Abstract
Background
Pronounced intrafamilial variability is unusual in Menkes disease and its variants. We report two unrelated families featuring affected members with unusually disparate clinical and biochemical phenotypes and explore the underlying molecular mechanisms.
Methods
We measured biochemical markers of impaired copper transport in five patients from two unrelated families and used RNase protection, quantitative reverse transcription (RT)‐PCR, Western blot analysis and yeast complementation studies to characterise two ATP7A missense mutations, A1362D and S637L.
Results
In two brothers (family A) with A1362D, RNase protection and Western blot analyses revealed higher amounts of ATP7A transcript and protein in the older, mildly affected patient, who also had a higher plasma copper level and lower cerebrospinal fluid dihydroxyphenylalanine : dihydroxyphenylglycol ratio. These findings indicate greater gastrointestinal absorption of copper and higher activity of dopamine‐β‐hydroxylase, a copper‐dependent enzyme, respectively. In family B, three males with a missense mutation (S637L) in an exon 8 splicing enhancer showed equally reduced amounts of ATP7A transcript and protein by quantitative RT‐PCR and western blot analysis, respectively, despite a more severe phenotype in the youngest. This patient's medical history was notable for cardiac arrest as a neonate, to which we attribute his more severe neurodevelopmental outcome.
Conclusions
These families illustrate that genetic and non‐genetic mechanisms may underlie intrafamilial variability in Menkes disease and its variants.
Keywords: Menkes disease; intrafamilial variation; gene expression, ATP7A; residual copper transport
Menkes disease (MD) is an X‐linked recessive neurodegenerative disorder caused by mutations in ATP7A, a copper‐transport gene.1,2,3 Patients with this disorder are unable to adequately absorb and use copper, a micronutrient required for energy metabolism, catecholamine biosynthesis and connective tissue formation. In the classic form of MD, patients have severe neurodegeneration, leading to death by 3 years of age.4 They develop little or no motor skills, do not acquire language skills and many experience seizures.5 Affected individuals also manifest kinky, steel‐coloured hair and tortuous blood vessels. Biochemical markers of the disease include low serum levels of copper and caeruloplasmin4 and altered cerebrospinal fluid catechol levels.6
Complete loss‐of‐function mutations in ATP7A result in MD, whereas less severe defects produce the allelic disorder occipital horn syndrome (OHS).7 The latter phenotype involves mainly connective tissue problems, including skin and joint laxity, tortuous blood vessels and hernias. These individuals may also have dysautonomia, related to reduced activity of dopamine‐β‐hydroxylase, a copper‐dependent enzyme. However, they show normal or only mildly delayed cognitive abilities, rather than the hallmark neurodegeneration associated with MD. Some patients with OHS have mutations that result in aberrant splicing of ATP7A mRNA and substantially reduced levels of normal transcript.7,8
Profound intrafamilial variation is rare in MD and OHS,4 and when reported, the precise cause has not been clear.7,9 In this paper, we report on two families in which substantial intrafamilial variation is evident and investigate potential molecular and clinical explanations for this phenomenon.
Methods
Patients
The study was approved by the institutional review boards of the National Institute of Child Health and Human Development and the National Institute of Neurological Disorders and Stroke. Informed consent was obtained from the patients' parents.
Family A
Family A consists of two brothers (figure 1A, B), current ages 19 (A‐1) and (A‐2) 13 years, who reside in Trinidad. Patient A‐1 walked at 2 years of age and attended normal schools. He suffers from chronic diarrhoea and occasional dizziness upon rising. With achievement of major motor and cognitive milestones, the phenotype of A‐1 is consistent with OHS. In contrast, his younger brother (A‐2) shows a phenotype intermediate between classic, severe and mild MD. He shows profound neurodevelopmental delay; he is unable to sit, roll, stand or walk and developed seizures at 6 years of age.
