Abstract
Background
The 3243A→G MTTL1 mutation is the most common heteroplasmic mitochondrial DNA (mtDNA) mutation associated with disease. Previous studies have shown that the percentage of mutated mtDNA decreases in blood as patients get older, but the mechanisms behind this remain unclear.
Objectives and method
To understand the dynamics of the process and the underlying mechanisms, an accurate fluorescent assay was established for 3243A→G heteroplasmy and the amount of mtDNA in blood with real‐time polymerase chain reaction was determined. The amount of mutated and wild‐type mtDNA was measured at two time points in 11 subjects.
Results
The percentage of mutated mtDNA decreases exponentially during life, and peripheral blood leucocytes in patients harbouring 3243A→G are profoundly depleted of mtDNA.
Conclusions
A similar decrease in mtDNA has been seen in other mitochondrial disorders, and in 3243A→G cell lines in culture, indicating that depletion of mtDNA may be a common secondary phenomenon in several mitochondrial diseases. Depletion of mtDNA is not always due to mutation of a nuclear gene involved in mtDNA maintenance.
The 3243A→G MTTL1 gene mutation of mitochondrial DNA (mtDNA) is the most common heteroplasmic pathogenic mtDNA mutation and is found in approximately 1 in 6000 of the general population.1 Although first described in mitochondrial encephalomyopathy with lactic acidosis and stroke‐like episodes (MELAS), the phenotypic spectrum is extremely diverse, including isolated diabetes and deafness, hypertrophic cardiomyopathy and retinitis pigmentosa.2 The clinical variability can be explained partly by tissue‐specific differences in the percentage of mutated mtDNA.3,4
Intriguingly, the percentage of mutated mtDNA is consistently lower in peripheral blood than in post‐mitotic tissues such as skeletal muscle and brain.3,5 Serial measurements in the same subject have shown that the percentage of the 3243A→G mutation in blood decreases over time,6,7 but the reasons for this are not clear. One possibility is that vegetative segregation in rapidly proliferating leucocyte precursors leads to high percentages of mutated mtDNA in some cells. This causes a biochemical defect of the respiratory chain, which either impairs the further proliferation of that cell lineage or leads to cell death.7 This would ultimately lead to a decrease in the percentage of mutated mtDNA in the daughter cells present in the peripheral blood. However, it is currently not known whether the biochemical defect is primarily because of high amounts of mutated mtDNA,8 low amounts of wild‐type mtDNA9 or a combination of both.
To advance our understanding of this process, we developed and validated a highly sensitive fluorescent assay to measure the changes in heteroplasmy over time, and also measured the absolute amount of mutated and wild‐type mtDNA in 11 subjects known to harbour 3243A→G.
Methods
Subjects and control samples
Sequential venous blood samples between 2.0 and 7.8 years apart were collected from 11 subjects known to harbour the 3243A→G mtDNA mutation (five men and six women; table 1), and single blood samples were taken from 10 healthy controls. For all the samples, the peripheral blood leucocyte and platelet counts were within the normal range, with a normal proportion of each leucocyte subset. Skeletal muscle biopsy specimens were available for 9 of the 11 subjects with the 3243A→G mutation and 6 controls for comparison. Total genomic DNA was extracted using a standard procedure.
Table 1 Percentage of 3243A→G mutation and the mean amount of mitochondrial DNA (copy number) in peripheral blood for 11 subjects.
