Abstract
Background
Transient neonatal diabetes (TND) is a rare form of diabetes usually present in the first few days after birth that resolves within 1 year but that has a tendency to recur later in life. It can be associated with chromosome 6 paternal uniparental disomy (UPD), paternal duplications or loss of maternal methylation at the 6q24 imprinted locus.
Objective
To report on a cohort of 13 sporadic TND cases, including five with birth defects (congenital abnormalities of heart, brain and bone) and eight without.
Results
The hallmarks of diabetes were similar in patients with or without 6q24 defects. The chromosome 6 abnormalities in our patients (n = 13) included 2 of 13 (approximately 15.4%) cases of paternal UPD6, 2 of 11 (approximately 18%) cases of complete and 3 of 11 (approximately 27%) cases of partial loss of the maternal methylation signature upstream of ZAC1‐HYMAI imprinted genes in non‐UPD cases, and 1 of 13 (approximately 7.7%) cases of hemizygotic deletion.
Conclusion
The deletion was found in a patient with severe congenital abnormalities. This genetic lesion was not reported previously. The hypothesis of an effect on regulatory elements critical for imprinting and tissue‐specific gene expression in early development by the deletion is raised. The data presented here may contribute to the diagnosis and the understanding of imprinting in the region.
Keywords: transient neonatal diabetes, genetics, imprinting, methylation, deletion
Transient neonatal diabetes (TND; MIM 601410) is a developmental disease of insulin production with an incidence of about 1 in 400 000 in Europe. TND is characterised by hyperglycaemia occurring within the initial days after birth, glycosuria, dehydration, severe intrauterine growth retardation and decreased adipose tissue. Patients require insulin treatment, but diabetes usually resolves within 6 months. More than 50% of patients with TND are prone to relapse in adolescence.1 In most (about 90%) TND cases, chromosome 6 defects are involved, including paternal uniparental disomy (UPD) and paternal duplications. About one fifth of the patients have a loss of maternal imprinting marks at 6q24, the differentially methylated region (DMR).2,3,4,5,6 Two overlapping maternally imprinted genes, ZAC1/PLAGL1 (zinc finger apoptosis, cell cycle arrest or pleiomorphic adenoma of the salivary gland gene‐like 1) and HYMAI (hydatidiform mole‐associated and imprinted transcript), are located in the 6q24 region.3,5,7,8ZAC1 encodes a zinc finger transcription factor involved in cell cycle arrest, apoptosis and autocrine control of insulin secretion in pancreatic islets via activation of the type 1 PACAP receptor.7,8HYMAI is an untranslated gene with unknown function.5 Overexpression of neighbouring genes due to the presence of two unmethylated alleles at this locus is expected.
The expression of imprinted genes is regulated in a developmental and tissue‐specific manner. Deregulation of imprinted genes has been implicated in abnormal development, in several growth and behavioural syndromes in humans and in cancer progression. The occurrence of phenotypic abnormalities such as macroglossia, umbilical hernia,9 pancreatic β cell agenesis,10 heart and skeletal defects10,11,12,13,14 that were observed in some TND cases remains poorly explained. We describe the phenotype and a molecular characterisation of a cohort of 13 patients with TND, including five cases with congenital abnormalities, one of which also has a hemizygotic deletion at the 6q24 imprinted locus.
Materials and methods
Patients and DNA samples
In all, 13 patients with TND (five females and eight males) were included in this study. All the patients were of Caucasian origin, except patient P7 from Guyana. Written consent from all participants was obtained in accordance with national ethics rules. Data for human leucocyte antigen typing and genotypes established using 22 chromosome 6 microsatellite markers were reported earlier.14,15 Two patients, P1 and P3, had paternal UPD6; the other patients had biparental transmission of microsatellite markers and no other chromosomal defect.14,15 Table 1 summarises the hallmarks of the disease for patients without and with chromosome 6q24 defects: the mean (standard error of mean (SEM)) birth weight was 2373 (237) v. 2146 (230) g for those without versus those with defects; the mean (SEM) age at diagnosis of diabetes was 11 (6.1) v. 4 (1.96) days and the mean (SEM) duration of insulin treatment was 193 (100.3) v 78 (24) days.
Table 1 Characteristics of patients with transient neonatal diabetes without and with 6q24 defects*.
| Without 6q24 defects (n = 6) | With 6q24 defects (n = 7) | Total (n = 13) | |
|---|---|---|---|
| Birth weight (g) | 2373 (237) (1520–3200) | 2146 (230) (1602–3160) | 2302 (165.4) (1520–3200) |
| Age at the diagnosis of diabetes (days) | 11 (6.1) (1–26) | 4 (1.96) (1–13) | 7.63±2.94 (1–26) |
| Insulin treatment (days) | 193 (100.3) (50–489) | 78 (24) (6–150) | 124.2 (43.37) (6–489) |
| Patients with congenital abnormalities* | 2 (33.3%) | 3 (42.8%) | 5 (38.5%) |
Data are mean (SEM), followed by ranges in parentheses.