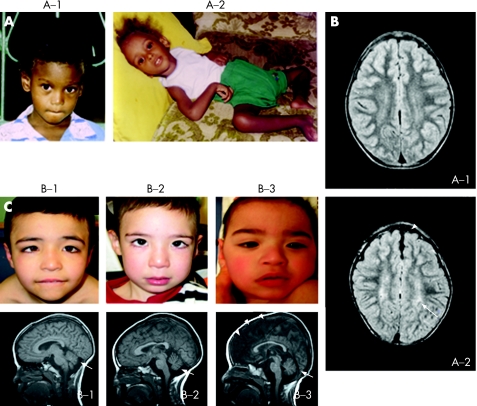
Figure 1 Intrafamilial variability in clinical phenotypes. (A) Two brothers in family A with A1362D mutation in ATP7A at 9 (A‐1) and 4 years (A‐2). Note hair depigmentation in both, and profound hypotonia in A‐2. (B) Axial flair brain MRI scans in family A show normal brain volume and myelination in patient A‐1. In patient A‐2, cerebral cortical atrophy (arrowhead) and delayed myelination (increased white matter signal, arrow) are present. (C) Ocular and cerebellar abnormalities in three siblings from family B with S637L mutation. Patients B‐1 (8 years old), B‐2 (4 years) and B‐3 (2 years) all exhibit ocular strabismus (top panels). Patient B‐3 is cortically blind. Cerebellar hypoplasia (arrows) is evident on T1‐weighted MRI scans of the brain in all three brothers (bottom panels). Patient B‐3 also shows prominent cerebral degeneration (arrowheads). Parental/guardian informed consent was obtained for publication of this figure.
Family B
Family B consists of three brothers (figure 1C) of Hispanic descent. Patient B‐1 was born full‐term and weighed 3.4 kg. He is currently 8 years old and has no history of seizures or other medical problems. He did not walk independently until 3.5 years of age. His speech is currently limited to two or three words, although his overall cognitive level is estimated as equivalent to 4 years of age. Patient B‐2 was born at 7.5 months gestation and weighed 3 kg. Tetralogy of Fallot was noted at birth and repaired at 1 year of age. He is currently 4 years old and has no history of other medical problems, including seizures. He walked at 3 years of age and has very limited speech. Patient B‐3 was born at 7.5 months gestation and weighed <2.3 kg. He had jaundice, episodes of apnoea and a cardiac arrest during the neonatal period. Seizures were noted at 6 months of age. At his current age (2 years) he is able to roll from back to front but exhibits no independent crawling, sitting, or walking and has no functional speech or vision. He receives phenobarbital and topiramate for seizures and is fed via a gastrostomy tube that was inserted at 18 months of age.
By virtue of achieving ambulation, a major motor milestone, patients B‐1 and B‐2 are considered to have mild MD, whereas B‐3 shows the classical MD phenotype.4
Cerebrospinal fluid catechols
Cerebrospinal fluid catechol levels were measured as described previously.6
Mutation analysis
Mutation analysis in family A was performed as described previously,7 and by automated DNA sequencing of ATP7A exon 8 in family B.
Cell culture and cell lines
Patient and normal control lymphoblast and fibroblast cell lines were cultured as described previously.7 Two normal control lymphoblast cell lines were used: GM32017 and another that was established from a healthy man. We used two normal control fibroblast cell lines: GM5659 (Coriell Institute Cell Repository, Camden, New Jersey, USA) and one that was established from a young woman. Fibroblast cell lines used were at or before passage 7.
RNase protection assays
High specific activity (32P‐UTP) antisense riboprobes were generated by in vitro transcription using a 698 bp Menkes cDNA fragment or a β‐actin fragment, as described previously.7
Isolation of microsomal cell membranes
Microsomal membranes were isolated from cultured fibroblasts and lymphocytes. For fibroblasts, 1.0×105 cells were plated in 150 mm diameter tissue culture dishes with Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 100 U/ml penicillin and streptomycin at 37°C in 5% CO2, and grown until confluent. Cell lysis buffer (2 ml) containing protease inhibitor cocktail (Pierce Biotechnology Inc., Rockford, Illinois, USA) was added directly to the plates after washing the cells with sterile phosphate‐buffered saline (PBS). The cells were then homogenised with a Dounce homogeniser. The homogenate was centrifuged at 10 000 g for 5 min at 4°C to remove cell debris and nuclear fractions. The supernatant was re‐centrifuged to isolate microsomal proteins at 100 000 g for 60 min. The microsomal pellet was then suspended in lysis buffer with 5% sodium dodecyl sulphate. Protein concentration was determined using a microplate protein assay (Bio‐Rad, Hercules, California, USA).
For lymphocytes, 100 ml of cell culture was centrifuged at 200 g for 10 min to obtain cell pellets weighing approximately 2 mg. The pellets were washed with PBS, homogenised and microsomal fractions prepared, according to the procedures described above.