| Patient demographics | Last cycle fluorescent PCR 3243A→G mutation (%) | Real‐time PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Time between samples (years) | Age at sample (years) | mtDNA | nDNA | ΔCt | Mean mtDNA copy number/cell | |||||
| Mean Ct | SD | Mean Ct | SD | Total copy number | Wild‐type copy number | Mutated copy number | |||||
| 1 | 7.8 | 18.9 | 47.9 | 17.1 | 0.153 | 21.8 | 0.115 | −4.6 | 49.6 | 25.9 | 23.8 |
| 26.7 | 36.2 | 19.7 | 0.208 | 25.6 | 0.173 | −5.9 | 122.2 | 78.0 | 44.2 | ||
| 2 | 5.3 | 59.1 | 7.7 | 22.6 | 0.153 | 28.4 | 0.289 | −5.8 | 111.4 | 102.9 | 8.6 |
| 64.4 | 6.9 | 14.9 | 0.115 | 21.5 | 0.200 | −6.6 | 189.6 | 176.5 | 13.1 | ||
| 3 | 6.1 | 40.0 | 37.2 | 17.2 | 0.173 | 22.7 | 0.100 | −5.5 | 90.5 | 56.8 | 33.7 |
| 46.1 | 34.2 | 14.9 | 0.115 | 21.5 | 0.200 | −6.6 | 189.6 | 124.7 | 64.8 | ||
| 4 | 5.7 | 24.9 | 39.2 | 18.4 | 0.058 | 23.0 | 0.252 | −4.7 | 50.8 | 30.9 | 19.9 |
| 30.6 | 32.4 | 15.7 | 0.058 | 20.6 | 0.100 | −4.9 | 58.4 | 39.4 | 18.9 | ||
| 5 | 3.2 | 47.8 | 9.8 | 17.2 | 0.058 | 22.7 | 0.265 | −5.5 | 88.4 | 79.8 | 8.7 |
| 51.0 | 8.6 | 16.0 | 0.361 | 21.4 | 0.153 | −5.4 | 82.5 | 75.4 | 7.1 | ||
| 6 | 5.4 | 48.0 | 14.8 | 18.0 | 0.153 | 23.0 | 0.100 | −5.0 | 65.5 | 55.8 | 9.7 |
| 53.4 | 12.7 | 15.6 | 0.265 | 20.9 | 0.200 | −5.3 | 78.8 | 68.8 | 10.0 | ||
| 7 | 5.5 | 38.0 | 15.6 | 19.6 | 0.115 | 23.8 | 0.115 | −4.2 | 36.8 | 31.0 | 5.7 |
| 43.5 | 13.8 | 16.5 | 0.153 | 21.2 | 0.100 | −4.7 | 53.2 | 45.9 | 7.3 | ||
| 8 | 5.7 | 35.0 | 13.2 | 26.2 | 0.289 | 30.5 | 0.000 | −4.3 | 40.3 | 35.0 | 5.3 |
| 40.7 | 12.6 | 16.1 | 0.153 | 21.9 | 0.115 | −5.7 | 106.4 | 93.0 | 13.4 | ||
| 9 | 2.0 | 43.5 | 27.9 | 15.8 | 0.153 | 21.5 | 0.306 | −5.6 | 99.3 | 71.6 | 27.7 |
| 45.5 | 27.1 | 17.2 | 0.058 | 22.7 | 0.321 | −5.5 | 90.5 | 66.0 | 24.5 | ||
| 10 | 5.1 | 44.0 | 15.8 | 15.3 | 0.153 | 20.7 | 0.208 | −5.3 | 80.6 | 67.9 | 12.7 |
| 49.1 | 10.5 | 16.1 | 0.208 | 21.6 | 0.231 | −5.4 | 86.4 | 77.3 | 9.1 | ||
| 11 | 7.0 | 60.7 | 12.1 | 16.8 | 0.100 | 21.6 | 0.153 | −4.8 | 54.4 | 47.9 | 6.6 |
| 66.7 | 5.5 | 25.9 | 0.115 | 29.2 | 0.100 | −3.3 | 20.2 | 19.1 | 1.1 | ||
mtDNA, mitochondrial DNA; nDNA, nuclear DNA; PCR, polymerase chain reaction; ΔCt, difference between the threshold cycles for nDNA and mtDNA.
Mean percentage of mutated mtDNA was determined by the last cycle fluorescent PCR (undiluted, mean of three measurements from the genomic sample). Copy number measurements were determined by real‐time PCR using iQ Sybr Green on the BioRad ICycler (mean of three measurements from the genomic sample). mtDNA: using a target template spanning from nucleotides 3459 to 3569 of the mtDNA ND1 gene; nDNA: using a target template spanning from nucleotide 804 to 903 of the single‐copy nuclear gene GAPDH. Wild‐type and mutated mtDNA copy numbers were determined from the mean total mtDNA copy number per cell, and the mean percentage mutated mtDNA. Relative copy number was calculated from the ΔCt value, 2(2−ΔCt), to account for the two copies of glyceraldehyde‐3‐phosphate dehydrogenase in each mononuclear peripheral blood cell.