*Paternal UPD6, 6q24 methylation loss and hemizygotic deletion.
Five patients presented with developmental defects (approximately 38.5%).14 The main features (table 2) included mitral valve insufficiency (patient P3), interrupted aortic arch, ventricular septal defect, bronchodysplasia (patient P5), bone maturation delay, metaphyseal and epiphyseal dysplasia (patients P3, P5, P7 and P8), and genital defects (patient P11). Muscular hypotonia (patients P3, P5, P7 and P11), seizures (patients P5 and P8) and motor retardation (patients P3 and P11) were also observed.
Table 2 Phenotypical features in patients with transient neonatal diabetes with congenital abnormalities.
| Patients | |||||
|---|---|---|---|---|---|
| P3 | P5 | P7 | P8 | P11 | |
| Features | |||||
| Gestational age (weeks) | 42 | 40 | 38 | 40 | 41 |
| Birth weight (g) | 2800 | 1750 | 1850 | 3200 | 2360 |
| Centile rank* | <5 | <5 | <5 | 50–90 | <5 |
| Age diabetes diagnosed (days) | 2 | 1 | 1 | 26 | 26 |
| Glucose at presentation (mmol/l) | 12.2 | 11.06 | NA | 30.41 | NA |
| Insulin treatment (months) | 2.5 | 4 | 3.5 | 16 | 3.3 |
| Congenital heart defect | +(1) | +(2) Bronchodysplasia | No | No | No |
| Bone defect | +(3) | +(4) | +(3,4) | +(3) | No |
| Genitourinary defect | No | No | No | Left kidney agenesis, right mega urethra | + |
| Brain anomalies | No | No | No | Subcortical and cortical atrophy | No |
| Hypotonia | + | + | +(5) | No | + |
| Motor defects | No | +(6) | No | +(7) | +(8) |
| Other | Major β thalassaemia hypothyroidism | High blood pressure | Anaemia | Anaemia | Hyperthyroidism |
| Chromosome 6 defect | Pat UPD6 | Hemizygotic deletion† | Methylation loss | No | No |
NA, not available; Pat, paternal; UPD, uniparental disomy; (1) mitral valve insufficiency; (2) interrupted aortic arch, interventricular communication (ventricular septal defect), hypoplasia annular aortic; (3) delayed bone maturation; (4) epiphyseal and metaphyseal dysplasia; (5) axial hypotonia at birth, peripheric at 6 months; (6) delayed sitting (11 months); (7) delayed sitting (12 months) and walking (22 months); (8) delayed sitting and walking (21 months).
*Centile of birth weight according to term.
†Parental origin of the deleted allele could not be identified.
Genomic DNA was extracted from peripheral blood samples.15 DNA from non‐affected patients was obtained from the CEPH (Centre d'Etude des Polymorphismes Humains, Paris, France) and from our laboratory collection.
Methylation studies, sodium bisulphite DNA treatment
We treated 1 μg of denatured DNA under mineral oil with 3 mol/l sodium bisulphite/hydroxyquinone (Sigma, Missouri, USA; 3.1 mol/l/0.5 mmol/l final) for 16 h at 50°C in the dark.16 The reaction was desalted with a Wizard clean‐up column (Promega, Wisconsin, USA) suspended in 50 μl water, incubated in 0.3 mol/l sodium hydroxide for 5 min, neutralised and ethanol precipitated. Modified DNA was resuspended in 25 μl of water.
Bisulphite genomic sequencing
Bisulphite‐treated DNA was cloned and sequenced to analyse the methylation pattern of CpG sites at the DMR upstream of the ZAC1/HYMAI genes, and at five other CpG islands (CGi) spanning 450 kb genomic DNA at the TND locus (fig 1A). Bisulphite‐treated DNA was amplified on a 9600 thermal cycler (Applied Biosystems, Foster City, California, USA). Primers and amplification conditions are available in supplementary material at http://jmg.bmjjournals.com/supplemental (table A). Polymerase chain reaction (PCR) products were purified using Gene‐Clean (Bio101 Systems, La Jolla, California, USA) and cloned into the pMOSBlue plasmid (Amersham Biosciences, Buckinghamshire, UK). In all, 10–20 colonies were isolated and insert size was checked by PCR using the U‐19 and T7 primers. The correct PCR amplicons were purified on Sephadex G50 fine (Bio‐Rad, Hercules, California, USA), sequenced on an ABI 3100 sequencer and analysed using ABI PRISM sequencing software. At least 20 clones were sequenced for each individual, two independent bisulphite treatments were done for each DNA sample and two independent PCR amplifications were done for each patient.