Western blot analysis
Microsomal lysates were denatured by boiling for 5 min in 5× loading buffer (Quality Biological Inc., Gaithersburg, Maryland, USA), separated using Tris‐glycerine sodium dodecyl sulphate polyacrylamide gel electrophoresis (4–12% Novex; Invitrogen Corp., Carlsbad, California, USA) and transferred to polyvinylidine fluoride membranes. Membranes were incubated at 4°C overnight in blocking buffer (0.9% NaCl, 20 mmol Tris‐HCl pH 7.5, 0.5% SDS, 0.1% Tween 20, Tris‐buffered saline (TBS)) containing 5% non‐fat milk powder. Blots were washed three times for 5 min each with TBS, then incubated for 3 hrs with a 1:1000 dilution of a rabbit anti‐ATP7A antibody raised to the carboxyterminal 18 amino acids (NH2‐DKHSLLVGDFREDDDTAL‐OH) of human ATP7A (Antibody Solutions, Palo Alto, CA, USA). After washing, membranes were incubated with anti‐rabbit IgG horseradish peroxidase conjugate (1:2000, Santa Cruz Biotechnology, Santa Cruz, California, USA) for 1 h at room temperature, washed and developed (SuperSignal West Pico Luminol/Enhancer Solution; Pierce) according to the manufacturer's instructions.
After membrane stripping, β‐actin was detected by incubation with a primary mouse anti‐β‐actin monoclonal antibody conjugated with horseradish peroxidase (Santa Cruz Biotechnology) and developed using an enhanced chemiluminescent reagent, as above.
Densitometry
Western blot results were scanned to create digital greyscale images and imported into ImageJ software (http://rsbweb.nih.gov/ij/download.html) for densitometric analysis. ATP7A levels were normalised to β‐actin.
Yeast complementation and timed growth assays
Site‐directed mutagenesis to generate the mutant ATP7A alleles A1362D and S637L, transformation of the Saccharomyces cerevisiae copper transport mutant, ccc2Δ, yeast complementation experiments and timed growth assays were all performed as described previously.10 Western blot analysis was used to confirm equivalent expression for the mutant alleles A1362D and S637L.
Reverse transcription‐PCR and quantitative reverse transcription‐PCR
Total RNA was extracted from primary fibroblast cultures (Qiagen RNeasy Kit; Qiagen, Valencia, California, USA) and first strand synthesis performed (Enhanced Avian RT First Strand Kit; Sigma‐Aldrich Corp. St. Louis, MO, USA). Reverse transcription (RT)‐PCR for mRNA species analysis was carried out with primers to amplify exons 5–138 or exons 8–11 (forward: 5′‐GAAGGATCGGTCAGCAAGTC‐3′; reverse: 5′‐CATCCACTGGAAATTTGCCTCC‐3′). Quantitative RT‐PCR was carried out using SYBR Green (JumpStart Taq ReadyMix; Sigma) on an Opticon instrument (MJ Research) using primers for ATP7A (forward, 5′‐CGAGAAATAAGACAATGGAGACG‐3′; reverse, 5′‐GGTTGCCAGCACAATCAGTA‐3′), hCTR1 (forward, 5′‐TGGACTCCAACAGTACCATGC‐3′; reverse, 5′‐CTGACTTGTGACTTACGCAGC‐3′) and ATOX1 (forward, 5′‐TGTGCTGAAGCTGTCTCTCGG‐3′; reverse, 5′‐CTGCTACTCAAGGCCAAGGTAGG‐3′) and GAPDH (forward, 5′‐CCACCCATGGCAAATTCCATGGCA‐3′; reverse, 5′‐TCTAGACGGCAGGTCAGGTCCACC‐3′). Experiments were performed in triplicate and, for certain comparisons, Student's t test (two‐tailed) was used to assess statistical significance.
Mosaicism assays
Genomic DNA was isolated from whole blood or cultured lymphoblasts (Wizard Genomic DNA Purification Kit; Promega Corporation, Madison, Wisconsin, USA). The region of the family A mutation (A1362D) was amplified with forward primer 5′‐TTCACATATTACCCTAAGGTAGACG‐3′ and reverse primer 5′‐ACTAATAGTAAGAAGACAGGCTTCC‐3′, the PCR product digested with MboI, and the resulting fragments separated by agarose gel electrophoresis. The region of the family B mutation (S637L) was amplified with forward primers 5′‐TCTCATGAATTTCCTTAGAGCTTAGG‐3′ and reverse 5′‐CTCGTTTATGATCTAAGTGACCAGCT‐3′, the PCR product digested with PvuII, and the resulting fragments separated by agarose gel electrophoresis.
