Abstract
Background
Patients with interferon‐γ receptor 1 (IFNγR1) deficiency show selective susceptibility to intracellular pathogens such as mycobacteria. IFNγR1 deficiency is an inherited immunodeficiency disorder, which can be either recessive or dominant. Dominant forms of IFNγR1 deficiency are known to be associated with mutations that introduce a premature stop codon in the intracellular domain of IFNγR1. One such mutation, 818del4, is believed to be the most common type. Although these mutations are presumed to exert a dominant‐negative effect on IFNγ signal transduction, the underlying molecular mechanism is unresolved.
Objective
We characterised the 774del4 mutant of IFNγR1 using a gene‐expression system to examine the effects of this mutation on IFNγ signal transduction.
Results
We identified a novel dominant mutation in IFNGR1, designated 774del4, which produced a truncated form of IFNγR1 in a patient with recurrent mycobacterial infections. IFNγR1 was overexpressed on the surfaces of CD14‐positive cells from the peripheral blood of this patient, and STAT1 phosphorylation in response to high doses of IFNγ was partially deficient. We expressed two truncated forms of IFNγR1, 774del4 and 818del4, in HEK 293 cells using transient transfection and found that these mutants overexpressed IFNγR1 on the cell surface because of impaired receptor stability, which resulted in a dominant‐negative effect on IFNγ signal transduction.
Conclusion
Like the 818del4 mutation, 774del4 produces a truncated form of IFNγR1, which has a dominant‐negative effect on IFNγ signal transduction through altered receptor stability.
Keywords: congenital immunodeficiency, IFNγ receptor 1, IFNGR1 , mycobacterium, osteomyelitis
Mendelian susceptibility to mycobacterial diseases (MSMD; OMIM 209950) is a rare congenital disorder characterised by susceptibility to infection by poorly virulent intracellular pathogens such as bacillus Calmette‐Guérin (BCG) and non‐tuberculosis mycobacterium (NTM), without any other marked immunodeficiency.1 MSMD is caused by an underlying genetic defect in the IL‐12/23–IFNγ circuit that enhances the pathogenesis of BCG and NTM. To date, six genes (IFNGR1, IFNGR2, IL12B, IL12RB1, STAT1, and NEMO) are known to be associated with this disorder.2,3,4,5,6,7,8,9,10,11 Interferon‐γ receptor 1 (IFNγR1) deficiency, caused by mutations in IFNGR1, is included in this disease category and is clinically characterised by disseminated BCG and severe NTM infections. This inherited immunodeficiency exists in both recessive and dominant forms, with a high degree of allelic heterogeneity.2,3,4 The dominant forms of IFNγR1 deficiency are frequently associated with a mutation in the intracellular domain of IFNGR1; in particular, the mutant known as 818del4 involves a 4 bp deletion in exon 6. In addition, the 811del4, 817insA, 818delT and G832T mutations have been reported.12,13,14,15 In this study, we identified a novel dominant mutation in IFNGR1, designated 774del4, in a patient with multifocal osteomyelitis due to mycobacterial infection. Although these mutations are presumed to exert a dominant‐negative effect on IFNγ signal transduction, the exact molecular mechanism is unknown. Therefore, we used a gene‐expression system in heterologous cells to examine the effects of the mutant receptors on IFNγ signalling. We found that the truncated forms of IFNγR1 are overexpressed on the cell surface because of altered receptor stability, resulting in a dominant‐negative effect on IFNγ signal transduction.
Materials and methods
Case report
The subject was a Japanese girl who was vaccinated with BCG at 4 months of age. She developed regional lymph node enlargement 2 months after vaccination, and lymph‐node biopsies revealed a tuberculoid granuloma. She was diagnosed with BCG lymphadenitis and treated with isoniazid for 3 years. At 12 years of age, she developed pain in her left knee and back, and persistent low‐grade fever. Radiography indicated multifocal osteomyelitis. Bone biopsies revealed tuberculoid granulomas, and PCR detected the presence of Mycobacterium avium. The patient was treated with isoniazid, rifampicin, and clarithromycin or azithromycin, and her symptoms gradually improved.
We obtained blood samples from the patient, her relatives and healthy adult controls, after obtaining informed consent. This study was approved by the ethics committee and internal review board of Hiroshima University.
Molecular genetics
Genomic DNA was extracted from peripheral blood leucocytes. Exon 6 of IFNGR1 and its flanking introns were amplified by PCR using the following primers: sense 5′‐GTGTTCTTTAAAACTCTGGCC‐3′ and antisense 5′‐TGGTAGACTGACTGATTGATG‐3′. The products were sequenced directly and then further analysed by cycle sequencing (BigDye Terminator v3.1 cycle sequencing kit; Applied Biosystems, Foster City, California, USA) and run on an automated analyser (ABI PRISM 310 Genetic Analyzer; Applied Biosystems).
