Abstract
Background
Autism is a common childhood neurodevelopmental disorder with a possible genetic background. About 5–10% of autism cases are associated with chromosomal abnormalities or monogenic disorders. However, the role of subtle genomic imbalances in autism has not been delineated. This study aimed to investigate a hypothesis suggesting autism to be associated with subtle genomic imbalances presenting as low‐level chromosomal mosaicism.
Methods
We surveyed stochastic (background) aneuploidy in children with/without autism by interphase three‐colour fluorescence in situ hybridisation. The rate of chromosome loss and gain involving six arbitrarily selected autosomes and the sex chromosomes was assessed in the peripheral blood cells of 60 unaffected children and 120 children with autism.
Results
Of 120 analysed boys with autism, 4 (3.3%) with rare structural chromosomal abnormalities (46,XY,t(1;6)(q42.1;q27); 46,XY,inv(2)(p11q13); 46,XY,der(6),ins(6;1)(q21;p13.3p22,1)pat; and 46,XY,r(22)(p11q13)) were excluded from further molecular cytogenetic analysis. Studying <420 000 cells in 60 controls and 116 children with idiopathic autism, we determined the mean frequency of stochastic aneuploidy in control and autism: (1) autosome loss 0.58% (95% CI 0.42 to 0.75%) and 0.60% (95% CI 0.37 to 0.83%), respectively, p = 0.83; (2) autosome gain 0.15% (95% CI 0.09 to 0.21%) and 0.22% (95% CI 0.14 to 0.30%), respectively, p = 0.39; and (3) chromosome X gain 1.11% (95% CI 0.90 to 1.31%) and 1.01% (95% CI 0.85 to 1.17%), respectively, p = 0.30. A frequency of mosaic aneuploidy greater the background level was found in 19 (16%) of 116 children with idiopathic autism, whereas outlier values were not found in controls (p = 0.0019).
Conclusions
Our findings identify low‐level aneuploidy as a new genetic risk factor for autism. Therefore, molecular cytogenetic analysis of somatic mosaicism is warranted in children with unexplained autism.
Keywords: autism, aneuploidy, low‐level chromosomal mosaicism, molecular cytogentics, subtle genomic abnormalities
Autism is among the most devastating psychiatric disorders in childhood and is characterised by impaired social interaction and communication and restricted repetitive and stereotypic behaviour, interest and activities. Autism occurs more frequently in males (2–3/1000) with a male:female ratio estimated at around 4:1. Numerous biological and environmental factors with strong evidence for genetic influences have been suggested to play a role in the pathogenesis of autism. The diagnosis of autism is almost exclusively based on clinical examination, as diagnostic procedures using biologic or genetic markers are lacking. Identification of genetic factors underlying predisposition to autism is among current challenges in basic and applied medical science. In some cases, autism is associated with chromosomal abnormalities and monogenic disorders (so‐called “essential” autism), but in the majority of cases (90–95%), the cause of autism remains unexplained.1,2,3 A wide range of numerical and structural chromosome abnormalities (aneuploidy and large structural rearrangements, including translocations, deletions and duplications) visible in banding cytogenetic techniques has been reported in autistic patients.2,3,4 However, cryptic chromosomal imbalances including low‐level chromosomal mosaicism have not been thoroughly evaluated. Advances in genomics and molecular cytogenetics, such as microarray‐based comparative genomic hybridisation (array CGH), genotyping, genome scanning and fluorescence in situ hybridisation (FISH), have opened prospects for screening genomic (chromosomal) imbalances at unprecedented high resolution.4,5,6 However, the ability of these technologies to detect and evaluate low to medium levels of chromosomal mosaicism has neither been used or approved for studying idiopathic autism.