Results
Family A
Biochemical analysis of family A
Serum copper levels and cerebrospinal fluid dihydroxyphenylalanine : dihydroxyphenylglycol (DOPA:DHPG) ratios were determined for both patients (table 1) and showed marked differences, which correlated with disease severity.
Table 1 Clinical and biochemical findings in five patients from families A and B.
| Patient | Age (years) | Motor development | Language development | Seizure history | Serum copper (μmol/L) | DOPA:DHPG* |
|---|---|---|---|---|---|---|
| A‐1 | 19 | Walked at 2 years | Normal | No | 11.3 | 0.87 |
| A‐2 | 9 | Never walked | None | Yes | 6.4 | 2.1 |
| B‐1 | 8 | Walked at 3.5 years | 2–3 words | No | 8.6 | 0.57 |
| B‐2 | 4 | Walked at 3 years | None | No | 8.0 | 0.81 |
| B‐3 | 2 | 4 month level | None | Yes | 4.9 | 0.91 |
| Normal reference ranges6,7 | 11 to 23.6 | 0 to 0.3 | ||||
| Menkes disease6,7 | 1.6 to 8.3 | 1.2 to 4.6 | ||||
*In cerebrospinal fluid.
Magnetic resonance imaging analysis of family A
Brain MRI scans of the two siblings (figure 1B) indicated normal structure and myelination in A‐1 and cortical atrophy with delayed myelination in A‐2.
Family A mutation analysis
In both siblings, we detected a cytosine to adenine transversion at base 4230 in exon 21 of ATP7A (data not shown) that predicts substitution of aspartate (D) for alanine (A) at amino acid residue 1362.
Intrafamilial variation in ATP7A expression in family A
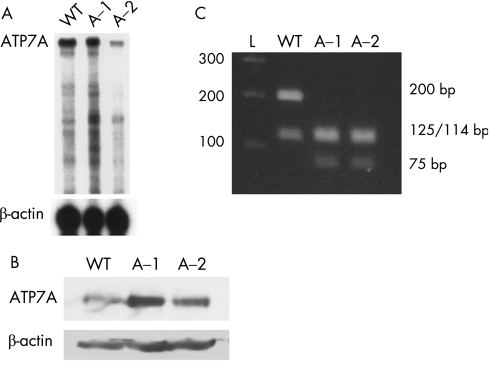
To determine if differences in the level of ATP7A expression between the two siblings contributed to the intrafamilial variation, we performed RNase protection assays using lymphoblast RNA. A normal control and the older, less affected sibling, A‐1, had substantially more ATP7A transcript than the more severely affected sibling A‐2 (figure 2A). Consistent with this result, western blotting (figure 2B) and densitometric assessment indicated that cultured lymphoblasts from patient A‐1 contained more ATP7A than did those from patient A‐2 (144% vs. 93% of a normal lymphoblast cell line). Quantitative RT‐PCR of fibroblast RNA showed that A‐1 had 242% and 249% ATP7A transcript compared with two normal control fibroblast cell lines, whereas patient A‐2, the severely affected sibling, had 117% and 121% compared with the same normal fibroblast controls (table 2). Thus, the level of ATP7A expression between these two siblings differed consistently in both lymphoblasts and fibroblasts, with higher expression in patient A‐1, correlating with his milder biochemical and clinical phenotype.
Figure 2 Molecular characterisation of family A with A1362D mutation. (A) RNase protection assay indicated disparate ATP7A expression between patients A‐1 and A‐2. A wild‐type control (WT) and the mildly affected sibling (A‐1) have more ATP7A transcript than the severely affected sibling (A‐2). β‐actin transcript, measured with a β‐actin riboprobe in the same assay (lower panel), indicates similar amounts in each sample. (B) Western analysis indicates increased ATP7A expression in A‐1 compared with A‐2. Densitometry, with correction for loading, indicated that ATP7A in A‐1 was 144% of WT and A‐2, 93% of WT. (C) Mosaicism assay. Using a restriction enzyme (MboI) assay, we were unable to detect the wild‐type allele in either sibling, suggesting that mosaicism does not explain phenotypic variability in this family. The wild‐type allele yields two bands (200 and 114 bp) whereas the mutant allele yields three bands (125, 114 and 75 bp).
