnle U, Wu L L, Schwinger E, Hinkel G K. Evidence for a critical region for penoscrotal inversion, hypospadias, and imperforate anus within chromosomal region 13q32.2q34. Am J Med Genet 199665218–221. [DOI] [PubMed] [Google Scholar]
- 21.Brown L Y, Hodge S E, Johnson W G, Guy S G, Nye J S, Brown S. Possible association of NTDs with a polyhistidine tract polymorphism in the ZIC2 gene. Am J Med Genet 2002108128–131. [DOI] [PubMed] [Google Scholar]
- 22.Inoue T, Hatayama H, Tohmonda T, Itohara S, Aruga J, Mikoshiba K. Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev Biol 2004270(1)146–162. [DOI] [PubMed] [Google Scholar]
- 23.Grinberg I, Millen K J. The ZIC gene family in development and disease. Clin Genet 200567290–296. [DOI] [PubMed] [Google Scholar]
- 24.Gul A, Cebeci A, Erol O, Ceylan Y, Basaran S, Yuksel A. Prenatal diagnosis of 13q‐syndrome in a fetus with Dandy‐Walker malformation. Obstet Gynecol 20051051227–1229. [DOI] [PubMed] [Google Scholar]
- 25.Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns W B, Millen K J. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy‐Walker malformation. Nat Genet 2004361053–1055. [DOI] [PubMed] [Google Scholar]