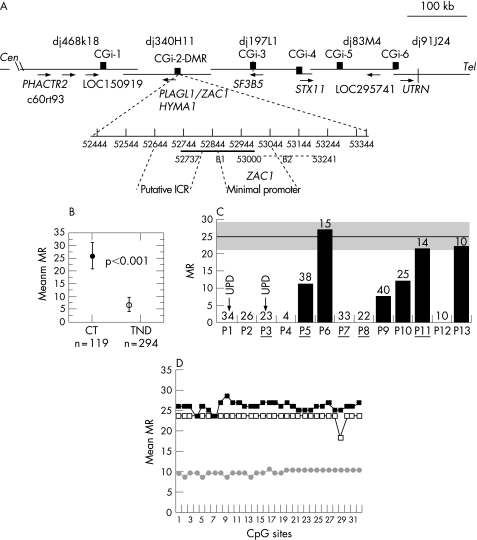
Figure 1 Schematic representation of the 6q24.2 transient neonatal diabetes (TND) locus and methylation analysis of 6q24 differentially methylated region (DMR) in controls and in patients with TND. (A) PAC contig, gene order and direction of transcription (arrows), compiled from http://www.Sanger.ensembl.org,http://www.ncbi.nlm.nih.gov/Entrez/. Black boxes show the physical location of CpG islands (CGi). Positions in GenBank accessions: CGi‐1, 24852–25585 in AL049844; CGi‐2 (DMR) 52444–53375, B1 52737–53000, B2 52976–53241 in AL109755; CGi‐3, 50830–51669 in AL031390; CGi‐4, 3282–4229 in AL512354; CGi‐5, 35362–36232 in AL135917; CGi‐6, 29334–31687 in AL024474. CGi‐2DMR (931 bp) with corresponding positions in AL109755. The ZAC1 transcription start sites are 52907 and 52934,8 and the LOT1 transcription start sites are 52919 and 53080. ICR is the putative imprinting control region5 and ZAC1 the minimal promoter region at which methylation down regulates ZAC1 expression.8 The DMR regions previously analysed are (53085–53362), (52734–53157) and (52977–53241).3,5,6 (B) Overall methylation at the DMR B1 region: methylation ratio (MR) values are shown for control (CT) DNAs (n = 119 clones) and patients with TND (n = 294 clones). p, Fisher's exact test. (C) MRs for individual patients. The arrows indicate patients with paternal uniparental disomy (UPD)6. The number of clones sequenced for each patient is indicated above the bar. The grey area corresponds to the range of control MR values: mean (SEM) MR = 25.9 (5). (D) Quantitative CpG methylation patterns at each CpG site, MR values for pooled clones (Materials and methods), (▪) controls (n = 119), (□) patients with a normal methylation range (n = 38), (•) patients with a partial methylation loss (n = 103); n is the number of sequences analysed.
To avoid biased amplification of methylated alleles and estimation of one allele type, we designed primers with no CpG dinucleotides and used an optimal amplicon size and sequenced both DNA strands. The methylation ratio (MR) was assessed for each sample. The methylation ratio is calculated as
(methylated allele or CpG sites/total alleles or total CpG sites counted)×100.
The methylation ratio was used to calculate the standard deviation (SD) and standard error of the mean (SEM) of each sample. The samples were considered significantly different if p<0.05 (Fisher's exact test done using StatView software V.5.0).
Another method was also used to analyse methylation. Bisulphite treatment was followed by duplex PCR of methylated and unmethylated alleles using specific primers (340B1M, 340B1UM) with a common universal reverse primer (340B1R; primer sequences are listed in the supplementary material at http://jmg.bmjjournals.com/supplemental). PCR products were analysed using silver‐stained acrylamide gels (Amersham Biosciences, UK) and signal intensity was quantified using V.1.63 of SCION software (NIH, Bethesda, Maryland, USA).
Determination of gene copy number
A semiquantitative method was used to determine gene copy number, based on duplex PCR amplification of five loci mapping to 6q24 (TIGR‐A003N06), SGC33367 (PHACTR2 gene), stSG9563 (PLAGL1 gene), stSG300079 and SHC‐37610 (in PAC dj91J24), each coamplified with the reference locus stSG51062 on chromosome 8 (fig 3B). Primer sequences were from the National Center for Biotechnology Information database, and the forward primer of each set was labelled with the fluorophore 6‐fluorouracil, doxorubicin, mitomycin C. Duplex PCR was carried out on four DNA concentrations: 150–18.75 ng in a final volume of 15 μl with an optimal combination of primer concentration and magnesium chloride (1–2.5 mmol/l), 2.5 mmol/l deoxyribonucleotide triphosphate and 1 U of Taq Platinum (Invitrogen, Paisley, UK). PCR was done in a Stratagene 96 gradient thermocycler (94°C for 5 min; 94°C for 30 s, 47–58°C for 30 s and 72°C for 30 s for 21 cycles). PCR products were mixed with a size standard (ABI GS500ROX), loaded onto a 6% denaturing acrylamide gel and run on an ABI 377 automated sequencer. Data were analysed using Genotyper V.1.1 software (Applied Biosystems, USA). Three control DNA samples were used for each PCR, and two independent experiments were carried out in duplicate for each DNA sample.