Mosaic aneuploidy is a relatively frequent occurrence in genetic disorders, even though subtle mutant cell lineages are rather difficult to detect.7 Moreover, low and medium levels of somatic chromosomal mosaicism are usually overlooked, probably owing to them usually being considered insignificant because of an apparent lack of phenotypic effect.8 Chromosomal abnormalities and numerous genetic and epigenetic mechanisms leading to somatic mosaicism can cause subtle alterations in gene expression and contribute to disease phenotype.9 Recently, it was hypothesised that mosaic aneuploidy affecting the brain rather than other tissues could be involved in the pathogenesis of common mental illnesses such as schizophrenia and autism.7 However, experimental proof of this hypothesis still requires the analysis of cryptic somatic mosaicism in different tissues to delineate the role it plays in intercellular diversity and human disease. To fill this gap in the knowledge concerning the effect of intercellular genomic variations in health and disease, we addressed the frequency of mosaic aneuploidy in peripheral blood lymphocytes of normal controls and children with unexplained autism using interphase three‐colour FISH assay.10,11,12
Methods
Children with autism (all boys) were drawn from a general clinical population at the Department of Clinical Genetics, Institute of Pediatrics and Pediatric Surgery, Moscow and the Department of Child Psychiatry, Group of Autism and Rett syndrome, Research Center of Mental Health, RAMS, Moscow, Russia. Ethics committee approval and informed consent from the families of the children were obtained. Two child psychiatrists performed the diagnosis of autism according to diagnostic behavioural criteria from the Diagnostic and statistical manual of mental disorders, 4th edition (DSM‐IV)13 and results from the Autism Diagnostic Interview (ADI‐R).14 Children with clinically recognised “essential” autism associated with chromosomal syndromes, fragile X syndrome and Rett syndrome (confirmed by molecular genetic analyses) were excluded from the study. No affected siblings were identified in the cohort of autistic children (>85% of families analysed had only one child; non‐affected children from these families were not studied). Thus, cytogenetic and molecular cytogenetic studies were carried out on a cohort of 120 children with idiopathic autism (mean age 7 years) and 60 unaffected controls (mean age 8 years).
All cytogenetic preparations were made from cultured peripheral blood cells using established protocols. Standard G‐banding karyotyping at the 550 band level and analysis of 20–30 metaphases were applied. A three‐colour FISH protocol and the criteria for aneuploidy scoring and quantification of FISH signals applied were used as detailed previously.10,11,12 Samples were assessed for aneuploidy with chromosome‐enumeration DNA probes for arbitrary selected autosomes (chromosomes 1, 9, 15–18) and the sex chromosomes, obtained from the original DNA probe collection.10,15 Specific probes for chromosomes 1 (D1Z1), 9 (D9Z1), 15 (D15Z1), 16 (D16Z3), 17(D17Z1), 18 (D18Z1), X (DXZ1) and Y (DYZ3), labelled either with FluorX (green), Cy3 (red), or diethylaminocoumarine (blue) were used. The probe combinations were 1+X+Y, 1+9+16, 1+15+17 and 1+9+18. For each sample/chromosome, no fewer than 500 nuclei were scored in double‐blinded interphase FISH studies (no fewer than 3000 nuclei per autosome were scored in each child from cohorts of 60 controls and 60 boys with autism). In total, no fewer than 3000 nuclei per 6 autosomes were analysed for each sample. The mean frequency, 95% CI and the cut‐off levels for outlier values (mean (3SD)) for autosomes were determined for 60 controls and 60 randomly selected affected children (altogether, 360 000 nuclei were scored for each cohort of controls and boys with autism). The same parameters for scoring of aneuploidy were determined for the sex chromosome for the controls (n = 60) and all children with idiopathic autism (n = 116), making an additional 120 000 nuclei scored (total number of nuclei scored was thus around 420 000). Outlier frequencies of aneuploidy over the cut‐off (threshold) value were considered to be cases of low‐level chromosome‐specific mosaicism. Comparison between groups was performed with the Yates‐corrected χ2 test.