Total RNA was extracted from the peripheral blood mononuclear cells (PBMCs) using Isogen extraction (Nippon Gene Co., Tokyo, Japan), and cDNA was synthesised from 5 μg of total RNA using a first‐strand synthesis system for RT‐PCR (SuperScript; Invitrogen, Carlsbad, California, USA). PCR of the wild‐type (WT) and 774del4 mutant alleles was performed with primers that span the entire coding region of IFNγR1 (5′‐GGATCCCGCAGGCGCTCGGGGTTGGA‐3′ and 5′‐CTCGAGGAAAACTGTCCAGGAAAATCAGACT‐3′). The PCR products were cloned into pGEM‐T Easy vector (Promega, Madison, Wisonsin, USA). To generate the 818del4 mutant, we performed PCR‐based mutagenesis of the WT construct using the following mismatched PCR primers: sense 5′‐TTTATATTAAGAAAATCCATTGAAGGAAAA‐3′ and antisense, 5′‐TTTTCCTTCAATGGATTTTCTTAATATAAA‐3′. These fragments were subcloned into the BamHI and XhoI sites of a mammalian expression vector pcDNA (Invitrogen).
Flow cytometry
PBMCs were stimulated with 20 μg/L lipopolysaccharide (LPS) (Sigma‐Aldrich, St. Louis, Missouri, UK) and 100 μg/L macrophage colony‐stimulating factor (M‐CSF) (Kyowa Hakko Kogyo, Tokyo, Japan) for 48 h. Portions of the cells were stained with phycoerythrin‐conjugated monoclonal anti‐human IFNγR1 antibody (GIR‐94; Santa Cruz Biotechnology, Santa Cruz, CA) or the isotype control, and analysed by gating in a FACS Calibur apparatus (Becton Dickinson, Franklin Lakes, New Jersey, USA). The remaining cells were recultured for an additional 18 h under starvation conditions and then left untreated, or treated with 50 μg/L LPS and 10 000 U/mL IFNγ for 30 min at 37°C. After stimulation, the cells were analysed by flow cytometry using an anti‐Stat1 antibody (BD Phosflow anti‐Stat1 (pY701); Becton Dickinson).
Analysis of gene expression
The HEK 293 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS) (HyClone, Logan, Utah, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin. At 24 h before transfection, the cells were harvested by trypsinisation and replated at a density of 1×105 cells/mL in 100 mm culture dishes. Plasmid DNA (10 μg per plate) carrying the WT, 774del4 or 818del4 alleles of IFNGR1 was introduced into the HEK 293 cells by calcium phosphate‐mediated transfection.
At 24 h after transfection, a portion of the cells was used for Western blotting16 to detect the IFNγR1 protein using anti‐human IFNγR1 antibodies. The remaining cells were treated with 20 μg/ml cycloheximide for 3 h and analysed by flow cytometry and Western blotting.
Primary cell separation and cytokine measurements
CD14‐positive cells and CD3‐positive cells were purified from PBMCs using a cell separation system (IMag; Becton‐Dickinson) according to the manufacturer's instructions. The purity of the CD14‐positive cell population was >90%, and that of the CD3‐positive cell population was >97% (data not shown). The CD14‐positive cells were cultured for 48 h in RPMI 1640 (Sigma‐Aldrich) supplemented with 15% FCS, with the addition of 500 ng/mL LPS and various concentrations (102–105 IU/mL) of IFNγ (Genentech, South San Francisco, California, USA). The cell‐culture supernatants were harvested, and the concentration of tumour necrosis factor (TNF)‐α was measured in duplicate using a human TNFα antibody bead kit (Biosource International, Camarillo, California, USA) and Luminex system (Luminex, Austin, Texas, USA). The CD3‐positive cells were incubated for 48 h with 1 μg/mL phytohaemagglutinin (PHA) (Sigma‐Aldrich) and various concentrations (1–105 pg/mL) of IL‐12. The concentration of IFNγ was measured as described above using Luminex.
Results
Sequence analysis
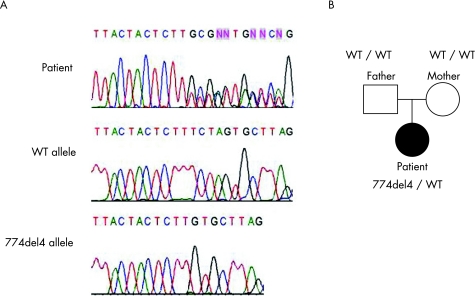
High molecular weight DNA was extracted from the peripheral blood samples, and the exons and flanking introns of IFNGR1 were amplified by PCR. Sequence analysis revealed that the patient had a heterozygous mutation, 774del4 (TCTA), in exon 6 of IFNGR1 (figure 1A). This mutation was not detected in the subject's parents or in 30 healthy Japanese controls (figure 1B).
Figure 1 Sequence analysis of exon 6 of IFNGR1. (A) The genomic DNA of IFNGR1 exon 6 from the patient was amplified by PCR, and the sequence was analysed by direct sequencing. The PCR product was cloned into the pGEM‐T Easy vector. Sequencing analysis detected the presence of the wild‐type (WT) and 774del4 alleles and that the patient had a heterozygous mutation, 774del4 (TCTA), in exon 6 of IFNGR1. (B) Pedigree of this family with the dominant form of IFNγR1 deficiency. Filled symbol, affected person; open symbols, healthy family member. The 774del4 mutation was not detected in either of the parents.