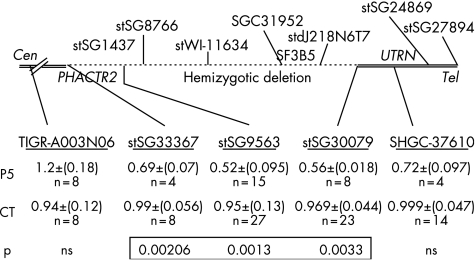
Figure 3 Results of gene copy number determination at the 6q24 transient neonatal diabetes (TND) locus in patient P5 and controls (CT) and location of the hemizygous deletion. Location of the anonymous markers and STS loci used for determination of gene copy number are underlined. Mean (SEM) values of ratios H1:H2 for patient P5 and for control DNA. H1 values represent peak height of 6q24 loci and H2 the peak height values of reference locus stSG51062 on chromosome 8. The p values of unpaired Student t‐test of H1:H2 ratios of patients versus H1/H2 ratios of controls were significant for three loci. The loci with no significant difference from controls (ns) are indicated. n, number of measurements.
Quantitative real‐time PCR with the SYBR‐Green dye method was also used for the ZAC1 gene locus (stSG9563) and a reference marker on chromosome 8 (stSG51 062). Five dilutions of genomic DNA (1 to 0.005 ng/μl) from patients and controls were used. We used the SYBR‐Green PCR Core Reagent Kit (95°C for 10 min, 35 cycles of 95°C for 15 s and 60°C for 1 min, Applied‐Biosystems). The ABI PRISM 7700 sequence detection system (Perkin Elmer ABI, Wellesley, Massachusetts, USA) was used for quantification by the cycle threshold (ct) method with ABI PRISM sequence detection software. Samples were normalised on the basis of the reference for chromosome 8 (stSG51 062). Gene doses were determined by the following ratio:
(copy number of target gene (ZAC1)/copy number of reference genomic region (StSG51 062)) in patients divided by (ZAC1 copy number/reference locus copy number) in control DNA
Normalised gene doses were determined for each dilution of DNA, and the corresponding means and standard deviation were calculated. A non‐deleted DNA sequence should give a ratio of 1, a heterozygotic deleted DNA should give a ratio of 0.5 and duplicated DNA should give a ratio of 1.5.17 Fisher's exact test was carried out using StatView software V.5.0 for statistical analysis.
Results
DNA methylation imprinting signature
Figure 1A shows the organisation of the 6q24.2 TND locus and the location of the CpG islands analysed. Cloned PCR products from amplified bisulphite modified DNA were sequenced to determine the methylated cytosine residues that were not converted to uracil in the DMR B1 region (fig 1a). We sequenced 294 clones from patients with TND and 119 from controls. A significant (p<0.001) overall reduction of the methylation ratio was found in TND patients (TND mean (SEM) methylation ratio 6.9 (2.83) v control mean (SEM) methylation ratio 26.9 (5); fig 1B). Quantitative analysis of methylation for individual patients showed that patients P1 and P3 with paternal UPD6 had the expected paternal unmethylated status (MR = 0; fig 1C). Among the eleven cases with biparental chromosome 6 inheritance, five patients (P2, P4, P7, P8 and P12) had complete methylation loss (MR = 0). Patients P5 (MR = 12.5), P9 (MR = 7.5) and P10 (MR = 12) had partial methylation loss. The methylation ratio values of three patients, P6 (27), P11 (21.4), P13 (22.2), were within the control methylation ratio range. The bisulphite sequencing method measures methylation at each CpG site, allowing quantitative comparison of DMR methylation patterns in patients and controls. The mean methylation ratio at each CpG site was determined for pooled sequences, as described in Materials and methods. Most DNA strands (clones) were present in an all‐or‐none manner in controls and in patients without UPD for the 31 CpG sites in the DMR B1 segment (fig 1D). Comparison of pooled sequences from patients with partial methylation loss (n = 103) or patients with normal methylation (n = 65) and controls (n = 119) did not show significant mean methylation ratio differences across CpG sites, suggesting no sporadic methylation in patients.