Table 2 Expression of copper transport genes by quantitative reverse transcription PCR.
| Patient | Tissue | ATP7A, percentage of normal | Percentage of normal | |
|---|---|---|---|---|
| hCTR1 | ATOX1 | |||
| A‐1 | F | 246* | ND | ND |
| A‐2 | F | 119* | ND | ND |
| B‐1 | F | 8 | 128 | 102 |
| B‐2 | F | 12 | 132 | 129 |
| B‐3 | F | 10 | 131 | 118 |
F, fibroblast; ND, not done.
*Mean % of two normal controls.
Residual copper transport function of family A mutation (A1362D)
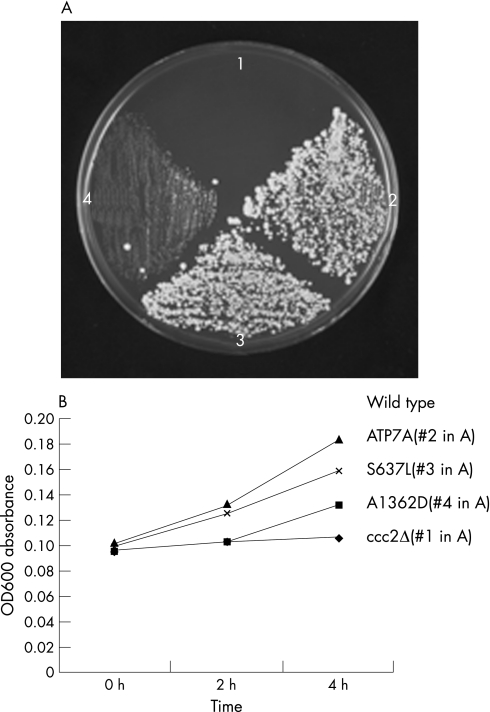
To determine if the A1362D mutation affected ATP7A function, we performed yeast complementation studies, using the S. cerevisiae copper transport mutant, ccc2Δ. The A1362D mutant allele complemented the ccc2Δ strain (figure 3A, quadrant 4) and showed functional copper transport of around 17% compared with wild‐type ATP7A, as measured in a timed growth assay (figure 3B).
Figure 3 Yeast complementation and timed growth assays indicate residual copper transport function by the A1362D and S637L mutant alleles. (A) Complementation of ccc2Δ on copper/iron‐deficient medium. Quadrant 1, mock‐transformed (empty vector) ccc2Δ; quadrant 2, ccc2Δ transformed with wild‐type ATP7A; quadrant 3, ccc2Δ transformed with S637L mutant allele; quadrant 4, ccc2Δ transformed with A1362D mutant allele. Growth in quadrants 3 and 4 indicates residual copper transport activity associated with S637L and (somewhat less) with A1362D. (B) Timed growth assay. The mean percentage growth in copper/iron‐limited media was 73% for ccc2Δ transformed with S637L and A1362D, and 17% for ccc2Δ transformed with wild‐type ATP7A. The mock‐transformed ccc2Δ strain (quadrant 1) failed to grow.
Evaluation for mosaicism in family A
To test for mosaicism, genomic DNA isolated from the patients' blood was amplified in the region of the A1362D mutation and the PCR products subjected to MboI restriction enzyme digest (figure 2C). Neither sibling showed evidence of the wild‐type digest pattern, suggesting that mosaicism was not contributing to intrafamilial variation.
Family B
Biochemical analysis of family B
Serum copper levels in all three siblings from family B were lower than normal, with the most severely affected patient (B‐3) having the lowest levels (table 1). Cerebrospinal fluid DOPA:DHPG ratios were also raised in these patients and the most severely affected sibling showed the highest ratio.
MRI analysis of family B
All three siblings in family B were evaluated by MRI to ascertain the degree of neurodegeneration. T1‐weighted MRI images showed cerebellar hypoplasia in all three (figure 1C). The youngest individual (B‐3) also has substantial cortical cerebral degeneration, consistent with his more severe clinical presentation.