Response to IFNγ
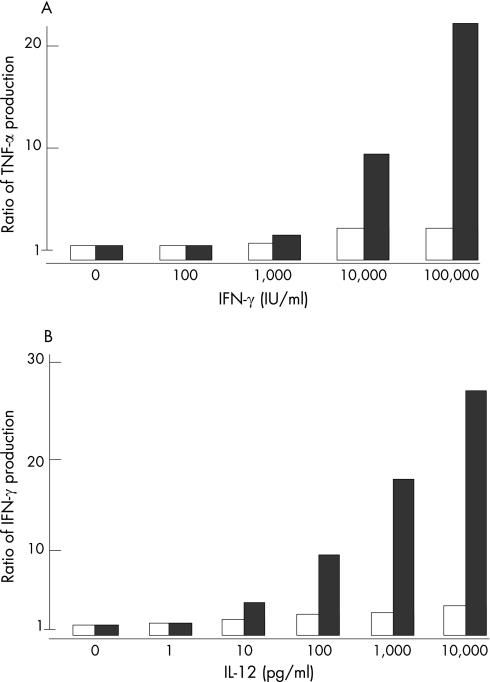
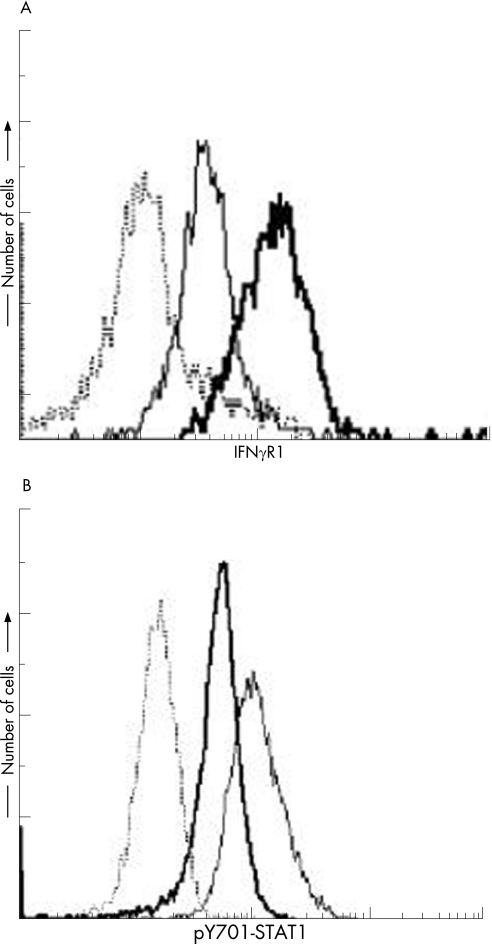
To ascertain the cellular response to IFNγ, we purified CD14‐positive cells from the PBMCs of the patient and controls. The CD14‐positive cells from the patient produced a lower level of TNFα in response to IFNγ stimulation, regardless of the dose (figure 2A). Once activated, the IFNγ receptor induces STAT1 homodimerisation via phosphorylation of tyrosine residue 701, and these homodimers enter the nucleus, where they act as transcription factors. Therefore, STAT1 Tyr701 phosphorylation (p‐Y701‐STAT1) in response to IFNγ was examined in PBMCs from the patient. PBMCs were incubated with M‐CSF and LPS for 48 h and then recultured for an additional 18 h under starvation conditions. The cells were then treated with or without LPS and IFNγ for 30 min or left untreated, and the level of p‐Y701‐STAT1 was analysed by flow cytometry. Although STAT1 phosphorylation was detected in the patient, it was significantly reduced (figure 3B).
Figure 2 Analysis of cytokine production. (A) CD14‐positive mononuclear cells in the peripheral blood samples of a patient and normal controls were stimulated with LPS and various concentrations of interferon (IFN)‐γ for 48 hours. The purity levels of the CD14‐positive cell populations were >90%. Cell‐culture supernatants were harvested, and the concentrations of tumour necrosis factor (TNF)‐α were measured by Luminex. The data represent the ratios of the levels of TNFα produced in response to LPS plus IFNγ to the levels of TNFα produced in response to LPS alone. Open columns, data from the patient; filled column, representative data from control subjects. (B) CD3‐positive mononuclear cells in the peripheral blood samples of a patient and normal controls were stimulated with purified hemagglutinin (PHA) 1 μg/ml and various concentrations of interleukin (IL)‐12 for 48 hours. The purity levels of CD3‐positive cells populations were >95%. Cell‐culture supernatants were harvested, and the concentrations of IFNγ were measured by Luminex. The data represent the ratios of IFNγ produced in response to PHA plus IL‐12 to the levels of IFNγ produced in response to PHA alone. Open columns, data from the patient; filled columns, representative data from the control subjects.
