Abstract
Background
Reports of differential mutagen sensitivity conferred by a defect in the mismatch repair (MMR) pathway are inconsistent in their conclusions. Previous studies have investigated cells established from immortalised human colorectal tumour lines or cells from animal models.
Methods
We examined primary human MSH2‐deficient neonatal cells, bearing a biallelic truncating mutation in MSH2, for viability and chromosomal damage after exposure to DNA‐damaging agents.
Results
MSH2‐deficient cells exhibit no response to interstrand DNA cross‐linking agents but do show reduced viability in response to irradiation. They also show increased chromosome damage and exhibit altered RAD51 foci kinetics after irradiation exposure, indicating defective homologous recombinational repair.
Discussion
The cellular features and sensitivity of MSH2‐deficient primary human cells are broadly in agreement with observations of primary murine cells lacking the same gene. The data therefore support the view that the murine model recapitulates early features of MMR deficiency in humans, and implies that the variable data reported for MMR‐deficient immortalised human cells may be due to further genetic or epigenetic lesions. We suggest caution in the use of radiotherapy for treatment of malignancies in individuals with functional loss of MSH2.
Keywords: homozygous MSH2 deficiency, Irradiation, RAD51, primary cells
The DNA mismatch repair (MMR) system reduces spontaneous mutations by correcting the misinsertion errors of replicative DNA polymerases. In addition to the recognition and processing of replication errors, MMR proteins have been implicated in the response to several other forms of DNA damage (reviewed by Stojic et al1). However, evidence of sensitivity to various DNA damaging agents is contradictory.2,3,4,5,6,7,8,9 Frequently cited explanations for the discrepancies between reports are the differences between the cell clones, the methods used, and the repair and death‐promoting pathways in mouse and human cell types and in tissue culture adaptation. These pathways are particularly problematic in MMR‐deficient cells, which acquire more mutations per division than MMR‐proficient cells. Key to understanding the likely effect of MMR deficiency is the examination of non‐adapted primary cells. These have been studied since the development of an Msh2‐deficient mouse; however, until now it was not known to what extent the murine system reflected the human cellular phenotype. Here we report, for the first time to our knowledge, the examination of the DNA damage and cell viability response of primary non‐tumour cells from a family with members homozygous and heterozygous for a mutation in the MMR gene, MSH2.
Methods and results
We identified a biallelic MSH2 226C→T mutation in a male child of Arab ethnicity (figure 1A, child 2) with T‐cell non‐Hodgkin lymphoma (NHL) whose parents were both heterozygous for the mutation (figure 1A,B). His brother had died from a similar T‐cell NHL ((figure 1A, child 1).
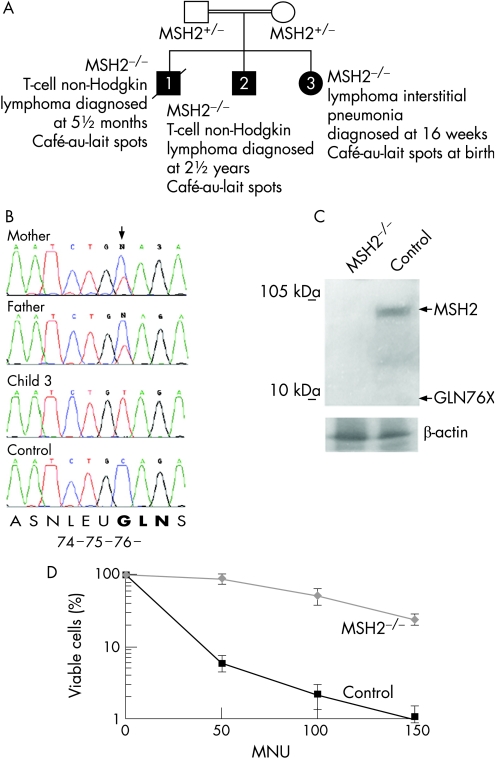
Figure 1 Homozygous MSH2 226C→T cells are MSH2‐deficient and tolerant to N‐methyl‐N1‐nitrosurea (MNU). (A) Partial pedigree of the MSH2 family. Genotypes and ages at diagnosis are shown. The extended family had numerous affected members with colon cancer, and members with cancer of the oesophagus, bowel, stomach and optic glioma. (B) Partial sequence of MSH2 exon 2 from genomic DNA isolated from peripheral blood lymphocytes (PBLs) of child 3, the child's parents and a normal control (sequence identifier: NM_000251). (C) Immunoblot of MSH2 protein (PC57, Ab‐3; Oncogene Research Products, Calbiochem, Darmstadt, Germany) from fibroblasts derived from a biopsy of child 3 and age‐matched control fibroblasts. β‐actin loading control is shown (ab8227; Abcam, Cambridge, UK). Expected sizes of full‐length MSH2 and the truncated Gln76X protein are indicated by arrows. (D) Tolerance of MSH2 226C→T homozygous fibroblasts from child 3, (diamonds) and age‐matched control fibroblasts (squares) to MNU at micromolar concentrations shown. In this analysis, as throughout this report, the primary fibroblast cells were examined at passage 10 or before (<30 doublings). Experiments were performed in triplicate, and the standard deviation about the mean is shown.