Family B mutation analysis
We found a cytosine to thymine transition at base 2055 of the ATP7A coding sequence in all three siblings (data not shown), predicting substitution of leucine for serine at amino acid residue 637. The S637L mutation lies within a splice enhancer motif11 in exon 8 and aberrant splicing of ATP7A was described previously in an patient with OHS with this alteration.8
Alternate splicing of ATP7A in family B
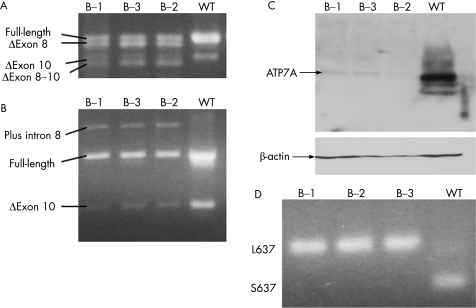
ATP7A mRNA species were analysed in family B by RT‐PCR using primers from exons 5 to 13,8 and exon 7 to 11 (figure 4A). The products of RT‐PCR were gel‐purified and sequenced. A splicing pattern similar to that previously reported8 was observed (figure 4A). In the wild‐type control, two bands were present. The major band contained exons 7–11 (the full‐length transcript). The minor band resulted from deletion of exon 10, a naturally occurring isoform.12 All three patients also showed these bands, though at lower intensity than in the wild‐type control. In addition, the three patients produced mutant transcript bands containing deletions of exon 8 and exons 8–10 (figure 4A), as well as exons 7–10 and exons 8–9 (data not shown).
Figure 4 Molecular characterisation of family B with S637L mutation. (A, B) RT‐PCR assays reveal multiple mutant transcripts and subnormal amounts of properly spliced transcript in B‐1, B‐2 and B‐3 compared with a wild‐type control (please see text for further details). (C) Western blot indicates reduced quantities of full‐length ATP7A protein relative to wild type. Densitometric analysis of ATP7A and β‐actin (to normalise for loading differences) indicated equivalent reduction in the amounts of ATP7A protein produced by all three brothers (10% in B‐1 and 11% in B‐2 and B‐3). (D) Mosaicism assay. Using a restriction enzyme (PvuII) digest assay, we failed to detect the wild‐type allele (S637) in any of the family B siblings, suggesting that mosaicism does not explain their phenotypic variability.
We found a novel mutant transcript when performing RT‐PCR with primers from exons 8 and 10 (figure 4B). Sequencing of this product demonstrated inclusion of intron 8, a transcript not previously described. This transcript and the other abnormal transcripts all produce shifts in the ATP7A translational reading frame, or delete transmembrane domains 1–4 and seem unlikely to produce functional copper transport molecules.
Quantitation of mRNA transcripts in family B
To measure the amount of properly spliced ATP7A transcript, we performed quantitative RT‐PCR, using primers specific for mRNAs containing exons 8 and 10, using fibroblasts from each patient. All three patients showed similar levels of full‐length transcript, approximately 8–12% of normal (table 2). Consistent with this, western blot analysis (figure 4C) showed that all three brothers had equivalent reductions in the amounts of the correct‐sized (≈178 kDa) ATP7A protein produced (10% of normal in B‐1; 11% of normal in B‐2 and B‐3).
We also measured expression of hCTR1 and ATOX1 by quantitative RT‐PCR (table 2). These copper transport proteins are important for importing copper into cells and delivering it to ATP7A, respectively.13 Interestingly, we found that hCTR1 levels were consistently higher in the three siblings compared with wild type, suggesting that this gene could be upregulated in response to reduced biologically available copper in MD/OHS cells.4,13 However, these differences compared with normal proved statistically significant in only one of the siblings (p<0.05, patient B‐3). Levels of hCTR1 and ATOX1 did not vary significantly among the three siblings, implying that differences in expression of these genes are not responsible for the intrafamilial variation.
Copper transport function of the S637L mutation
To determine if the S637L mutation substantially affected function of ATP7A, we performed yeast complementation analysis. The S637L mutant rescued the ccc2Δ strain (figure 3A, quadrant 3) and showed functional copper transport approximately 73% of normal, as measured in a timed growth assay (figure 3B).