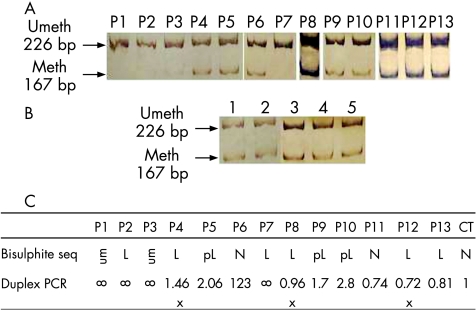
Duplex PCR amplification confirmed the results obtained by bisulphite sequencing. In patients P1, P2, P3 and P7 maternal methylation was not detected, whereas the other patients and controls (fig 2A,B) had both methylated and unmethylated alleles. The ratios of unmethylated/methylated PCR band signal intensities were ∞ in patients with complete methylation loss; 2.06, 1.7 and 1.8 for patients with partial methylation loss (P5, P9 and P10); 1.23, 0.74 and 0.81 for patients with normal methylation (P6, P11 and P13); and 1 for the controls (fig 2C). Three patients (P4, P8 and P12) had contradictory results, showing methylation loss based on bisulphite sequencing but no loss based on duplex PCR. This discrepancy may have been because of the small number of sequences analysed or because of the different sensitivity of methods used. We found normal methylation ratio values at the adjacent B2 DMR, based on bisulphite sequencing. We therefore considered these patients (P4, P8 and P12) to be normally methylated. We used bisulphite‐sequencing to verify that the imprint at the conserved B1 DMR region in mouse blood is stable with age, with methylation ratios of approximately 30, as in humans (not shown).
Figure 2 Results of semiquantitative determination of differentially methylated region methylation by duplex polymerase chain reaction (PCR) in (A) patients and (B) controls. Methylated (meth, 167 bp) and unmethylated (umeth, 226 bp) alleles. (C) Comparison of methylation results obtained by bisulphite sequencing and duplex PCR. L, methylation loss; pL, partial methylation loss; x, contradictory results resolved by finding normal methylation in the B2 region.
Methylation at non‐DMR CpG islands
We looked for methylation defects at other CpG islands in the region, and methylation ratios at centromeric CGi‐1 and distal CGi 3–6 were quantified by the bisulphite sequencing method for a total of 115 independent CpG sites (fig 1A). There was no detectable difference in methylation between patients and controls (results in supplementary material table B available at http://www.jmg.bmjjournals.com/supplemental). The CGi located in the promoter region of the genes PHACTR2 (CGi‐1), STX11 (CGi‐4) and UTRN (CGi‐6), and the intergenic CGi‐3 had no methylated sites in any of the clones sequenced in either controls or patients. CGi‐5 in exon 2 of the STX11 gene had extensive methylation in all clones, with few unmethylated CpG sites randomly distributed in controls and in all patients. Methylated and unmethylated clones were not detected, indicating that the five non‐DMR CGi analysed contain no imprinting marks in blood.
Genomic copy number
We determined the allele copy number to validate methylation findings in patients, as the region analysed for methylation lies between two microsatellite markers. A duplex semiquantitative gene dosage assay was first used at five loci mapping to 6q24 (fig 3) for patients with biparental inheritance of chromosome 6 microsatellite markers. All patients except one had ratios (mean of three independent experiments) within the normal values expected for two gene copies, ranging from 0.94 to 1.11 as in controls at all loci analysed. Patient P5 had reproducible and significant mean (SD) ratios of 0.69 (0.074) at SGC33367 (p = 0.002), 0.52 (0.095) at stSG9563 (p = 0.001) and 0.56 (0.02) at stSG30079 (p = 0.003), consistent with a heterozygotic microdeletion delimited by stSG33367 and stSG‐30079, spanning at least 365 kb. The centromeric D6S1684 and D6S1699 and the telomeric D6S1703 (EPM2A) microsatellite markers were heterozygotic. The hemizygosity was confirmed by quantitative PCR using the SYBR Green technology with a mean (SD) ratio of 0.564 (0.132) for the ZAC1 locus (stSG9563) versus the reference locus on chromosome 8, and also with alternative reference locus on chromosome 16p12 (DOK4‐2); mean ratio P5: 0.66 v 1.16 for control DNA (two independent experiments). No deletion was found in 25 control DNAs, and the mean (SD) ratio was 1.14 (0.175).