During the mother's third pregnancy the fetus showed evidence at 20 weeks of a mediastinal mass and at 16 weeks after birth, a thoracotomy was carried out. No discrete lesion was identified, and histopathological analysis of biopsied material showed lymphoid interstitial pneumonia, a diffuse pulmonary disorder characterised by an interstitial infiltrate of mature lymphocytes, including T‐cells.10 Sequencing of peripheral blood lymphocyte (PBL) DNA from child 3 showed homozygosity for the MSH2 mutation 226C→T (figure 1B). The transition introduces a stop codon in glutamine 76 of exon 2 and is predicted to result in nonsense‐mediated decay of the transcript.11 Any escaping transcript would produce a severely truncated protein (deletion of amino acids 76–934), lacking all known functional domains of MSH2.
Characterisation of MSH2‐deficient cells
Biopsied tissue from child 3 was cut into small explants and culktured in Ham's F10 culture medium containing 15% fetal calf serum. As controls, neonate fibroblasts were derived from a skin biopsy of a 3‐month‐old boy with a suspected mitochondrial metabolic disorder (although none was found on investigation).
We examined protein expression in fibroblasts cultured from the biopsy and age‐matched skin fibroblast controls (MSH2 is reported to be expressed in all tissues including the lung and skin12,13). No MSH2 protein, full‐length or truncated, was detected in lysate from MSH2 226C→T homozygous fibroblasts, whereas full‐length MSH2 expression was evident in the control cells (figure 1C).
To establish whether the MSH2‐deficient cells exhibited features of a MMR defect, we examined them for resistance to the DNA‐methylating agent N‐methyl‐N1‐nitrosourea (MNU). Viable cells were identified using trypan blue (Sigma Chemical Co., Minneapolis, Minnesota, USA) exclusion and counted. Cells were exposed for 1 hour to MNU at 50, 100 and 150 μmol/l before further culture in complete medium for 14 days. These steps were performed in the presence of O6‐benzylguanine (20 μmol/l) to inhibit endogenous methyltransferase activity that might otherwise remove the methyl groups added by MNU. MMR‐deficient cells are resistant to the toxic effects of DNA methylating agents such as MNU, whereas MMR‐proficient cells undergo replication arrest and cell death (thought to occur either via ‘futile MMR', and/or signalling to the apoptotic machinery).14 Control neonatal fibroblasts were sensitive to MNU, whereas resistance was evident in MSH2‐deficient cells (figure 1D), indicating a defect in processing DNA‐methylation products, consistent with a fault in MMR.
We also examined the DNA of MSH2 226C→T cells (passage 8) for evidence of microsatellite instability (MSI) (using Bat‐25, Bat26 and D5S346 as markers). Although MSI is often associated with MMR deficiency in solid tumours arising in adults, it is not detected in non‐tumour tissue of biallelic MSH2 mutation carriers in humans or mouse.15,16,17 DNA from a colon tumour, negative for MLH1 expression, showed MSI positivity, whereas this was not detected in the DNA from MSH2 226C→T homozygous fibroblasts (supplementary figure 1 (available online at http://jmg.bmj.com/supplemental) and data not shown), consistent with their non‐tumour and non‐clonal status.