Figure 3 Analysis of cell surface IFNγR1 and STAT1 phosphorylation in response to IFNγ stimulation. (A) Peripheral blood mononuclear cells were cultured with RPMI 1640 containing 15% FCS, 50 μg/l LPS and 100 μg/l M‐CSF for 48 hours. The cells were stained with phycoerythrin (PE)‐conjugated anti‐interferon γ receptor 1 (IFNγR1) antibody and fluorescein isothiocyanate (FITC)‐conjugated anti‐CD14 antibody and analysed by flow cytometry. The data represent the levels of expression of IFNγR1 on CD14‐positive cells. Bold line, patient; solid line, control. The dotted line indicates binding of the appropriate isotype control antibody. (B) Peripheral blood mononuclear leukocytes were incubated in RPMI 1640 that was supplemented with 15% fetal calf serum (FCS), 100 μg/l macrophage colony‐stimulating factor (M‐CSF), and 20 μg/l lipopolysaccharide (LPS) for 48 hours. The harvested cells were washed and then cultured for an additional 18 hours in RPMI 1640 medium alone. Then cells were treated with or without 50 μg/l LPS and 10 000 U/ml IFNγ for 30 minutes, and the cells were then stained with anti‐STAT1 (pY701) antibody and analysed by flow cytometry. Bold line, patient sample with cytokine stimulation; solid line, normal subject sample with cytokine stimulation; dotted line, normal subject sample without cytokine stimulation.
Expression of the IFNγR1 mutant proteins
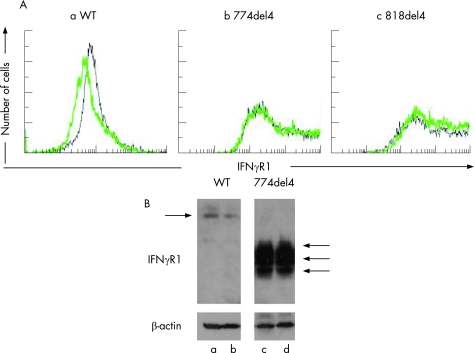
The 774del4 mutation was presumed to cause a frameshift that would lead to a premature stop codon at nucleotide positions 827–829, thereby producing a truncation in the cytoplasmic domain of the receptor.4,17,18,19 To characterise the 774del4 mutant protein, PBMCs from the patient and healthy controls were isolated and stimulated with LPS and M‐CSF for 48 h, and surface expression of IFNγR1 on the activated CD14‐positive mononuclear cells was analysed using flow cytometry. Increased expression of IFNγR1 was detected in the patient (figure 3A).
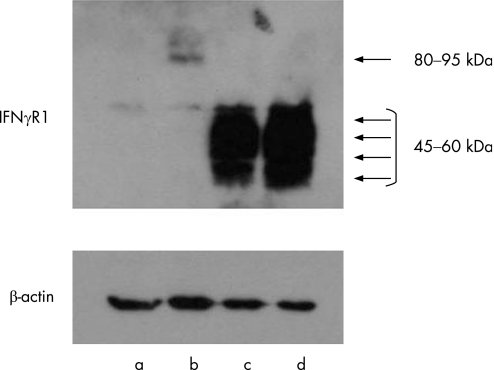
To characterise the 774del4 and 818del4 mutant versions of IFNγR1 further, we cloned the cDNAs for these mutants into pcDNA and transiently transfected each construct into HEK 293 cells, using calcium phosphate‐mediated transfection. The gene products were analysed by Western blot analysis (figure 4). The 774del4 and 818del4 mutant proteins had apparent molecular weights in the range of 45–60 kDa, in contrast to the WT molecular weight of 80–95 kDa. These results indicate that the 774del4 and 818del4 mutant alleles of IFNGR1 encode truncated forms of IFNγR1 that accumulate on the cell surface, as predicted by the primary structures of the mutants.
Figure 4 Expression of 774del4 and 818del4 mutant IFNγR1 proteins by transfected cells. Expression constructs carrying the wild‐type (WT), 774del4, and 818del4 forms of IFNGR1 were transiently transfected into HEK 293 cells using the calcium phosphate method. IFNγR1 protein was detected by Western blotting. The WT IFNγR1 (B) was detected with an apparent molecular weight of 80–95 kDa. The 774del4 (C) and 818del4 (D) mutant forms of IFNγR1 gave strong signals with an apparent molecular weight of 45–60 kDa; this band was not detected in the WT‐transfected cell lysate. IFNγR1 was not detected in the total lysate of cells transfected with the mock vector (A).
IFNγR1 stability
As cell‐surface overexpression of the 774del4 truncated form of IFNγR1 was detected in the patient, we established HEK 293 cells that transiently expressed either 774del4 or 818del4 and examined the stability of the truncated proteins. HEK 293 cells transiently transfected with either of the vectors were treated with cycloheximide for 3 h, and IFNγR1 expression was analysed by Western blotting (figure 5B) and flow cytometry (figure 5A). The 774del4 and 818del4 mutant versions of IFNγR1 were consistently detected, even after the inhibition of translation by cycloheximide (see supplementary figure, available online at http://jmg.bmj.com/supplemental). This result indicates that these mutant proteins are stably expressed on the cell surface due to alteration of receptor stability.
