Abstract
Background
Recently, conflicting reports have been published on the potential role of genetic variants in the α‐T catenin gene (VR22; CTNNA3) on the risk for Alzheimer's disease. In these papers, evidence for association is mostly observed in multiplex families with Alzheimer's disease, whereas case–control samples of sporadic Alzheimer's disease are predominantly negative.
Methods
After sequencing VR22 in multiplex families with Alzheimer's disease linked to chromosome 10q21, we identified a novel non‐synonymous (Ser596Asn; rs4548513) single nucleotide polymorphism (SNP). This and four non‐coding SNPs were assessed in two independent samples of families with Alzheimer's disease, one with 1439 subjects from 437 multiplex families with Alzheimer's disease and the other with 489 subjects from 217 discordant sibships.
Results
A weak association with the Ser596Asn SNP in the multiplex sample, predominantly in families with late‐onset Alzheimer's disease (p = 0.02), was observed. However, this association does not seem to contribute substantially to the chromosome 10 Alzheimer's disease linkage signal that we and others have reported previously. No evidence was found of association with any of the four additional SNPs tested in the multiplex families with Alzheimer's disease. Finally, the Ser596Asn change was not associated with the risk for Alzheimer's disease in the independent discordant sibship sample.
Conclusions
This is the first study to report evidence of an association between a potentially functional, non‐synonymous SNP in VR22 and the risk for Alzheimer's disease. As the underlying effects are probably small, and are only seen in families with multiple affected members, the population‐wide significance of this finding remains to be determined.
In the October 2005 issue of the Journal of Medical Genetics, Martin et al1 reported evidence of a genetic association between risk for Alzheimer's disease and one of 11 investigated common single‐nucleotide polymorphisms (SNPs) in the gene encoding α‐T catenin (VR22, aka CTNNA3). In addition to this paper, association data had also been published for seven independent case–control samples2,3,4—all of which failed to show an association between the investigated markers and the risk for Alzheimer's disease—after a report of an association between plasma Aβ42 levels and two non‐coding SNPs (ie, rs7070570 (“4360”) or rs12357560 (“4783”), both located in intron 13) in a small sample of multiplex families with Alzheimer's disease.5 Notably, none of the follow‐up studies, including the one by Martin et al, detected evidence of an association with the two originally associated intron 13 markers. This is in line with meta‐analyses from our group based on published genotype data (except from Martin et al, who did not provide their genotype or allele frequencies), in which the summary odds ratio currently fails to show evidence for a considerable effect.6 Instead, the supportive evidence of Martin et al's paper stems from a novel SNP located slightly 5′ in the same intron (ie, rs7074454 (“SNP6”); fig 1). The major C allele of this SNP showed marginal association in a sample of predominantly multiplex families with Alzheimer's disease (p = 0.01), and—after stratification on apolipoprotein‐ε4 genotype—in an independent case–control population (p = 0.047). Taken together, we are left with a situation that is quite typical for genetic association studies of complex diseases: a highly significant association in the initial report, largely followed by a lack of confirmation, leading to overall insignificant effects in unbiased meta‐analyses. After the initial paper, the only instance supporting the α‐T catenin gene as a risk gene for Alzheimer's disease is with a different SNP than originally reported, and has emerged after testing nearly a dozen individual variants.1
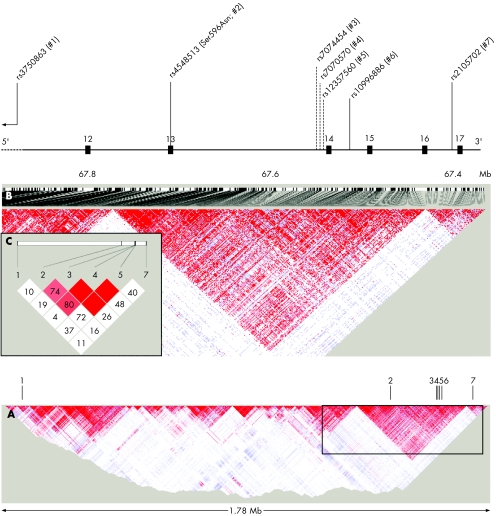
Figure 1 Localisation of tested single‐nucleotide polymorphisms (SNPs) in the VR22 gene and underlying linkage disequilibrium patterns. (A) Overview of linkage disequilibrium pattern of 3190 SNPs genotyped for the International HapMap Project14 in 30 trios from the Centre d'Etude du Polymorphisme Humaine Utah sample (release #20/phase II, January 2006). Vertical lines with indices represent the location of the five SNPs genotyped here and elsewhere. Box indicates inlet magnified in (B). VR22 is depicted in the 5′→3′ orientation, which represents qter → pter on chromosome 10. (B) Details of approximately 500 kb encompassing exons 12–17 of the VR22 gene including SNPs analysed here (numbers 1, 2, 4, 6, 7). The following SNPs have been associated with the risk for Alzheimer's disease (rs7074454 (#3)1) or plasma Aβ‐levels (rs7070570 (#4) and rs12357560 (#5)5) in previous studies (broken lines). As can be seen, all SNPs currently reported to show association, including the Ser596Asn variant (bold) identified here, are located within an approximately 300 kb region of extensive linkage disequilibrium. (C) Pairwise linkage disequilibrium analyses of six of the seven SNPs based on the most current HapMap release suggest the presence of pronounced linkage disequilibrium between the Ser596Asn SNP (#2) and variants previously associated with Alzheimer's disease and Aβ‐levels (numbers 3–5). Note that rs10996886 (#6) is currently not listed on HapMap.