Discussion
We analysed allelic dosage and methylation in 13 patients with TND. We found altered maternal methylation in the presence of chromosome 6 biparental inheritance; 2 of 11 patients (about 18%) had complete methylation loss and 3 of 11 patients (about 27%) had partial methylation loss. The five non‐DMR CpG islands in the region had similar methylation patterns in patients and controls. No difference in birth weight was noted in patients with TND with or without chromosome 6 defects; hallmarks of diabetes were similar to those in the literature.18 However, patients with no chromosome 6 defects had insulin treatment for a longer time than patients without. Imprinting abnormalities at 6q24 affecting about 20% of TND cases4,5,6 may occur in the maternal germline at imprinting erasure establishment, during imprinting maintenance5,19 processes. These anomalies may in turn concern other imprinted loci.18,20 Otherwise, the partial methylation loss observed in three patients may represent mosaicism after a zygotic event.18,19
We detected allele loss at 6q24 STS loci and thus identified a hemizygotic deletion in one patient of at least 365 kb encompassing the genes PHACTR2, ZAC1‐PLAGL1, HYMAI, SF3b5 and STX11. To our knowledge, this is the first patient with TND with severe congenital anomalies described to have a 6q24 deletion. The deleted region, as STS reference loci, was not prone to copy number polymorphism21 or chromosomal imbalance (http://www.sanger.uk). The deletion was confirmed by two methods: it was not found in either 25 controls or in 100 patients and controls from previous reports,4,5,6 and no heterozygotic polymorphisms were found in the ZAC1 promoter and coding sequence. This patient had classical TND; insulin treatment for hyperglycaemia was initiated on the first day after birth and discontinued at four months. He had severe congenital heart defects, bronchodysplasia, delayed bone maturation, and died at 6 years of age due to heart failure. A 4.5‐Mb 6q24 de novo interstitial paternal deletion with a distal breakpoint at the GRM1 locus harboring gene copy number variations21 was associated with intrauterine growth retardation and mild phenotypic abnormalities.22 Different regulatory elements in these regions with opposite effects could be involved. This contrasts with Zac1 transgenic or knockout studies in mice showing that paternal transmission, not maternal, has phenotypic consequences.23,24
The parental origin of the deleted allele could not be determined. For a constitutional deletion, we should expect either fully methylated or fully unmethylated alleles. Although DNA from tissues other than blood were not available, the possibility of mosaicism with deletion in blood lineages and two copies in other tissues cannot be excluded. The partial methylation loss and reduced number of methylated alleles observed in patient P5 may support this hypothesis. The parents were healthy and there was no history of diabetes in the family. This would reconcile with the wide acceptation of increased 6q24 gene dosage as a cause of TND.2,3,4,5,6
Developmental abnormalities, not previously highlighted, occurred in five unrelated patients in whom sequencing of the ZAC1 gene had not showed any mutation (Diatloff‐Zito unpublished results). The delayed bone maturation, epiphyseal dysplasia and cardiac defects of the patient with the 6q24 deletion were also observed in patients with paternal UPD6 (P3) or with methylation loss (P7). The nature of the defects (heart and bone) suggested that the events responsible may have arisen at early stages of development. Congenital anomalies associated with TND include umbilical hernia and macroglossia,9,11,13 pancreatic β cell agenesis,10 ventricular septal defect,13 cardiomegaly12 and dysmorphy.14ZAC1 and HYMAI genes are expressed in fetal brain, heart, kidney, muscle and lung.3 Expression patterns at early stages of embryonic development in mice3,5 of Zac1 throughout the neural tube,25 in heart walls of the outflow tract, atria and ventricles,26 in myocardium and great vessels,25 and in progenitor cells involved in chondrogenic differentiation27 imply tightly regulated switches. It can be proposed that the observed deletion at 6q24 imprinted region would affect regulatory elements critical for execution of the developmental expression programme. As in the model of pseudo hypoparathyroidism (PHP‐Ib), at 20q GNAS imprinted a locus at which a deletion distant from the ICE can impact methylation status in the involved locus.28
Key points
The combination of congenital abnormalities of heart, brain and delayed bone maturation is observed in four patients with transient neonatal diabetes.
A unique case with hemizygous deletion at the 6q24 imprinted region and severe congenital abnormalities raises the hypothesis of a possible effect on a control element for gene expression at early developmental stages.
Moderately activating mutations of the KATP channel (KCNJ11 gene), which account for approximately 8% of all TND,29 were not found in patients without chromosome 6 defects and in patients with neurological manifestations (hypotonia, motor delay). SUR1 activating mutations giving neurological complications30 should be screened. Other non‐6q24 aetiology could be present in the two patients in whom no chromosome 6 anomaly was found (mutations in GCK, PDX1, NeuroD1‐beta2, EIF2aK3 and ZAC1 genes were excluded).
The abnormalities associated with TND in 13 patients included paternal UPD6 (2/13, or approximately 15.4%), loss of the maternal methylation signature in the ZAC1/HYMAI DMR in about 45.4% of non‐UPD cases and hemizygotic deletion in one patient (1/13, or about 7.7%). The combination of congenital abnormalities of heart, brain and delayed bone maturation observed in four patients could be partly explained by epigenetic deregulation affecting signal(s) for temporal or tissue‐specific gene expression dependent on the parent of origin at the 6q24 ZAC‐HYMAI locus. One patient with severe congenital abnormalities had a hemizygotic deletion in the 6q24 region. This defect had not been detected previously in TND, raising a new hypothesis for imprinting of the region. Our findings extend the phenotypic spectrum of TND, and may contribute to a better understanding of the mechanism of imprinting at the 6q24.2 region and the pathogenic consequences of its deregulation.