Establishing an accurate fluorescent 3243A→G heteroplasmy assay
A range of heteroplasmic samples was generated by mixing mutated and wild‐type cloned fragments containing 3243 nucleotides generated from two siblings, one with a high percentage of the 3243A→G mutation and one with a low percentage 3243A→G, but who otherwise had an identical mtDNA sequence. Genomic DNA from each sibling was amplified across the region containing the MTTL1 gene and cloned using the pGEM‐T Easy vector system (Promega, Southampton, UK), according to the manufacturer's instructions. Plasmids from transformed JM109‐competent Escherichia coli cells were purified using the Qiaprep spin mini prep kit (Qiagen, Crawley, UK) and quantified by ultraviolet spectrophotometry (Eppendorf, Cambridge, UK). Mutated and wild‐type cloned DNA were mixed to generate heteroplasmic samples of 0%, 5%, 10%, 25% and 50%.
The percentage of mutated mtDNA was determined in triplicate using the gold standard technique of last cycle hot polymerase chain reaction (PCR). A 154‐bp region encompassing the MTTL1 gene was amplified using standard cycling conditions and the following primers: nucleotides 3200–3218 forward, nucleotides 3353–3334 reverse. After the addition of 5 μCi [α‐32P]dCTP (3000 Ci/mmol) to the last PCR cycle, products were precipitated and digested at 37°C with HaeIII (10 U; New England Biolabs, Ipswich, UK). Restriction fragments were separated on a 12% non‐denaturing polyacrylamide gel, dried on to a support, and exposed to a PhosphorImager cassette. The level of mtDNA heteroplasmy was quantified using ImageQuant TL software (Amersham Biosciences, Buckinghamshire, UK).10
The last cycle hot PCR method was adapted to a fluorescent analyser (Beckman CEQ 8000; Beckman Coulter, Buckinghamshire, UK) using modified primers (nucleotides 3155–3171 forward, nucleotides 3353–3334 reverse) and optimised PCR conditions. Fluorescent‐labelled forward primer was added in the last PCR cycle instead of the radioactive label, followed by HaeIII digestion (10 U). Serial dilutions (in water) of the last cycle fluorescent PCR restriction fragment length polymorphism (RFLP) products were analysed in triplicate using a Beckman Coulter CEQ 8000 fluorescent DNA fragment analyser and fragment analysis software (V.8.0).
Real‐time PCR
The total amount of mtDNA (referred to as the mtDNA copy number) in homogenate DNA samples from patients and controls was determined by real‐time PCR using iQ Sybr Green on the BioRad ICycler (BioRad, Hercules, California, USA) to a target template spanning from nucleotide 3459 to nucleotide 3569 of the mtDNA ND1 gene. This was normalised using a nuclear‐encoded template for the glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) gene spanning from nucleotide 804 to nucleotide 903 (NCBI, NM_002046.2). Relative copy number was calculated from the threshold cycle value, ΔCt value, where the mean amount of mtDNA/cell = 2(2−ΔCt), to account for the two copies of GAPDH in each cell nucleus. Each reaction was optimised and confirmed to be linear over an appropriate concentration range using genomic DNA standards. Samples were analysed in triplicate for both assays, enabling calculation of the average mtDNA to nDNA ratio. As the overwhelming majority of blood mtDNA is present in the leucocyte population, this assay provides a good estimation of the average amount of mtDNA within the blood leucocyte population.
Statistical analysis
Statistical analyses were carried out using Minitab (V.14), including correlations analysis with Spearman's correlation coefficient (R2), and two‐sample t tests for the comparison of the amount of mtDNA.
Results
Comparison of the radioactive and fluorescent last cycle assays
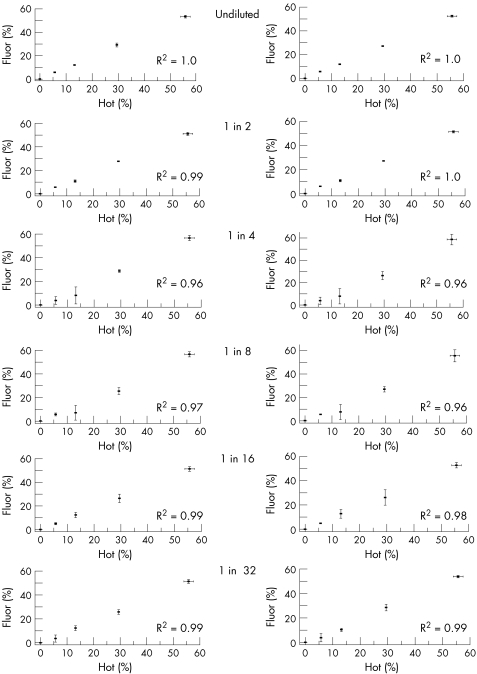
The last cycle hot PCR and last cycle fluorescent PCR methods were compared directly by analysing the mixed cloned DNA. Each assay was performed in triplicate from the mixed clone templates. Fluorescent‐labelled products can be analysed in two ways: measuring the area under the curve or measuring the peak height amplitude. Both measurements are likely to be reliable when the signal to noise ratio is high, but the point at which they become unreliable has not been formally studied. We therefore compared both measurements over a range of dilutions with the current gold standard technique of the last cycle hot PCR‐RFLP (fig 1).