Exclusion of mosaicism as the cause of intrafamilial variation in family B
We also considered the possibility of somatic mosaicism, previously observed to lessen the severity of other diseases,14 as an explanation for intrafamilial variation in family B; however, we found no evidence for any of the three patients in this family, suggesting that mosaicism does not underlie the intrafamilial variation among these siblings.
Discussion
We report on a striking intrafamilial variation in association with mutations in the MD/OHS copper‐transporting ATPase gene, ATP7A. Family A is perhaps the most dramatic example of intrafamilial variation in MD reported in the literature; the older sibling (A‐1) has a phenotype consistent with mild OHS whereas the younger sibling (A‐2) has a phenotype not unlike the classic form of MD. Consistent with these phenotypes, the younger patient has a lower serum copper level and higher cerebrospinal fluid catechol ratio. The source of these variations appears to be increased expression of the mutant ATP7A protein in the less affected individual, which may compensate for reduced copper transport function by the A1362D allele, as indicated by the yeast complementation and timed growth assay results (figure 3). The cause of this increased expression is unclear and warrants further investigation. A difference in the activities of ATP7A transcription factors is one possible explanation.
A1362 is a highly conserved residue in the seventh transmembrane domain of ATP7A.1 Substitution of a charged amino acid residue (aspartic acid) for a non‐polar residue (alanine) presumably alters the structure of this domain. Ambrosini and Mercer15 reported a patient with a mild MD phenotype and substitution of valine for alanine at this position. In cell biology experiments to characterise this defect, the A1362V mutant allele did not localise to the plasma membrane under high copper conditions, but rather remained localised in the perinuclear region. Owing to the mild neurological effects in the patient, the authors hypothesised that some copper efflux and trafficking activities were retained by the mutant molecule. Although we did not formally evaluate cellular localisation of the A1362D mutant protein, the level of functional copper transport, as measured in yeast complementation experiments, was approximately 17% of wild‐type ATP7A. Coupled with the above‐normal ATP7A expression in patient A‐1 documented by western blot analysis (144%) and quantitative RT‐PCR (approximately two‐fold compared with wild type), we estimate that the level of residual copper transport in this individual may range between 25% and 35%, which agrees with findings in patients with mild OHS studied previously.7,10 In contrast, patient A‐2 is predicted to have residual copper transport in a range between 15% and 20% of normal.
The clinical and biochemical heterogeneity in family B is not as marked as in family A. The most severely affected child (B‐3) has a slightly lower serum copper level and slightly higher cerebrospinal fluid catechol ratio compared with his siblings (B‐1, B‐2). The cause of this variation is not clear, as ATP7A levels appear to be similar for all three patients in family B, and our studies, including analysis of other genes involved copper transport, failed to uncover molecular differences that correlated with disease severity. We believe that his premature birth (32 weeks gestation) and neonatal medical problems are probably responsible for the more severe neurodevelopmental outcome in B‐3.
Examples of intrafamilial variability have implications for human studies evaluating the efficacy of treatments for MD. Usually, neurological outcomes in untreated, affected family members may be safely assumed to represent the natural history for a particular ATP7A mutation. However, as these unusual families exemplify, disease trajectory among untreated first‐degree relatives is, in rare instances, not closely comparable. Evaluation of treatment outcomes in MD should therefore be cautious and include comparisons of relevant biochemical and molecular data among treated and untreated siblings, when feasible.16
In summary, we report two cases of substantial intrafamilial variation for MD/OHS, with different causes. In the first case, there is a clear relationship to differences in expression of the mutant ATP7A between the more and less affected individuals, whereas in the second family, it appears that medical problems during the neonatal period, including possible hypoxia due to cardiac arrest, contributed significantly to a more severe clinical outcome in one sibling.
Acknowledgements
We gratefully acknowledge Dr Jasmine Ramcharan, Ms Nisi Philip and staff at The University of The West Indies, St Augustine, Trinidad and Tobago, Dr Rosita Kirshman and the late Dr Peter Mamunes for expert care of family A and Dr Soma Das for mutation analysis of the proband (B‐3) in family B. This work was supported by the intramural research program of the National Institute of Child Health and Human Development, National Institutes of Health.
Abbreviations
DOPA:DHPG - dihydroxyphenylalanine: dihydroxyphenylglycol
MD - Menkes disease
OHS - occipital horn syndrome
PBS - phosphate‐buffered saline
RT - reverse transcription
TBS - Tris‐buffered saline
Footnotes
Competing interests: None declared.