Response of primary MSH2‐deficient cells to DNA‐damaging agents
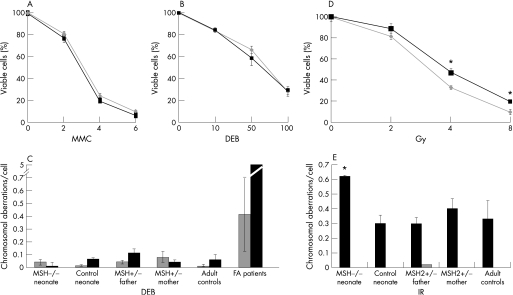
We assessed the survival of neonatal fibroblasts after treatment with ionising radiation (IR) and interstrand cross‐linking agents. PBLs from family members and controls were examined for chromosomal aberrations after exposure to IR. The survival of MSH2‐deficient fibroblasts in the presence of interstrand cross‐linking agents, diepoxybutane (DEB) and mitomycin C (MMC), did not differ significantly from controls (figure 2A,B). Furthermore, when PBLs were examined for chromosome abnormalities after exposure, no significant differences were seen between MSH2‐deficient cells and normal controls. Cells from individuals with Fanconi anaemia are highly sensitive to cross‐linking agents (reviewed by Kennedy and d'Andrea18) and in the same assay, PBLs from these patients showed increased DNA damage in response to DEB (figure 2C). We similarly assessed the effect of IR on cell survival and DNA integrity. A modest, but significant, loss of viability was evident in MSH2‐deficient fibroblasts (figure 2D) compared with control cells (p<0.001, t‐test). Consistent with this, an increase in chromosome damage after IR was also apparent in PBLs of the neonate homozygous for the MSH2 mutation (child 3), but not in PBLs from heterozygous family members, an age‐matched control or adult controls (figure 2E).
Figure 2 Sensitivity and DNA damage after exposure to interstrand cross‐linking agents and ionising radiation. Survival, expressed relative to untreated cells, of homozygous MSH2 226C→T fibroblasts (grey lines and diamonds) and control neonate fibroblasts (black squares and lines) after exposure to micromolar concentrations shown of (A) mitomycin C (MMC), (B) 1,2,3,4‐diepoxybutane (DEB) or (D) ionising radiation. Each experiment was performed in triplicate and the standard deviation about the mean for three experiments is shown. *Significant difference for cell viability between groups (p<0.001, t test). The average number of chromosome aberrations per cell in peripheral blood lymphocytes (PBLs) from family members and controls is shown in the absence of treatment (grey) or after exposure to DNA‐damaging agents (black). (C) 0.1 μg/ml DEB, (E) 3 Gy ionising radiation: 25 metaphase spreads were examined for each case and standard error about the mean for two experiments is shown for family member samples. The average and standard error is shown for aberrations/cell from PBLs of (C) 38 normal adult and 28 Fanconi anaemia patients, and (E) 10 normal adult controls. *Significant difference in chromosome aberrations after treatment (p<0.05, unpaired Mann‐Whitney test).
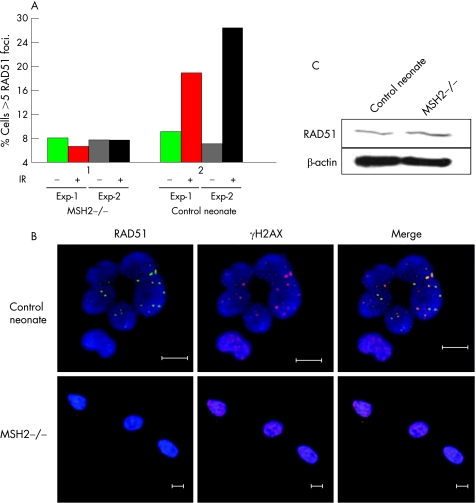
To establish whether DNA repair factors were perturbed in primary human MSH2‐deficient cells, we examined BRCA1 and RAD51 “foci” by microscopy. These areas are thought to represent sites of DNA‐damage repair. An hour after exposrue to 10 Gy IR, the expected increased localisation of RAD51 into foci was evident in control neonatal fibroblasts, but was not in the MSH2‐deficient cells (figure 3A and B). These observations were not due to loss of RAD51 expression, which was equally present in MSH2‐deficient and control cells (figure 3C), suggesting altered RAD51 formation in focal sites. No difference in recruitment of BRCA1 to foci was observed (data not shown).