In our ongoing efforts to pinpoint the putative gene(s) causing Alzheimer's disease underlying one or several linkage region(s) on chromosome 10q identified by us7,8 and others,9,10 we are assessing several positional candidate genes in two large and well‐characterised family samples. In addition to genotyping the most strongly associated SNP (ie, rs7070570 (“4360”)) of the original paper suggesting VR22 as a putative gene causing Alzheimer's disease, we sequenced VR22 in families showing the most pronounced linkage to the approximately 80–90 cM region around marker D10S1225. This effort led to the discovery of a hitherto unknown non‐synonymous coding change in exon 13 (Ser596Asn), approximately 175 kb 5′ of rs7070570. Although this amino acid is not particularly well conserved across species, it is located in an evolutionarily highly conserved vinculin domain, and it is conceivable that the codon 596 substitution could lead to changes in α‐T catenin function. Early in 2003, this SNP was deposited in the public domain as rs4548513 (http://www.ncbi.nlm.nih.gov/SNP/). In addition to Ser596Asn and rs7070570, we tested three other SNPs upstream and downstream of the exon/intron 13 region (table 1 and fig 1) in a sample of 1439 members from 437 multiplex families with Alzheimer's disease recruited as part of the National Institute of Mental Health (NIMH) Genetics Initiative Alzheimer's Disease Study Sample (all procedures are described in detail in Bertram et al11; primer sequences for the VR22 SNPs are available on request). Of the five SNPs analysed, only the Ser596Asn variant showed evidence of association in the unstratified sample (p = 0.04). This association was most pronounced in families with age of onset ⩾65 years (p = 0.02; table 1). In contrast with the findings of Martin et al,1 we did not detect an interaction between this variant and the apolipoprotein‐ε4 allele (p = 0.4). Further, analysis of the genome screen linkage data, after excluding families showing the strongest association with Ser598Asn, reduced the chromosome 10q linkage signal only slightly, suggesting that the VR22 Ser596Asn association is not the major underlying cause of the overall linkage observed in this region (fig 2). Finally, we assessed whether the Ser596Asn association is present in our second, independent sample of 489 subjects from 217 families, mostly consisting of discordant sibships, recruited as part of the Consortium on Alzheimer's Genetics (CAG).11 However, we found no significant association in this sample, and in fact there was a modest overtransmission of the opposite allele from that found associated with the risk for Alzheimer's disease in the NIMH families (table 1). The power to detect odds ratios of approximately 2 at disease allele frequencies of approximately 0.4 (ie, the values suggested for SNP rs7074454 in Martin et al) is >80% in both samples analysed here (using Pedigree‐Based Association Test v3.112; for details on how these power analyses were performed, see Bertram et al13).
Table 1 Association analyses of five single nucleotide polymorphisms in α‐T catenin (VR22) in two independent family samples.
| All families | z score (p) | ||||
|---|---|---|---|---|---|
| rs3750863 | rs4548513 (Ser596Asn) | rs7070570 | rs10996886 | rs2105702 | |
| NIMH (n = 437) | −0.2 (0.9) | −2.0 (0.04) | 0.8 (0.4) | 0.9 (0.4) | 0 (1) |
| CAG (n = 217) | ND | 0 (1) | ND | ND | ND |
| Late‐onset | |||||
| NIMH (n = 320) | 0.1 (0.9) | −2.3 (0.02) | 1.5 (0.1) | 1.6 (0.1) | −0.4 (0.7) |
| CAG (n = 155) | ND | 1.0 (0.3) | ND | ND | ND |
CAG, Consortium on Alzheimer's Genetics; ND, not done; NIMH, National Institute of Mental Health.