Figure 1 Comparison of the last cycle fluorescent polymerase chain reaction‐ restriction fragment length polymorphism (PCR‐RFLP) method with the last cycle hot PCR‐RFLP method to quantify the percentage of 3243A→G mutation in mixed clones (see Methods). y Axis: percentage of 3243A→G mutation determined from the relative fluorescence units (standard deviation (SD) from three independent measurements) of either the peak height amplitude (left‐hand graphs) or the area under the curve. x Axis: percentage of 3243A→G mutation determined by the last cycle hot PCR (SD from three independent measurements). Each pair of graphs shows the results from the serial dilution (from undiluted to 1:32 in water) of the last cycle fluorescent PCR‐RFLP products run independently on the fluorescent genetic analyser in triplicate. R2 = Spearman correlation coefficient using the individual data points (ie not the mean values).
In each case, there was a strong correlation between the different measures (R2>0.95), but the undiluted and 1:2 dilution of the last cycle fluorescent‐labelled assay had the strongest correlation with the last cycle hot assay (fig 1). For greater dilutions, the last cycle fluorescent assay showed the most variation at lower percentage levels of heteroplasmy. We found no obvious difference between the area under the curve or peak height amplitude methods.
Sequential blood heteroplasmy quantification
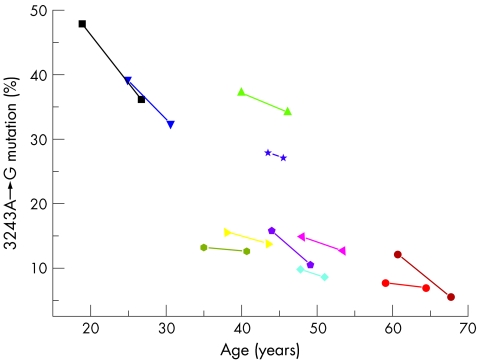
The percentage of mutated mtDNA was determined in each sample from the 11 subjects harbouring the 3243A→G mutation using the last cycle fluorescent PCR‐RFLP. Measurements were made from the original genomic DNA sample on three independent occasions. The last cycle fluorescent PCR‐labelled HaeIII digestion products were not diluted. In each subject, the percentage of 3243A→G decreased over time (fig 2). The overall mean change was a decrease of 0.6% per year (standard deviation (SD) 0.5).
Figure 2 Relationship between the percentage of the 3243A→G mitochondrial DNA (mtDNA) mutation in blood and age (years) for 11 subjects measured by last cycle fluorescent polymerase chain reaction (PCR). Each patient has a different symbol and colour, with serial measurements on the same patient connected by a straight line. PCR products were not diluted before loading.