Figure 3 Reduced DNA repair intermediates in MSH2‐deficient cells. (A) Percentage of untreated (grey) and irradiated (10 Gy) (black) MSH2‐deficient and age‐matched control fibroblasts exhibiting >5 RAD51 foci; 100 γH2AX positive nuclei per experiment were evaluated for the presence of RAD51 foci. (B) Representative immunohistochemical staining of MSH2‐deficient and control cells. The presence of γH2AX foci (red) was used as a marker of double‐stranded DNA damage. Immunostaining of RAD51 (ab213, Abcam) is shown in green and nuclei labelled with Hoechst stain are blue. Bar: 10 μm. (C) Immunoblot of RAD51 protein expression in MSH2‐deficient and age‐matched control fibroblasts. β‐actin loading control is shown.
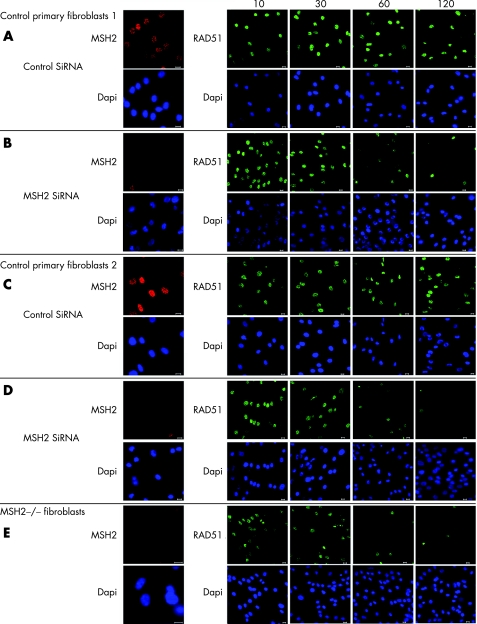
To test that the effect was not due to another gene mutation in the family under study, we next undertook MSH2 small interfering (si)RNA knock‐down in primary fibroblasts from two healthy neonates. In addition we examined RAD51 foci formation at various times after exposure to IR. In cells treated with control siRNA RAD51 foci were evident 10 minutes after irradiation and remained detectable after 2 hours (figure 4 A,C). In cells transfected with the MSH2 siRNA and in fibroblasts from child 3, RAD51 foci were also evident 10–30 minutes post‐irradiation. However after 1 hour, few cells still exhibited foci (figure 4 B,D,E). These data indicate that the kinetics of RAD51 foci formation after irradiation are regulated at least in part by MSH2.
Figure 4 Altered RAD51 foci kinetics in MSH2‐deficient primary fibroblasts after irradiation. Imunohistochemical examination of RAD51. Primary skin fibroblasts from an 18‐month‐old boy (A,B) and 16‐month‐old girl (C,D), where transfected for 96 hours with MSH2 or control siRNA (OnTarget; Dharmacon, Lafayette, Colorado, USA), according to the manufacturers' instructions. These cells, together with fibroblasts from child 3 (E), were then exposed to irradiation (10 Gy) and recovered for the time shown. They were then fixed in 4% paraformal‐dehyde, permeablised in 0.1% Triton‐X and incubated with anti‐MSH2 or anti‐RAD51 antibodies, appropriate secondary flourophore‐conjugated antibodies and stained for DNA with Hoechst‐33342 (shown in blue). All bars are 10 μm.