Acknowledgements
We thank the patients and their relatives who provided samples for our analysis, and the doctors who referred their patients. We thank Pr Claudine Junien for her support, Dr Catherine Turleau for critical reading of the manuscript, Dr Laurence Colleaux for providing DOK4 reference and the Centre National de la Recherche Scientifique (CNRS), the Centre National d'Etude du Polymorphisme Humain (CEPH), Paris, France.
Abbreviations
CGi - CpG islands
DMR - differentially methylated region
ICE - imprinting control element
PCR - polymerase chain reaction
TND - transient neonatal diabetes
UPD - uniparental disomy
Footnotes
Funding: This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (Inserm) and the Association Aide aux Jeunes Diabètiques (AJD).
Competing interests: None.
References
- 1.Shield J P, Temple I K, Sabin M, Mackay D, Robinson D O, Betts P R, Carson D J, Cavé H, Chevenne D, Polak M. An assessment of pancreatic endocrine function and insulin sensitivity in patients with transient neonatal diabetes in remission. Arch Dis Child Fetal Neonatal Ed 200489(4)341–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temple I K, Gardner R J, Robinson D O, Kibirige M S, Fergusson A W, Baum J D, Barber J C K, James R S J, Shield J P H. Further evidence for an imprinted gene for neonatal diabetes localized to chromosome 6q22‐q23. Hum Mol Genet 19965(8)1117–1121. [DOI] [PubMed] [Google Scholar]
- 3.Kamiya M, Judson H, Okazaki Y, Kusakabe M, Muramatsu M, Takada S, Takagi N, Arima T, Wake N. The cell cycle control gene ZAC/PLAGL1 is imprinted a strong candidate gene for transient neonatal diabetes. Hum Mol Genet 20009(3)453–460. [DOI] [PubMed] [Google Scholar]
- 4.Gardner R J, Mackay D J G, Mungall A J, Polychronakos C, Siebert R, Shield J P H, Temple I K, Robinson D O. An imprinted locus associated with transient neonatal diabetes mellitus. Hum Mol Genet 20009589–596. [DOI] [PubMed] [Google Scholar]
- 5.Arima T, Drewell R A, Arney K L, Inoue J, Makita Y, Hata A, Oshimura M, Wake N, Surani A. A conserved imprinting control region at HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum Mol Genet 200110(14)1475–1483. [DOI] [PubMed] [Google Scholar]
- 6.Mackay D J G, Temple I K, Shield J P, Robinson D O. Bisulphite sequencing of the transient neonatal diabetes mellitus DMR facilitates a novel diagnostic test but reveals no methylation anomalies in patients of unknown aetiology. Hum Genet 2005116255–261. [DOI] [PubMed] [Google Scholar]
- 7.Spengler D, Villalba M, Hoffman A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J 1997162814–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varrault A, Bilanges B, Mackay D J G, Basyuk E, Ahr B, Fernandez C, Robinson D O, Bockaert J, Journot L. Characterization of the methylation‐sensitive promoter of the imprinted ZAC gene supports its role in transient neonatal diabetes mellitus. J Biol Chem 200127618653–18656. [DOI] [PubMed] [Google Scholar]
- 9.Dacou‐Voutetakis C, Anagnostakis D, Xanthou M. Macroglossia, transient neonatal diabetes mellitus and intrauterine growth failure: a new distinct entity? Pediatrics 197555(1)127–130. [PubMed] [Google Scholar]
- 10.Abramowicz M J, Andrien M, Dupont E, Dorchy H, Parma J, Duprez L, Ledley F D, Courtens W, Vamos E. Isodisomy of chromosome 6 in a newborn with methylmalonic acidemia and agenesis of pancreatic beta cells causing diabetes mellitus. J Clin Invest 199494(1)418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Battin M, Yong C, Phang M, Daaboul J. Transient neonatal diabetes mellitus and macroglossia. J Perinatol 199616(4)288–291. [PubMed] [Google Scholar]
- 12.Zneimer S M, Ziel B, Bachman R. Partial trisomy of chromosome 6q: An interstitial duplication of the long arm. Am J Med Genet 199880(2)133–135. [PubMed] [Google Scholar]
- 13.Valerio G, Franzese A, Palmieri A, Mackay D J, Gardner R J, Temple I K. Central precocious puberty in a girl with triple X syndrome and neonatal diabetes mellitus associated with paternal isodisomy of chromosome 6. J Pediatr Endocrinol Metab 200114(7)897–900. [DOI] [PubMed] [Google Scholar]