mtDNA copy number in leucocytes
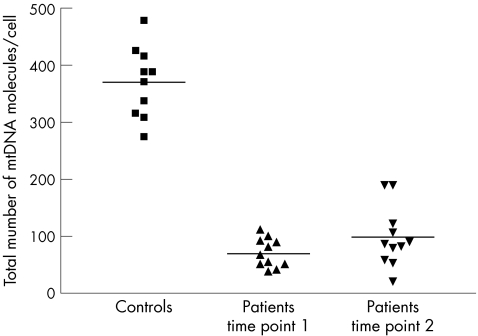
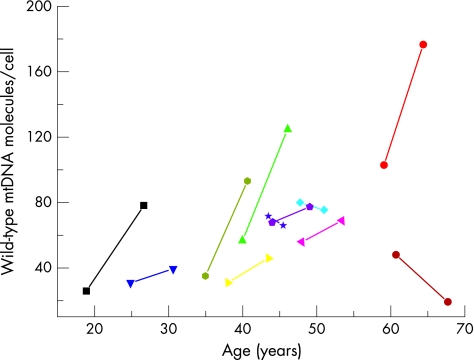
The mean mtDNA copy number in control blood leucocytes was 370.1 (SD 61.9) (fig 3, table 1). All the samples from subjects with the 3243A→G mutation had low levels of mtDNA (mean for sample set 1 = 69.8 (SD 25.4), two‐sample t test comparison with controls, t = −14.28, p<0.001; mean for sample set 2 = 98.0 (SD 52.8), two‐sample t test comparison with controls t = −10.79, p<0.001). In eight of the subjects with the 3243A→G mutation, the mean amount of mtDNA per leucocyte increased over time. In three subjects, the mean amount of mtDNA per leucocyte decreased over time. The overall mean change was an increase in the mean amount of mtDNA per leucocyte (4.4 molecules/cell/year (SD 7.6); fig 4). We found no clear correlation between the total amount of mtDNA and the age of the subjects, the initial percentage of mutated mtDNA or the absolute amounts of mutated and wild‐type mtDNA.
Figure 3 Total number of mitochondrial DNA (mtDNA) molecules per cell in 10 controls and 11 subjects with the 3243A→G mutation. Serial measurements are represented by time points 1 and 2. Total mtDNA (mtDNA copy number) was determined by real‐time polymerase chain reaction (PCR) using iQ Sybr Green on the BioRad ICycler. Each data point represents the mean of three independent measurements from the original genomic DNA sample. Solid horizontal lines represent the mean value for each dataset.
Figure 4 Mean amount of wild‐type mitochondrial DNA (mtDNA) in 11 subjects with the 3243A→G mutation on two occasions. y Axis: wild‐type mtDNA (mtDNA copy number) was determined by real‐time polymerase chain reaction (PCR) in triplicate using iQ Sybr Green on the BioRad ICycler to determine the total amount of mtDNA. The wild‐type mtDNA was calculated from the mean percentage determined by last cycle fluorescent PCR shown in fig 2. x Axis: age of the subject (years) at the time of sampling. Each patient has a different symbol and colour, with the serial measurements connected by a straight line and using the same colour code as fig 2.
Skeletal muscle mtDNA
In each patient, the percentage of 3243A→G was greater in muscle than in blood. The amount of mtDNA per nucleus was greater in the skeletal muscle of 3243A→G subjects than of the controls (supplementary table A, available online at www.jmg.bmjjournals.com/supplemental).
Discussion
Several conclusions can be drawn from this work. On the technical side, we have shown that the addition of a fluorescent‐labelled oligonucleotide primer in the last PCR cycle enables the accurate quantification of mtDNA heteroplasmy. Although the strongest correlation was between the undiluted and 1:2 diluted fluorescent products and the last cycle hot PCR, all the assays correlated well with an R2>0.95. We were surprised to find that the serial dilution of the fluorescent PCR products did not have a dramatic effect on the assay. This probably reflects the automated electrostatic injection methods used by most capillary‐based DNA analysers, which load a fixed amount of labelled DNA rather than a fixed volume of solution. It is possible that different results are obtained for manually loaded gel‐based systems. Arguably, the reproducibility of the fluorescent assay exceeds that of last cycle hot PCR (see the approximately 50% 3243A→G data in fig 1), and, given the inherent risks of using radiochemicals, last cycle fluorescent analysis should now be the benchmark for measuring mtDNA heteroplasmy.
Using the last cycle fluorescent PCR, we confirmed that the percentage of 3243A→G decreases in blood over time. The mean percentage decrease of 0.6% per year corresponds well to the largest previous study (0.69% per year; SD 0.61).6 In our study (fig 2), the greatest rate of decrease in the percentage of mutated mtDNA was seen in the younger subjects, consistent with an exponential loss of 3243A→G from the peripheral blood. With this in mind, reporting mean values for the percentage decrease could be misleading, and explains why the mean value in our adult study was significantly less than that found in one study of six subjects (1.12% per year; SD 0.65),7 which predominantly involved observations made over childhood (two‐sample t test comparing the data in table 1 with the data in Rahman et al7: t = 3.53, p = 0.004).