Figure 5 Stability of interferonγ receptor 1 (IFNγR1) in HEK 293 cells transfected with the wild‐type (WT), 774del4 or 818del4 forms of IFNGR1. The WT, 774del4 and 818del4 IFNGR1 cDNAs were introduced into the HEK 293 using the calcium phosphate method. At 24 hours after transfection, the cells were treated with 20 μg/ml cycloheximide for 3 hours. The expression levels of IFNγR1 were analysed by flow cytometry and Western blotting using monoclonal mouse antibodies against human IFNγR1. (A) IFNγR1 analysed by flow cytometry. (a) WT; (b) 774del4; (c) 818del4; solid line, cycloheximide‐treated cells; dotted line, untreated cells. (B) The lysates of cells transfected with WT and 774del4 IFNγR1 were analysed by Western blotting. The cells were treated with CHX (a, c) or untreated (b, d).
Cellular response to IL‐12
To examine the cellular response to IL‐12, we purified CD3‐positive cells from the PBMCs of the patient and the controls. Compared with the CD3‐positive cells of the controls, those of the patient produced very little IFNγ in response to IL‐12 (figure 2B).
Discussion
IFNγ and IL‐12 play important roles in initiating and sustaining the Th1 cytokine cascade. Antigen‐presenting cells (APCs), including macrophages and dendritic cells, produce IL‐12 in response to mycobacterial infection. IL‐12 stimulates T cells and natural killer cells through its receptor (a heterodimer of IL‐12 receptors β1 and β2) to produce IFNγ. IFNγ acts through its receptor (a heterodimer of IFNγR1 and IFNγR2) on macrophages to activate the defence against mycobacteria. IFNγ signalling in APCs increases microbiocidal activity and upregulates IL‐12 and TNFα production through STAT1 activation. Thus, IFNγ and IL‐12 work synergistically to induce cellular immunity,20,21 and impairment of the IL‐12/23‐IFNγ circuit results in MSMD. Recently, mutations were identified in the IFNGR1, IFNGR2, IL12B, IL12RB1, STAT1, and NEMO genes of patients with MSMD.2,3,4,5,6,7,8,9,10,11 In fact, 13 different genetic disorders associated with MSMD have been shown to be caused by mutations in these six genes.11 In particular, defects in IFNGR1, IFNGR2, and STAT1 are associated with impaired cellular responses to IFNγ, and defects in IL12B, IL12RB1, and NEMO are associated with IL‐12‐dependent IFNγ production.11 Further, an X‐linked recessive form of MSMD was recently identified and has been mapped to two candidate regions, Xp11.4‐Xp21.2 and Xq25‐Xq26.3.22 Patients bearing these mutant alleles share certain clinical features, including selective susceptibility to mycobacteria, Salmonellae and some viral infections, although with various sensitivity. In addition, they show no marked immune deficiencies against other pathogens.
In this study, we identified a novel mutation in IFNGR1, designated 774del4, in a patient with MSMD. Consistent with the results of a previous report,23 the CD14‐positive cells from this patient exhibited impaired production of TNFα in response to LPS and IFNγ, and the patient's CD3‐positive cells did not produce IFNγ effectively in response to PHA and IL‐12. It seems likely that the IFNγR1 deficiency not only impairs IFNγ signaling but also abolishes the Th1 cytokine cascade.
Based on these results, we examined the molecular basis for the dominant forms of IFNγR1 deficiency. We identified a 4 bp deletion, designated 774del4, in the IFNGR1 gene of a patient with multiple osteomyelitic lesions due to M. avium infection. This was considered to be a sporadic case, as the same mutation was not detected in either of the patient's parents. The deletion causes a frameshift that introduces a premature stop codon (TGA) at nucleotides 827–829; the resulting truncated version of IFNγR1 lacks the expected intracellular domain. The 818del4 allele, which is the most frequent mutation observed in patients with the dominant form of IFNγR1 deficiency, also introduces a frameshift and premature stop codon at nucleotides 827–829.4 Although the truncated protein encoded by 774del4 differs from that encoded by 818del4 in terms of the last 14 amino acids at the C‐terminal end, patients with these mutations have similar clinical features.5 Multifocal osteomyelitis caused by BCG or NTM in the absence of susceptibility to other infectious agents appears to be a characteristic feature of patients with either 818del4‐related or 774del4‐related IFNγR1 deficiency.