Minor allele frequencies in the NIMH (CAG) sample are rs3750863 (A) = 0.30 (ND), rs4548513 (Ser596Asn) (C (Ser)) = 0.36 (0.39), rs7070570 (G) = 0.29 (ND), rs10996886 (A) = 0.44 (ND), rs2105702 (C) = 0.31 (ND). z score for minor allele (positive values indicate overtransmission to affected individuals) using Family‐Based Association Test (V.1.5.5) after selecting an additive genetics model, the empirical variance function and an equal weight offset correction (see FBAT website for more details: http://www.biostat.harvard.edu/∼fbat/).
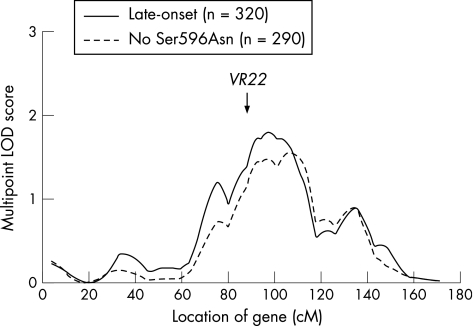
Figure 2 Results of genetic multipoint linkage analyses obtained in the original genome screen using Genehunter‐Plus in 320 NIMH families with Alzheimer's disease where all affected members showed an age of onset ⩾65 years (late onset8). ‘No Ser596Asn' indicates analyses after excluding families (n = 30) in which at least two alleles were transmitted to affected subjects (for details on these analyses, see Betram et al11). Around the location of VR22 (approximately 88 cM) the multipoint logarithm of the odds (LOD) score decreased only slightly from 1.39 to 1.15, suggesting that the Ser596Asn association does not make a major contribution to the overall linkage signal on this chromosome.
Although these results alone neither unequivocally support nor dismiss VR22 as a risk gene for Alzheimer's disease, they markedly extend two lines of evidence accrued in previous studies. First, the strongest and the most consistent association signals in the two published positive studies were observed with variants located in intron 13, a few hundred bases 5′ of exon 14. The Ser596Asn SNP found to be associated in one of our family samples here represents an amino‐acid changing variant in the exon 5′ of this intron (fig 1). On the basis of the current HapMap release (#20/phase II, January 2006; www.hapmap.org14), it is in linkage disequilibrium (D′ = 0.81, r2 = 0.13, p<0.001; fig 1C) with the rs7070570 SNP originally associated with plasma Aβ‐levels by Ertekin‐Taner et al,5 and with rs7074454 found to be associated with the risk for Alzheimer's disease by Martin et al1 (D′ = 0.74, r2 = 0.52, p<0.001; fig 1C), overall suggesting a region of extensive linkage disequilibrium extending from introns 12 to 14 (fig 1B). Thus, although the Ser596Asn SNP is located closer to exon 14, quite possibly the previously observed associations between non‐coding SNPs in intron 13 and risk for Alzheimer's disease or plasma Aβ were actually due to linkage disequilibrium with this previously unknown variant. Secondly, similar to previous studies, we only observed evidence of an association between Alzheimer's disease and VR22 in families with at least two affected first‐degree relatives—that is, those included in the NIMH Study. No association of this SNP was found in the independent CAG family sample. As the CAG probands were ascertained without requiring any additional affected family members, they are similar to the cases in conventional case–control samples, all but one of which also failed to show an association between polymorphisms in VR22 and the risk for Alzheimer's disease. Therefore, it is conceivable that Ser596Asn or other VR22 variants are actually moderate risk factors for Alzheimer's disease, but only (or predominantly) in cases with a family history of the disease. A similar argument has been made for common variants in the α2‐macroglobulin gene on chromosome 12, which also shows a discrepancy between negative results in case–control samples and meta‐analyses, and at least suggestive evidence of association in almost all family samples investigated to date.6 However, in both cases, the positive reports may also represent false‐positive findings, where none of the associated variants in these genes actually make a relevant contribution to the overall risk for Alzheimer's disease.
Nevertheless, we believe that our novel results with the Ser596Asn variant in Alzheimer's disease warrant follow‐up analyses in both family‐based and case–control samples. Of all previously investigated VR22 polymorphisms, this SNP may help to explain the varying association signals observed across earlier reports, and currently seems to be the most probable candidate to exert a relevant functional effect on the pathogenesis of Alzheimer's disease.
Acknowledgements
We thank all families for participating in this study.
Abbreviations
CAG - Consortium on Alzheimer's Genetics
NIMH - National Institute of Mental Health
SNP - single‐nucleotide polymorphism
Footnotes
Funding: This work was sponsored by grants from the NIMH, NIA (ADRC) and the Alzheimer Association. LB is supported by grants from the NIA (1R01 AG023667‐01) and a “Young Investigator Award” from the National Alliance for Research on Schizophrenia and Depression (NARSAD).