The most striking novel finding is the profound depletion of mtDNA in blood leucocytes in patients harbouring this mutation. Our control data for the mean mtDNA copy number/leucocyte are remarkably similar to other published values based on real‐time PCR and other techniques,11,12 confirming that we were using a reliable assay. How can we therefore explain these observations? Given that the peripheral blood count was normal in each case, a change in the number of cell nuclei or cell types probably does not account for the profound depletion we have observed. From first principles, the depletion of mtDNA could be due to decreased rates of mtDNA synthesis relative to the intense cellular proliferation in leucocyte precursors. An alternative (and non‐exclusive) explanation is an increased loss of mtDNA from the precursors of the mature cells.
The peripheral leucocyte population is continuously recycled. Neutrophils are the largest single component (up to 80% of circulating leucocytes), maturing in the bone marrow for 11–12 days before being released into the circulation, where they remain for 6–8 h.13 The entire neutrophil population in the peripheral blood (approximately 5×1010 cells) is therefore replenished on a daily basis, requiring a massive amount of mtDNA replication to maintain normal mtDNA levels. This is sustained by a limited number of stem cells (1 in 104 to 1 in 105 marrow cells), which turn over at a low rate and repopulate the blood through a hierarchical cascade of cell divisions.14 It is therefore far more probable that any effect on mtDNA replication would manifest during the massive expansion of daughter cells leading to the formation of mature leucocytes, rather than in the stem‐cell population itself. Even a subtle decrease in the replication rate could lead to the dramatic depletion that we have observed in the peripheral blood. For example, if the mtDNA replication rate was a fraction f of the normal replication rate, then after a series of n cell divisions the leucocyte precursors would be depleted by a factor fn. Twenty cell divisions (the number required for a million‐fold increase in cells) would give our observed depletion to 23% of the normal level with f = 0.93.
Could the bioenergetic defect be responsible for a relative decrease in rate of mtDNA synthesis in the leucocyte precursors? Although plausible, this explanation is difficult to reconcile with the increased amount of mtDNA seen in post‐mitotic tissues of subjects harbouring 3243A→G.15 In addition, if the loss of mtDNA were directly related to a defect of ATP synthesis, we would expect a closer relationship between the percentage level of mutated mtDNA and the degree of depletion. This was clearly not the case—for example, low amounts of mtDNA were found in a young subject with approximately 50% 3243A→G in blood and an older subject with approximately 5% 3243A→G in blood (figs 2 and 3). Recent in vitro studies on human NT2 teratocarcinoma cybrids harbouring 3243A→G (NT2.3243 cybrids) cast light on this issue.16 High percentages of 3243A→G were associated with a profound loss of mtDNA from the NT2.3243 cybrids. This seemed to be mutation specific, as the same depletion was not seen in NT2 cybrids homoplasmic for the 1555A→G mtDNA mutation. Moreover, the presence of a known suppressor mutation (12300G→A), which ameliorates the biochemical effects of 3243A→G at high levels, did not prevent the loss of mtDNA from the cybrid cell lines, indicating that the biochemical defect is not the principal factor behind the depletion. Remarkably, not all the cell lines with >99% 3243A→G lost their mtDNA, proving that the percentage of mutated mtDNA is not an absolute determinant of the depletion. Further work showed slow rates of segregation of the 3243A→G mutation in NT2.3243 cybrids, providing indirect evidence that functional or physical partitioning of mtDNA molecules contributes to the segregation process. These observations highlight the complex interplay between intercellular and intracellular signals that are involved in the mitotic segregation of the 3243A→G mutation.16 Similar mechanisms may be involved in rapidly dividing tissues in vivo, including the leucocyte population. It is intriguing that, as the percentage of mutated mtDNA in blood decreases in subjects harbouring the 3243A→G mutation (fig 2), there is an overall increase in the amount of mtDNA in blood, particularly wild‐type mtDNA (fig 4). This suggests a degree of recovery from the depletion as time passes, although the amount never reaches normal values.