Results
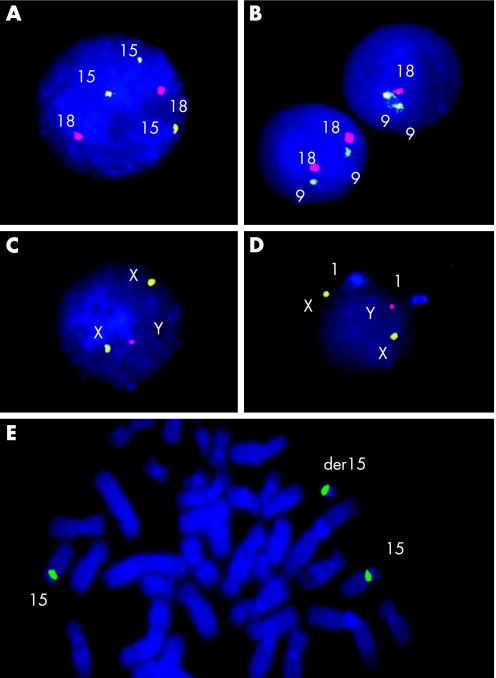
G‐banding revealed normal karyotypes in all controls (n = 60) and in the 116 children with presumably idiopathic autism. Of 120 boys with autism, 4 (3.3%) had the following rare structural chromosomal abnormalities: 46,XY,t(1;6)(q42.1;q27); 46,XY,inv(2)(p11q13); 46,XY,der(6),ins(6;1)(q21;p13.3p22,1)pat; and 46,XY,r(22)(p11q13). These cases were excluded from FISH studies as having “essential” autism associated with non‐mosaic chromosomal rearrangements. The rate of stochastic (background) autosome losses for in controls (n = 60; 30 000 nuclei scored) was 0.3–0.7% with cut‐off levels of 1.0–2.7%. The rate of stochastic autosome gains for in controls (n = 60; 30 000 nuclei scored) was 0.1–0.2% with cut‐off levels of 0.4–0.8%. The mean frequencies of stochastic losses and gains of autosomes in peripheral lymphocytes in the control group (n = 60; 180 000 nuclei scored for six different autosomes) were 0.58% (95% CI 0.42–0.75%) and 0.15% (0.09–0.21%), respectively. Chromosome X loss (nullisomy) was not registered. Chromosome X gain (disomy X) was registered with the frequency of 1.1% (0.9–1.3%) with a cut‐off level of 3.7% (n = 60; 30 000 nuclei scored). No outliers for chromosome losses and gains were detected in controls (table 1). Figure 1 illustrates FISH analysis of interphase nuclei and metaphase chromosomes from cultured peripheral blood lymphocytes of boys with autism, demonstrating mosaic aneuploidy.
Table 1 The frequency of low‐level aneuploidy (%) in controls and children with autism.
| Chromosome losses and gains | Stochastic aneuploidy in controls | Stochastic aneuploidy in autism | Low‐level mosaic aneuploidy in autism (outliers) | ||
|---|---|---|---|---|---|
| Mean (95%CI) | Cut‐off level | Mean (95%CI) | Cut‐off level | ||
| 1 loss | 0.7 (0.4–1.0) | 2.7 | 0.6 (0.4–0.8) | 1.5 | No |
| 1 gain | 0.1 (0.1–0.2) | 0.4 | 0.1 (0.1–0.2) | 0.4 | No |
| 9 loss | 0.5 (0.3–0.7) | 1.7 | 0.6 (0.3–0.8) | 1.8 | No |
| 9 gain | 0.2 (0.1–0.2) | 0.5 | 0.3 (0.2–0.5) | 0.9 | Trisomy 9 (2.1; 3.1) |
| 15 loss | 0.7 (0.4–0.9) | 1.8 | 0.6 (0.3–0.9) | 1.7 | Monosomy 15 (2.2; 2.6; 4.6) |
| 15 gain | 0.2 (0.1–0.3) | 0.8 | 0.2 (0.1–0.3) | 0.5 | Trisomy 15 (1.2) |
| 16 loss | 0.6 (0.3–0.8) | 2.1 | 0.4 (0.2–0.6) | 1.3 | Monosomy 16 (3.0) |
| 16 gain | 0.1 (0.1–0.3) | 0.4 | 0.2 (0.1–0.3) | 0.5 | No |
| 17 loss | 0.3 (0.1–0.2) | 1 | 0.4 (0.3–0.6) | 1.3 | No |
| 17 gain | 0.1 (0.1–0.2) | 0.4 | 0.2 (0.1–0.2) | 0.5 | No |
| 18 loss | 0.7 (0.3–0.9) | 1.6 | 1.0 (0.4–1.3) | 2.2 | Monosomy 18 (2.7) |
| 18 gain | 0.2 (0.1–0.3) | 0.8 | 0.3 (0.2–0.4) | 0.9 | No |
| X gain | 1.1 (0.9–1.3) | 3.7 | 1.0 (0.8–1.2) | 3.6 | Disomy X (4.0; 4.3; 4.5; 4.8; 5.6; 5.9; 6.2; 7.0; 8.5; 11.0; 11.6) |
| Y loss | 0.2 (0.0–0.3) | 0.5 | 0.2(0.0–0.3) | 0.8 | No |
| Y gain | 0.5 (0.3–0.7) | 1.5 | 0.4 (0.2–0.7) | 2 | No |
Figure 1 FISH with chromosome‐enumeration DNA probes in autism: (A) nucleus characterised by trisomy 15 (three green signals) and two copies of chromosome 17 (two red signals). (B) A nucleus with monosomy 18 (one red signal) and a normal nucleus with disomy 18 (two red signals). Two chromosomes 9 are present in each nuclei (two green signals). (C) A nucleus with disomy X (two green signals) and one chromosome Y (one red signal); (D) a nucleus with disomy X (two green signals), two chromosomes 1 (two light blue signals) and one chromosome Y (one red signal); (E) metaphase with additional chromosome der(15) and two normal chromosomes 15 (green signals at the centromeric regions of chromosomes 15).