- 14.Marquis E, Robert J J, Bouvattier C, Bellanne‐Chantelot C, Junien C, Diatloff‐Zito C. Major difference in aetiology and phenotypic abnormalities between transient and permanent neonatal diabetes. J Med Genet 200239(5)370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marquis E, Robert J J, Benezech C, Junien C, Diatloff‐Zito C. Variable features of transient neonatal diabetes mellitus with paternal isodisomy of chromosome 6. Eur J Hum Genet 20008137–140. [DOI] [PubMed] [Google Scholar]
- 16.Clark S J, Harrisson J, Paul C L, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 1994222990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurendeau I, Bahuau M, Vodovar N, Larramendy C, Olovi M, Bieche I, Vidaud M, Vidaud D. TaqMan PCR‐based gene dosage assay for predictive testing in individuals from a cancer family with INK4 locus haploinsufficiency. Clin Chem 199945982–986. [PubMed] [Google Scholar]
- 18.Mackay D J G, Hahnemann J M D, Boonen S E, Poerksen S, Bunyan D J, White H E, Durston VJ T N, Robinson D O, Shield J P H, Clayton‐Smith J, Temple K. Epimutation of the TNDM locus and the Beckwith‐Wiedemann syndrome centromeric locus in individuals with transient neonatal diabetes mellitus. Hum Genet 2006119179–184. [DOI] [PubMed] [Google Scholar]
- 19.Mann M R, Bartolomei M S. Maintaining imprinting. Nat Genet 200025(1)74–78. [DOI] [PubMed] [Google Scholar]
- 20.Arima T, Kamikihara T, Hayashida T, Kato K, Inoue T, Shirayoshi Y, Ishimura M, Soejima H, Mukai T, Wake N.ZAC, LIT1 (KCNQ1OT1) and p57KIP2(CDKN1C) are in an imprinted gene network that may play a role in Beckwith‐Wiedemann syndrome. Nucleic Acids Res 2005332650–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iafrate A J, Feuk L, Rivera M N, Listewnik M L, Donahoe P, Qi Y, Scherer S W, Lee C. Detection of large‐scale variation in the human genome. Nat Genet 200436949–951. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Cassidy S B, Romero L, Schwartz S. Molecular cytogenetics of e de novo interstitial deletion of chromosome arm 6q in a developmentally normal girl. Am J Med Genet 199986227–231. [DOI] [PubMed] [Google Scholar]
- 23.Ma D, Shield J P H, Dean w, Leclerc I, Knauf C, Burcelin R, Rutter G A, Kelsey G. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest 2004114339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan T, Hagan J P, Kozlov S V, Stewart C L, Muegge K. Lsh controls silencing of the imprinted Cdkn1c gene. Development 2005132(4)635–644. [DOI] [PubMed] [Google Scholar]
- 25.Tsuda T, Markova D, Wang H, Evangelisti L, Pan T C, Chu M L. Zinc finger protein Zac1 is expressed in chondrogenic sites of the mouse. Dev Dyn 2004229340–348. [DOI] [PubMed] [Google Scholar]
- 26.Alam S, Zinyk D, Ma L, Schuurmans C. Members of the Plag gene family are expressed in complementary and overlapping regions in the developping murine nervous system. Dev Dyn 2005234772–782. [DOI] [PubMed] [Google Scholar]
- 27.Valente T, Junyent F, Auladell C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1‐expressing cells during development. Dev Dyn 2005233667–679. [DOI] [PubMed] [Google Scholar]
- 28.Bastepe M, Fröhlich L F, Hendy G N, Indridason O S, Josse R G, Koshiyama H, Korkko J, Nakamoto J M, Rosenbloom A L, Slyper A H, Sugimoto T, Tsatsoulis A, Crawford J D, Jüppner H. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest 20031121255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gloyn A L, Reimann F, Girard C, Edghill E L, Proks P, Pearson E R, IK T. Mackay D JG, Shield J PH, Freedenberg D, Noyes K, Ellard S, Ashcroft F M, Gribble F M, Hattersley A T. Relapsing diabetes can result from moderately activating mutations in KCNJ11. Hum Mol Genet 200514925–934. [DOI] [PubMed] [Google Scholar]
- 30.Proks P, Arnold A L, Bruining J, Girard C, Flanagan S E, Larkin B, Colclough K, Hattersley A T, Ashcroft F M, Ellard S. A heterozygous activating mutation in the sulphonylurea receptor SUR1 (ABCC8) causes neonatal diabetes. Hum Mol Genet 200615(11)1793–1800. [DOI] [PubMed] [Google Scholar]