Discussion
The clinical features of the family in this study are consistent with previous case reports of human germline homozygosity or compound heterozygosity for MMR gene mutations. These features include café‐au‐lait spots and early onset of haematological malignancy or brain tumours, as reviewed by Ostergaard et al,19 who suggested that colorectal neoplasia might be the fully extended manifestation of biallelic MMR gene loss of function. The frequent occurrence of lymphoma in this group of patients is in agreement with observations of Msh2 knockout mice, which also develop early‐onset lymphomas.20,21
By examining primary cells from individuals of a rare pedigree, our data provide a new perspective in the controversy of differential mutagen sensitivity attributed to MMR deficiency. In our analysis of primary human material, passage numbers were kept at <10 to ensure tissue‐culture adaptation was restricted. We found that human MSH2 deficiency is not associated with overt sensitivity to DNA interstrand cross‐linking agents but is involved in the DNA damage response to IR. These data are also in agreement with observations from the cells of Msh2‐knockout mice, which exhibit sensitivity to DNA damage in response to IR, perturbed RAD51 foci formation8 and have aberrant homologous recombination.20,22 Together, these data suggest that lack of functional MSH2 results in constitutional IR sensitivity and deregulated homologous recombinational repair in mammalian cells. Hence the variability of response to DNA damaging agents in previous reports using cells of tumour origin or immortalised cell lines may be a result of further genetic or epigenetic lesions that lead to various levels of resistance or sensitivity to further agents.
Cells from individuals bearing mutations in specific DNA repair genes, for example in the Fanconi pathway and ATM, exhibit sensitivity to particular therapeutic agents (DNA interstrand cross‐linking agents and IR respectively). Here we have shown that normal primary cells from an individual lacking functional MSH2, exhibit increased sensitivity to IR. These findings suggest caution in the use of radiotherapy for treatment of malignancies arising in people with functional loss of MSH2 and possibly other MMR gene‐mutation backgrounds.
The supplementary figure can be viewed on the JMG website at http://jmg.bmj.com/supplemental
Acknowledgements
We thank the family for participating in this study, and C. Mathew, C. Whitehouse and D. Grimwade for critical reading of the manuscript. JRM is supported by MRC Grant No. G6900577, LP by the Generation Trust. JB and SH are supported by Cancer Research UK. AG, ZD and IK are supported by Special Trustees of Guy's and St Thomas' Hospital.
Abbreviations
DEB - diepoxybutane
IR - ionising radiation
MMC - mitomycin C
MMR - mismatch repair
MNU - N‐methyl‐N1‐nitrosourea
MSI - microsatellite instability
NHL - non‐Hodgkin lymphoma
PBL - peripheral blood lymphocyte
si - small interfering
Footnotes
Competing interests: None declared.
The first two authors contributed equally to this work.
Ethics approval was given by St George's Hospital Ethics Committee, London, UK.
The supplementary figure can be viewed on the JMG website at http://jmg.bmj.com/supplemental
References
- 1.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 200431091–1101. [DOI] [PubMed] [Google Scholar]
- 2.Davis T W, Wilson‐Van Patten C, Meyers M, Kunugi K A, Cuthill S, Reznikoff C, Garces C, Boland C R, Kinsella T J, Fishel R, Boothman D A. Defective expression of the DNA mismatch repair protein, MLH1, alters G2‐M cell cycle checkpoint arrest following ionising radiation. Cancer Res 199858767–778. [PubMed] [Google Scholar]
- 3.Bucci B, D'Agnano I, Amendola D, Citti A, Raza G H, Miceli R, De Paula U, Marchese R, Albini S, Felsani A, Brunetti E, Vecchione A. Myc down‐regulation sensitizes melanoma cells to radiotherapy by inhibiting MLH1 and MSH2 mismatch repair proteins. Clin Cancer Res 2005112756–2767. [DOI] [PubMed] [Google Scholar]
- 4.Aquilina G, Crescenzi M, Bignami M. Mismatch repair, G(2)/M cell cycle arrest and lethality after DNA damage. Carcinogenesis 1999202317–2326. [DOI] [PubMed] [Google Scholar]