Competing interests: None declared.
References
- 1.Martin E R, Bronson P G, Li Y J, Wall N, Chung R H, Schmechel D E, Small G, Xu P T, Bartlett J, Schnetz‐Boutaud N, Haines J L, Gilbert J R, Pericak‐Vance M A. Interaction between the alpha‐T catenin gene (VR22) and APOE in Alzheimer's disease. J Med Genet 200542787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomqvist M E, Andreasen N, Bogdanovic N, Blennow K, Brookes A J, Prince J A. Genetic variation in CTNNA3 encoding alpha‐3 catenin and Alzheimer's disease. Neurosci Lett 2004358220–222. [DOI] [PubMed] [Google Scholar]
- 3.Busby V, Goossens S, Nowotny P, Hamilton G, Smemo S, Harold D, Turic D, Jehu L, Myers A, Womick M, Woo D, Compton D, Doil L M, Tacey K M, Lau K F, Al‐Saraj S, Killick R, Pickering‐Brown S, Moore P, Hollingworth P, Archer N, Foy C, Walter S, Lendon C, Iwatsubo T, Morris J C, Norton J, Mann D, Janssens B, Hardy J, O'Donovan M, Jones L, Williams J, Holmans P, Owen M J, Grupe A, Powell J, van Hengel J, Goate A, Van Roy F, Lovestone S. Alpha‐T‐catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer's disease. Neuromolecular Med 20045133–146. [DOI] [PubMed] [Google Scholar]
- 4.Cellini E, Bagnoli S, Tedde A, Nacmias B, Piacentini S, Sorbi S. Insulin degrading enzyme and alpha‐3 catenin polymorphisms in Italian patients with Alzheimer disease. Alzheimer Dis Assoc Disord 200519246–247. [DOI] [PubMed] [Google Scholar]
- 5.Ertekin‐Taner N, Ronald J, Asahara H, Younkin L, Hella M, Jain S, Gnida E, Younkin S, Fadale D, Ohyagi Y, Singleton A, Scanlin L, de Andrade M, Petersen R, Graff‐Radford N, Hutton M. Fine mapping of the alpha‐T catenin gene to a quantitative trait locus on chromosome 10 in late‐onset Alzheimer's disease pedigrees. Hum Mol Genet 2003123133–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertram L, McQueen M, Mullin K, Blacker D, Tanzi R. The AlzGene Database Alzheimer Research Forum. http://www.alzgene.org (accessed 26 October 2006)
- 7.Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis M G, Go R C, Vekrellis K, Selkoe D J, Saunders A J, Tanzi R E. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science 20002902302–2303. [DOI] [PubMed] [Google Scholar]
- 8.Blacker D, Bertram L, Saunders A J, Moscarillo T J, Albert M S, Wiener H, Perry R T, Collins J S, Harrell L E, Go R C, Mahoney A, Beaty T, Fallin M D, Avramopoulos D, Chase G A, Folstein M F, McInnis M G, Bassett S S, Doheny K J, Pugh E W, Tanzi R E. Results of a high‐resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet 20031223–32. [DOI] [PubMed] [Google Scholar]
- 9.Ertekin‐Taner N, Graff‐Radford N, Younkin L H, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin S G. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late‐onset Alzheimer's disease pedigrees. Science 20002902303–2304. [DOI] [PubMed] [Google Scholar]
- 10.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze F W, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillystone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez‐Tur J, Williams J, Owen M J, Hardy J, Goate A M. Susceptibility locus for Alzheimer's disease on chromosome 10. Science 20002902304–2305. [DOI] [PubMed] [Google Scholar]
- 11.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson A J, Hsiao M Y, Elliott K J, Velicelebi G, Moscarillo T, Hyman B T, Wagner S L, Becker K D, Blacker D, Tanzi R E. Family‐based association between Alzheimer's disease and variants in UBQLN1. N Engl J Med 2005352884–894. [DOI] [PubMed] [Google Scholar]
- 12.Lange C, DeMeo D L, Laird N M. Power and design considerations for a general class of family‐based association tests: quantitative traits. Am J Hum Genet 2002711330–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertram L, Hsiao M, Lange C, Blacker D, Tanzi R E. Single‐nucleotide polymorphism rs498055 on chromosome 10q24 is not associated with Alzheimer disease in two independent family samples. Am J Hum Genet 200679183–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altshuler D, Brooks L D, Chakravarti A, Collins F S, Daly M J, Donnelly P. A haplotype map of the human genome. Nature 20054371299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]