An alternative explanation for the loss of one mtDNA genotype in blood is through immune surveillance mediated through an abnormal cell surface epitope. This hypothesis is supported by the description of a complex I (ND) gene variant in mice that forms a maternally transmitted major histocompatibility antigen.17 However, recent work on heteroplasmic mice has shown that immune‐mediated cell loss does not occur in heteroplasmic mice with the C57B/BALB genotype, and nuclear genes may be important.18
Finally, the depletion may be a secondary phenomenon, as in other mitochondrial disorders. Mutations in OPA1 cause the most common form of autosomal dominant optic atrophy. In a recent study, subjects with OPA1 mutations had markedly lower levels of mtDNA in leucocytes than controls.11 Given the central role of OPA1 in maintaining the structural integrity of mitochondria and mtDNA, it was suggested that the primary genetic defect led to the loss of both mitochondria and mitochondrial genomes. A similar structural disintegration of the mitochondrial network has been seen in subjects harbouring primary mtDNA defects, potentially explaining the observations we report here. By contrast, one study19 described a mild increase in the amount of mtDNA in leucocytes from subjects harbouring mtDNA mutations that cause Leber hereditary optic neuropathy. However, in contrast with the 3243A→G mutation, the biochemical defect in patients with Leber hereditary optic neuropathy is mild and sometimes undetectable.20,21 Further work will determine whether depletion of mtDNA is seen in other mtDNA disorders.
If the 3243A→G mutation is causing depletion through an effect on the structural integrity of mitochondria in leucocytes, why does this not occur in non‐dividing tissues such as skeletal muscle, where there tends to be a proliferation of mitochondria and mtDNA (supplementary table A online at www.jmg.bmjjournals.com/supplemental)? At present, we can only speculate, but recent work in our laboratory has shown that, although initially the amount of skeletal muscle mtDNA is high in subjects with mtDNA mutations, there is a progressive loss of mtDNA over time.22 This could be partly due to muscle deconditioning, but it may be due to a more general process related to mitochondrial fragility. These findings have important implications for the molecular diagnosis of mitochondrial disorders—the presence of depletion may be a secondary phenomenon and the primary molecular defect may be a mtDNA mutation or a nuclear gene mutation not directly involved in the synthesis of mtDNA.
Key points
The percentage of the 3243A→G MTTL1 mutation in blood decreases exponentially during life.
Peripheral blood leucocytes in patients harbouring the 3243A→G mutation are profoundly depleted of mitochondrial DNA (mtDNA).
Depletion of mtDNA is not always due to mutation of a nuclear gene involved in mtDNA maintenance.
Abbreviations
GADPH - glyceraldehyde‐3‐phosphate dehydrogenase
mtDNA - mitochondrial DNA
PCR - polymerase chain reaction
RFLP - restriction fragment length polymorphism
Footnotes
Funding: PFC also receives funding from Ataxia (UK), the Alzheimer's Research Trust, the Association Française Contre les Myopathies, and the United Mitochondrial Diseases Foundation, and the EU FP P
Competing interests: None declared.
References
- 1.Majamaa K, Moilanen J S, Uimonen S, Remes A M, Salmela P I, Karppa M, Majamaa‐Volti K A M, Rusanen H, Sorri M, Peuhkurinen K J, Hassinen I E. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: prevalence of the mutation in an adult population. Am J Hum Genet 199863447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciafaloni E, Ricci E, Shanske S, Moraes C T, Silvestri G, Hirano M, Simonetti S, Angelini C, Donati M A, Garcia C, Martinuzzi A, Mosewich R, Servidei S. MELAS: clinical features, biochemistry, and molecular genetics. Ann Neurol 199231391–398. [DOI] [PubMed] [Google Scholar]
- 3.Ciafaloni E, Ricci E, Servidei S, Shanske S, Silvestri G, Manfredi G, Schon E A, DiMauro S. Widespread tissue distribution of a tRNALeu(UUR) mutation in the mitochondrial DNA of a patient with MELAS syndrome. Neurology 1991411663–1664. [DOI] [PubMed] [Google Scholar]