The protein encoded by the 774del4 allele of IFNGR1 was overexpressed on the cell surface. To characterise this truncated version of IFNγR1, we cloned the WT allele and the 774del4 and 818del4 alleles of IFNGR1 into a mammalian expression vector. Cycloheximide treatment enabled us to assess the stability of IFNγR1 expression by flow cytometry and Western blotting. In contrast to the WT protein, which disappeared rapidly even after cycloheximide treatment, the 774del4 and 818del4 mutant proteins were stably retained on the cell surface. These observations suggest that both the 774del4 and 818del4 mutant forms of IFNγR1 are overexpressed on the cell surface, due to impaired receptor degradation. The intracellular domain of IFNγR1 contains two regions that are important for the receptor's function: the YDKPH sequence (residues 440–444), which is located near the C‐terminus of IFNγR1 and is required for IFNγ‐dependent cellular responses through STAT1 binding, and the membrane proximal region (residues 256–303), which is required for both receptor‐mediated ligand internalisation and subsequent degradation, and for the induction of the biological response.24,25 The importance of these regions is supported by radioligand binding analysis using both 125I‐labelled recombinant IFNγ and a series of IFNγR1 deletion mutants lacking the cytoplasmic region.26 Without this region, both the internalation and degradation of IFNγ is impaired, and it accumulates at the cell surface.18 Further analysis has revealed that a leucine–isoleucine motif (residues 270–271) near this region also plays an important role in receptor‐mediated ligand internalisation/degradation and that the LPKS sequence (residues 266–269) is required for biological responses through JAK‐1 binding.24,27,28
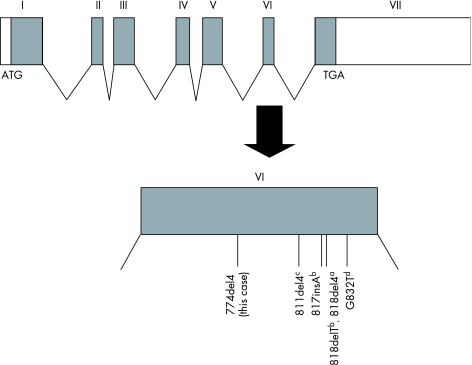
Jouanguy et al.4 have suggested that the 818del4 mutation impairs the recycling of IFNγR1 because of the lack of the membrane proximal region, which results in cell‐surface overexpression. This mutant receptor, which lacks both the LPKS and YDKPH motifs, is thought to impair IFNγ signalling via a dominant‐negative effect; however, this assumption has not yet been examined at the molecular level. In this study, we examined the molecular mechanisms through which the 774del4 and 818del4 mutants exert dominant‐negative effects on IFNγ signalling. In general, an mRNA with a premature stop codon tends to be rapidly degraded by the nonsense‐mediated mRNA decay pathway.29 However, both WT and mutant cDNAs were detected at similar ratios in the total RNA extracted from the peripheral blood of the patient, who carries both the WT and 774del4 mutant alleles (data not shown). Furthermore, we directly showed that the 774del4 and 818del4 mutant versions of IFNγR1 were stably overexpressed on the cell surface owing to alteration of receptor stability. Additionally, the CD14‐positive cells from the patient showed a weaker response to IFNγ than did those from the healthy controls. Similarly, STAT1 phosphorylation in response to IFNγ was significantly impaired in cells derived from the patient. Together, these observations suggest that the 774del4 version of IFNγR1 is non‐functional and has a dominant‐negative effect on IFNγ signal transduction. To our knowledge, all of the mutations identified to date that cause dominant forms of IFNγR1 deficiency are localised in exon 6 of IFNGR1 and lead to a premature stop in the intracellular domain (figure 6).
Figure 6 Summary of mutations in IFNGR1 associated with dominant forms of IFNγR1 deficiency. The primary genomic structure of IFNGR1, including its seven exons, is shown, and all of the mutations previously shown to be associated with dominant forms of IFNγR1 deficiency are indicated. Specifically, the 774del4 mutation described in this report is depicted, along with the alleles described by Jouanguy et al.a, Dorman et al.b, Sasaki et al.c, and Villella et al.d.4,12,13,14,15 All of the mutations are localised in exon 6. Unfilled boxes, non‐coding regions; filled boxes, IFNGR1 coding region.
IFNγR1 deficiency is a primary immunodeficiency characterised by severe mycobacterial infections. Multifocal osteomyelitis without other organ involvement is an outstanding clinical feature of individuals with this deficiency.5 Several patients with IFNγR1 deficiency who developed multiple osteomyelitis were initially misdiagnosed as having accompanying Langerhans cell histiocytosis and were treated with cytotoxic chemotherapy.30 Of 50 previously reported patients with dominant forms of IFNγR1 deficiency, most were found to have the 818del4 mutation.4,11 Sasaki et al reported four Japanese patients with dominant forms of IFNGR1 deficiency from three unrelated families.13 One of these patients was heterozygous for the 811del4 mutation, and the the other three were heterozygous for 818del4. The novel 774del4 mutation described in this study is the fifth allele shown to cause a dominant form of IFNGR1 deficiency in Japan. Given that adjunctive therapeutic administration of IFNγ is beneficial for patients with dominant forms of IFNγR1 deficiency, more attention should be paid to this deficiency, especially when treating patients with recurrent mycobacterial infections.