Parental/guardian informed consent was obtained for publication of fig 1.
References
- 1.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Isolation of a candidate gene for Menkes disease and evidence that it encodes a copper‐transporting ATPase. Nature Genet 199337–13. [DOI] [PubMed] [Google Scholar]
- 2.Chelly J, Tumer Z, Tonnesen T, Petterson A, Ishikawa‐Brush Y, Tommerup N, Horn N, Monaco A P.et al. Isolation of a candidate gene for Menkes disease that encodes a potential heavy metal binding protein. Nature Genet1993314–19. [DOI] [PubMed] [Google Scholar]
- 3.Mercer J F, Livingston J, Hall B, Paynter J A, Begy C, Chandrasekharappa S, Lockhart P, Grimes A, Bhave M, Siemieniak D. Isolation of a partial candidate gene for Menkes disease by positional cloning. Nat Genet 1993320–25. [DOI] [PubMed] [Google Scholar]
- 4.Kaler S G. Menkes Disease. Adv Pediatr 199441263–304. [PubMed] [Google Scholar]
- 5.White S R, Reese K, Sato S, Kaler S G. Spectrum of EEG findings in Menkes disease. Electroenceph Clin Neurophysiol 19938757–61. [DOI] [PubMed] [Google Scholar]
- 6.Kaler S G, Goldstein D S, Holmes C, Salerno J A, Gahl W A. Plasma and cerebrospinal fluid neurochemical pattern in Menkes disease. Ann Neurol 199333171–175. [DOI] [PubMed] [Google Scholar]
- 7.Kaler S G, Gallo L K, Proud V K, Percy A K, Mark Y, Segal N A, Goldstein D S, Holmes C S, Gahl W A. Occipital horn syndrome and a mild Menkes phenotype associated with splice site mutations at the MNK locus. Nature Genet 19948195–202. [DOI] [PubMed] [Google Scholar]
- 8.Ronce N, Moizard M P, Robb L, Toutain A, Villard L, Moraine C. A C2055T transition in exon 8 of the ATP7A gene is associated with exon skipping in an occipital horn syndrome family. Am J Hum Genet 199761233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borm B, Moller L B, Hausser I, Emeis M, Baerlocher K, Horn N, Rossi R. Variable clinical expression of an identical mutation in the ATP7A gene for Menkes disease/occipital horn syndrome in three affected males in a single family. J Pediatr 2004145119–121. [DOI] [PubMed] [Google Scholar]
- 10.Tang J, Robertson S P, Lem K E, Godwin S C, Kaler S G. Functional copper transport explains neurologic sparing in occipital horn syndrome. Genet Med 20068711–718. [DOI] [PubMed] [Google Scholar]
- 11.Fairbrother W G, Yeh R F, Sharp P A, Burge C B. Predictive identification of exonic splicing enhancers in human genes. Science 20022971007–1013. [DOI] [PubMed] [Google Scholar]
- 12.Kaler S G, Buist N R M, Holmes C S, Goldstein D S, Miller R C, Gahl W A. Early copper therapy in classic Menkes disease patients with a novel splicing mutation. Ann Neurol 199538921–928. [DOI] [PubMed] [Google Scholar]
- 13.Kuo Y M, Gybina A A, Pyatskowit J W, Gitschier J, Prohaska J R. Copper transport protein (Ctr1) levels in mice are tissue specific and dependent on copper status. J Nutr 200613621–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards M J, Wenstrup R J, Byers P H, Cohn D H. Recurrence of lethal osteogenesis imperfecta due to parental mosaicism for a mutation in the COL1A2 gene of type I collagen. The mosaic parent exhibits phenotypic features of a mild form of the disease. Hum Mutat 1992147–54. [DOI] [PubMed] [Google Scholar]
- 15.Ambrosini L, Mercer J F. Defective copper‐induced trafficking and localization of the Menkes protein in patients with mild and copper‐treated classical Menkes disease. Hum Mol Genet 199981547–1555. [DOI] [PubMed] [Google Scholar]
- 16.Kaler S G, Das S, Levinson B, Goldstein D S, Holmes C S, Patronas N, Packman S, Gahl W A. Successful copper therapy in Menkes disease associated with a mutant transcript containing a small in‐frame deletion. Biochem Mol Med 19965737–46. [DOI] [PubMed] [Google Scholar]