- 4.Chinnery P F, Howell N, Lightowlers R N, Turnbull D M. Molecular pathology of MELAS and MERRF. The relationship between mutation load and clinical phenotypes. Brain 1997120(Pt 10)1713–1721. [DOI] [PubMed] [Google Scholar]
- 5.Dubeau F, De Stefano N, Zifkin B G, Arnold D L, Shoubridge E A. Oxidative phosphorylation defect in the brains of carriers of the tRNAleu(UUR) A3243G mutation in a MELAS pedigree. Ann Neurol 200047179–185. [PubMed] [Google Scholar]
- 6.'t Hart L M, Jansen J J, Lemkes H H P J, de Knijff P, Maassen J A. Heteroplasmy levels of a mitochondrial gene mutation associated with diabetes mellitus decrease in leucocyte DNA upon aging. Hum Mutat 19967193–197. [DOI] [PubMed] [Google Scholar]
- 7.Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet 200168238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoubridge E A, Karpati G, Hastings K E. Deletion mutants are functionally dominant over wild‐type mitochondrial genomes in skeletal muscle fiber segments in mitochondrial disease. Cell 19906243–49. [DOI] [PubMed] [Google Scholar]
- 9.Bentlage H A C M, Attardi G. Relationship of genotype to phenotype in fibroblast‐derived transmitochondrial cell lines carrying the 3243 mutation associated with MELAS encephalomyopathy: shift towards mutant genotype and role of mtDNA copy number. Hum Mol Genet 19965197–205. [DOI] [PubMed] [Google Scholar]
- 10.McDonnell M T, Schaefer A M, Blakely E L, McFarland R, Chinnery P F, Turnbull D M, Taylor R W. Noninvasive diagnosis of the 3243A>G mitochondrial DNA mutation using urinary epithelial cells. Eur J Hum Genet 200412778–781. [DOI] [PubMed] [Google Scholar]
- 11.Kim J Y, Hwang J ‐ M, Ko H S, Seong M ‐ W, Park S S. Mitochondrial DNA content is decreased in autosomal dominant optic atrophy. Neurology 200464966–972. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Cooney D A, Sreenath A, Zhan Q, Agbaria R, Stowe E E, Fornace A J, Johns D J. Quantitation of mitochondrial DNA in human lymphoblasts by a competetive polymerase chain reaction method: application to the study of inhibitors of mitochondrial DNA content. Mol Pharmacol 1994461063–1069. [PubMed] [Google Scholar]
- 13.Handin R I, Lux S E, Stossel T P.Blood. The principles and practice of hematology. 2nd edn. Philadelphia: Lippincott, Williams and Wilkins, 2003463–464.
- 14.Quesenberry P, Levitt L. Hematopoietic stem cells. N Engl J Med 1979301755–761. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann P, Koga Y, Shanske S, Hirano M, DiMauro S, King M P, Schon E A. Mitochondrial DNA and RNA processing in MELAS. Ann Neurol 199640172–180. [DOI] [PubMed] [Google Scholar]
- 16.Turner C J, Granycome C, Hurst R, Pohler E, Juhola M K, Juhola M I, Jacobs H T, Sutherland L, Holt I J. Systematic segregation to mutant mitochondrial DNA and accompanying loss of mitochondrial DNA in human NT2 teratocarcinoma cybrids. Genetics 20051701879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loveland B, Wang C R, Yonekawa H, Hermel E, Lindahl K F. Maternally transmitted histocompatibility antigen of mice: a hydrophobic peptide of a mitochondrially encoded protein. Cell 199060971–980. [DOI] [PubMed] [Google Scholar]
- 18.Battersby B J, Redpath M E, Shoubridge E A. Mitochondrial DNA segregation in hematopoietic lineages does not depend on MHC presentation of mitochondrially encoded peptides. Hum Mol Genet 2005142587–2594. [DOI] [PubMed] [Google Scholar]
- 19.Nishioka T, Soemantri A, Ishida T. mtDNA/nDNA ratio in 14484 LHON mitochondrial mutation carriers. J Hum Genet 200449701–705. [DOI] [PubMed] [Google Scholar]
- 20.Carelli V, Ghelli A, Ratta M, Bacchilega E, Sangiorgi S, Mancini R, Leuzzi V, Cortelli P, Montagna P, Lugaresi E, Degli Esposti M. Leber's hereditary optic neuropathy: biochemical effect of 11778/ND4 and 3460/ND1 mutations and correlation with the mitochondrial genotype. Neurology 1997481623–1632. [DOI] [PubMed] [Google Scholar]
- 21.Carelli V, Ghelli A, Bucchi L, Montagna P, De Negri A, Leuzzi V, Carducci C, Lenaz G, Lugaresi E, Degli Esposti M. Biochemical features of mtDNA 14484 (ND6/M64V) point mutation associated with Leber's hereditary optic neuropathy. Ann Neurol 199945320–328. [PubMed] [Google Scholar]
- 22.Durham S E, Brown D T, Turnbull D M, Chinnery P F. Progressive depletion of mtDNA in mitochondrial myopathy. Neurology 200667502–504. [DOI] [PubMed] [Google Scholar]