Supplementary figure available on JMG website–http://jmg.bmj.com/supplemental
Abbreviations
APC - antigen‐presenting cell
BCG - bacillus Calmette‐Guérin
FCS - fetal calf serum
IFNγR1 - interferon gamma receptor 1
LPS - lipopolysaccharide
M‐CSF - macrophage colony‐stimulating factor
MSMD - mendelian susceptibility to mycobacterial diseases
NTM - non‐tuberculosis mycobacterium
PBMC - peripheral blood mononuclear cell
STAT1 - signal transducer and activator of transcription 1
TNF - tumour necrosis factor
WT - wild type
Footnotes
This work was supported by a grant‐in‐aid from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and a grant‐in‐aid from the Japanese Ministry of Health, Labor, and Welfare. The 774del4 mutation of IFNGR1 has been assigned GenBank accession number EF535103.
Competing interests: None declared.
Supplementary figure available on JMG website–http://jmg.bmj.com/supplemental
References
- 1.Hamosh A, Scott A F, Amberger J S, Bocchini C A, McKusick V A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 200533D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levin M. A mutation in the interferon‐gamma‐receptor gene and susceptibility to mycobacterial infection. N Engl J Med 19963351941–1949. [DOI] [PubMed] [Google Scholar]
- 3.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile J F, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova J L. Interferon‐gamma‐receptor deficiency in an infant with fatal bacille Calmette‐Guerin infection. N Engl J Med 19963351956–1961. [DOI] [PubMed] [Google Scholar]
- 4.Jouanguy E, Lamhamedi‐Cherradi S, Lammas D, Dorman S E, Fondaneche M C, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile J F, Ducoulombier H, Edgar D, Clarke J, Oxelius V A, Brai M, Novelli V, Heyne K, Fischer A, Holland S M, Kumararatne D S, Schreiber R D, Casanova J L. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet 199921370–378. [DOI] [PubMed] [Google Scholar]
- 5.Dorman S E, Holland S M. Mutation in the signal‐transducing chain of the interferon‐gamma receptor and susceptibility to mycobacterial infection. J Clin Invest 19981012364–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altare F, Lammas D, Revy P, Jouanguy E, Doffinger R, Lamhamedi S, Drysdale P, Scheel‐Toellner D, Girdlestone J, Darbyshire P, Wadhwa M, Dockrell H, Salmon M, Fischer A, Durandy A, Casanova J L, Kumararatne D S. Inherited interleukin 12 deficiency in a child with bacille Calmette‐Guerin and Salmonella enteritidis disseminated infection. J Clin Invest 19981022035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altare F, Durandy A, Lammas D, Emile J F, Lamhamedi S, Le Deist F, Drysdale P, Jouanguy E, Doffinger R, Bernaudin F, Jeppsson O, Gollob J A, Meinl E, Segal A W, Fischer A, Kumararatne D, Casanova J L. Impairment of mycobacterial immunity in human interleukin‐12 receptor deficiency. Science 19982801432–1435. [DOI] [PubMed] [Google Scholar]
- 8.de Jong R, Altare F, Haagen I A, Elferink D G, Boer T, van Breda Vriesman P J, Kabel P J, Draaisma J M, van Dissel J T, Kroon F P, Casanova J L, Ottenhoff T H. Severe mycobacterial and Salmonella infections in interleukin‐12 receptor‐deficient patients. Science 19982801435–1438. [DOI] [PubMed] [Google Scholar]
- 9.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, Holland S M, Schreiber R D, Casanova J L. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science 2001293300–303. [DOI] [PubMed] [Google Scholar]
- 10.Filipe‐Santos O, Bustamante J, Haverkamp M H, Vinolo E, Ku C L, Puel A, Frucht D M, Christel K, von Bernuth H, Jouanguy E, Feinberg J, Durandy A, Senechal B, Chapgier A, Vogt G, de Beaucoudrey L, Fieschi C, Picard C, Garfa M, Chemli J, Bejaoui M, Tsolia M N, Kutukculer N, Plebani A, Notarangelo L, Bodemer C, Geissmann F, Israel A, Veron M, Knackstedt M, Barbouche R, Abel L, Magdorf K, Gendrel D, Agou F, Holland S M, Casanova J L. X‐linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40‐dependent IL‐12 production. J Exp Med 20062031745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filipe‐Santos O, Bustamante J, Chapgier A, Vogt G, de Beaucoudrey L, Feinberg J, Jouanguy E, Boisson‐Dupuis S, Fieschi C, Picard C, Casanova J L. Inborn errors of IL‐12/23‐ and IFN‐gamma‐mediated immunity: molecular, cellular, and clinical features. Semin Immunol 200618347–361. [DOI] [PubMed] [Google Scholar]
- 12.Dorman S E, Picard C, Lammas D, Heyne K, van Dissel J T, Baretto R, Rosenzweig S D, Newport M, Levin M, Roesler J, Kumararatne D, Casanova J L, Holland S M. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet 20043642113–2121. [DOI] [PubMed] [Google Scholar]