- 5.O'Driscoll M, Martinelli S, Ciotta C, Karran P. Combined mismatch and nucleotide excision repair defects in a human cell line: mismatch repair processes methylation but not UV‐ or ionizing radiation‐induced DNA damage. Carcinogenesis 199920799–804. [DOI] [PubMed] [Google Scholar]
- 6.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, te Riele H, Nelson W G. Mouse embryonic stem cells carrying one or two defective Msh2 alleles respond abnormally to oxidative stress inflicted by low‐level radiation. Proc Natl Acad Sci U S A 19989511915–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritzell J A, Narayanan L, Baker S M, Bronner C E, Andrew S E, Prolla T A, Bradley A, Jirik F R, Liskay R M, Glazer P M. Role of DNA mismatch repair in the cytotoxicity of ionizing radiation. Cancer Res 1997575143–5147. [PubMed] [Google Scholar]
- 8.Franchitto A, Pichierri P, Piergentili R, Crescenzi M, Bignami M, Palitti F. The mammalian mismatch repair protein MSH2 is required for correct MRE11 and RAD51 relocalization and for efficient cell cycle arrest induced by ionizing radiation in G2 phase. Oncogene 2003222110–2120. [DOI] [PubMed] [Google Scholar]
- 9.Bignami M, Casorelli I, Karran P. Mismatch repair and response to DNA‐damaging antitumour therapies. Eur J Cancer 2003392142–2149. [DOI] [PubMed] [Google Scholar]
- 10.Kurosu K, Yumoto N, Rom W N, Takiguchi Y, Jaishree J, Nakata K, Tatsumi K, Mikata A, Kuriyama T, Weiden M D. Oligoclonal T cell expansions in pulmonary lymphoproliferative disorders: demonstration of the frequent occurrence of oligoclonal T cells in human immunodeficiency virus‐related lymphoid interstitial pneumonia. Am J Respir Crit Care Med 2002165254–259. [DOI] [PubMed] [Google Scholar]
- 11.Maquat L E. Nonsense‐mediated mRNA decay in mammals. J Cell Sci 20051181773–1776. [DOI] [PubMed] [Google Scholar]
- 12.Plevova P, Sedlakova E, Zapletalova J, Krepelova A, Skypalova P, Kolar Z. Expression of the hMLH1 and hMSH2 proteins in normal tissues: relationship to cancer predisposition in hereditary non‐polyposis colon cancer. Virchows Arch 2005446112–119. [DOI] [PubMed] [Google Scholar]
- 13.Shmueli O, Horn‐Saban S, Chalifa‐Caspi V, Shmoish M, Ophir R, Benjamin‐Rodrig H, Safran M, Domany E, Lancet D. GeneNote: whole genome expression profiles in normal human tissues. C R Biol 20033261067–1072. [DOI] [PubMed] [Google Scholar]
- 14.Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays 199416833–839. [DOI] [PubMed] [Google Scholar]
- 15.Whiteside D, McLeod R, Graham G, Steckley J L, Booth K, Somerville M J, Andrew S E. A homozygous germ‐line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe‐au‐lait spots. Cancer Res 200262359–362. [PubMed] [Google Scholar]
- 16.Bougeard G, Charbonnier F, Moerman A, Martin C, Ruchoux M M, Drouot N, Frebourg T. Early onset brain tumor and lymphoma in MSH2‐deficient children. Am J Hum Genet 200372213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohonen‐Corish M R, Daniel J J, te Riele H, Buffinton G D, Dahlstrom J E. Susceptibility of Msh2‐deficient mice to inflammation‐associated colorectal tumors. Cancer Res 2002622092–2097. [PubMed] [Google Scholar]
- 18.Kennedy R D, D'Andrea A D. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev 2005192925–2940. [DOI] [PubMed] [Google Scholar]
- 19.Ostergaard J R, Sunde L, Okkels H. Neurofibromatosis von Recklinghausen type I phenotype and early onset of cancers in siblings compound heterozygous for mutations in MSH6. Am J Med Genet A 200513996–105 discussion 96. [DOI] [PubMed] [Google Scholar]
- 20.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 199582321–330. [DOI] [PubMed] [Google Scholar]
- 21.de Wind N, Dekker M, van Rossum A, van der Valk M, te Riele H. Mouse models for hereditary nonpolyposis colorectal cancer. Cancer Res 199858248–255. [PubMed] [Google Scholar]
- 22.Elliott B, Jasin M. Repair of double‐strand breaks by homologous recombination in mismatch repair‐defective mammalian cells. Mol Cell Biol 2001212671–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]