- 13.Sasaki Y, Nomura A, Kusuhara K, Takada H, Ahmed S, Obinata K, Hamada K, Okimoto Y, Hara T. Genetic basis of patients with bacille Calmette‐Guérin osteomyelitis in Japan: identification of dominant partial interferon‐gamma receptor 1 deficiency as a predominant type. J Infect Dis 2002185706–709. [DOI] [PubMed] [Google Scholar]
- 14.Villella A, Picard C, Jouanguy E, Dupuis S, Popko S, Abughali N, Meyerson H, Casanova J L, Hostoffer R W. Recurrent Mycobacterium avium osteomyelitis associated with a novel dominant interferon gamma receptor mutation. Pediatrics 2001107e47. [DOI] [PubMed] [Google Scholar]
- 15.Dorman S E, Uzel G, Roesler J, Bradley J S, Bastian J, Billman G, King S, Filie A, Schermerhorn J, Holland S M. Viral infections in interferon‐gamma receptor deficiency. J Pediatr 1999135640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo P, Tresini M, Cristofalo V, Chen X, Saulewicz A, Gray M D, Banker D E, Klingelhutz A L, Ohtsubo M, Takihara Y, Norwood T H. Immortalization in a normal foreskin fibroblast culture following transduction of cyclin A2 or cdk1 genes in retroviral vectors. Exp Cell Res 2004294406–419. [DOI] [PubMed] [Google Scholar]
- 17.Arend S M, Janssen R, Gosen J J, Waanders H, de Boer T, Ottenhoff T H, van Dissel J T. Multifocal osteomyelitis caused by nontuberculous mycobacteria in patients with a genetic defect of the interferon‐gamma receptor. Neth J Med 200159140–151. [DOI] [PubMed] [Google Scholar]
- 18.Waibel K H, Regis D P, Uzel G, Rosenzweig S D, Holland S M. Fever and leg pain in a 42‐month‐old. Ann Allergy Asthma Immunol 200289239–243. [DOI] [PubMed] [Google Scholar]
- 19.Raszka W V, Trinh T T, Zawadsky P M. Multifocal M. intracellulare osteomyelitis in an immunocompetent child. Clin Pediatr 199433611–616. [DOI] [PubMed] [Google Scholar]
- 20.Reiner S L, Locksley R M. The regulation of immunity to Leishmania major. Annu Rev Immunol 199513151–177. [DOI] [PubMed] [Google Scholar]
- 21.Magram J, Connaughton S E, Warrier R R, Carvajal D M, Wu C Y, Ferrante J, Stewart C, Sarmiento U, Faherty D A, Gately M K. IL‐12‐deficient mice are defective in IFN gamma production and type 1 cytokine responses. Immunity 19964471–481. [DOI] [PubMed] [Google Scholar]
- 22.Bustamante J, Picard C, Fieschi C, Filipe‐Santos O, Feinberg J, Perronne C, Chapgier A, de Beaucoudrey L, Vogt G, Sanlaville D, Lemainque A, Emile J F, Abel L, Casanova J L. A novel X‐linked recessive form of Mendelian susceptibility to mycobaterial disease. J Med Genet 200744e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg J, Fieschi C, Doffinger R, Feinberg M, Leclerc T, Boisson‐Dupuis S, Picard C, Bustamante J, Chapgier A, Filipe‐Santos O, Ku C L, de Beaucoudrey L, Reichenbach J, Antoni G, Balde R, Alcais A, Casanova J L. Bacillus Calmette Guerin triggers the IL‐12/IFN‐gamma axis by an IRAK‐4‐ and NEMO‐dependent, non‐cognate interaction between monocytes, NK, and T lymphocytes. Eur J Immunol 2004343276–3284. [DOI] [PubMed] [Google Scholar]
- 24.Bach E A, Aguet M, Schreiber R D. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol 199715563–591. [DOI] [PubMed] [Google Scholar]
- 25.Farrar M A, Campbell J D, Schreiber R D. Identification of a functionally important sequence in the C terminus of the interferon‐gamma receptor. Proc Natl Acad Sci 19928911706–11710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrar M A, Fernandez‐Luna J, Schreiber R D. Identification of two regions within the cytoplasmic domain of the human interferon‐gamma receptor required for function. J Biol Chem 199126619626–19635. [PubMed] [Google Scholar]
- 27.Kaplan D H, Greenlund A C, Tanner J W, Shaw A S. 1996. Identification of an interferon‐gamma receptor alpha chain sequence required for JAK‐1 binding. J Biol Chem 19962719–12. [DOI] [PubMed] [Google Scholar]
- 28.Farrar M A, Schreiber R D. The molecular cell biology of interferon‐gamma and its receptor. Annu Rev Immunol 199311571–611. [DOI] [PubMed] [Google Scholar]
- 29.Maquat L E. Nonsense‐mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004589–99. [DOI] [PubMed] [Google Scholar]
- 30.Edgar J D, Smyth A E, Pritchard J, Lammas D, Jouanguy E, Hague R, Novelli V, Dempsey S, Sweeney L, Taggart A J, O'hara D, Casanova J L, Kumararatne D S. Interferon‐gamma receptor deficiency mimicking Langerhans' cell histiocytosis. J Pediatr 2001139600–603. [DOI] [PubMed] [Google Scholar]