The rate of stochastic aneuploidy for autosomes in idiopathic autism (n = 60; 30 000 nuclei were analysed for each autosome) was 0.4–1.0% (cut‐off levels 1.2–2.2%) for losses and 0.1–0.3% (cut‐off levels 0.4–0.9%) for gains. Eight outliers (low‐level chromosomal mosaicism) were detected for autosome losses and gains in children with autism. The mean frequencies of autosome losses and gains calculated for autism (n = 60; 180 000 nuclei scored for six different autosomes) were 0.60% (95% CI 0.37–0.83%) and 0.22% (0.14–0.30%), respectively. In total, 11 outliers for chromosome X gain (or low‐level chromosomal mosaics) with frequency >3.7% (the threshold level determined in the control group for chromosome X gain) were detected in the group of children with autism (table 1). Chromosome X gain (disomy X) in autism without outliers had a mean frequency of 1.0% (95%CI 0.8–1.2%) with cut‐off level of 3.6% (n = 101; 50 500 nuclei scored).
In total, we found 19 cases with outlier aneuploidy frequencies for autosomes (8 cases with gains of chromosomes 9 and 15 and losses of chromosomes 15, 16 and 18) and chromosome X (11 cases with chromosome X gain) among 116 males with idiopathic autism. These cases were considered to be low‐level mosaicism. Low‐level aneuploidy involving chromosomes 1, 17 and Y was not found, indicating that mosaic aneuploidy in autism is chromosome‐specific and the highest prevalence seems to be that of chromosome X gain. Estimated frequency of cases with mosaic aneuploidy (16%) in autistic children differed significantly from that in the controls (p<0.01; Yates corrected χ2 test). The rates of stochastic autosome losses and gains and of chromosome X gains (without outliers) was similar in the control and autism groups (p = 0.83, p = 0.39 and p = 0.30, respectively; t test for independent groups).
The scoring of aneuploidy by FISH with chromosome enumeration DNA probes also allowed the detection of mosaic aneuploidy in metaphase spreads; however, it was difficult to score a representative number of metaphase spreads for precise statistical evaluation of all autism cases. It should be noted that scoring of a standard number of metaphase spreads (n = 20–30) did not allow us to prove chromosomal mosaicism presence in all controls (n = 60) and children with autism (n = 120). Nevertheless, in a case of low‐level trisomy 9 detected by interphase FISH (2.1% with threshold level 0.5–09%), we found that all trisomic metaphase spreads (4 from 120 screened) had altered additional chromosome 9 or der(9)(pter→9q32:). Another case of low‐level mosaic trisomy involving chromosome 15 (frequency 1.2%, threshold level 0.5%) had a cell line with supernumerary chromosome der(15) (figure 1E). This marker chromosome der(15) was found in three metaphase spreads after extended analysis of 100 additional metaphases. These two cases refine the accuracy in detection of low‐level mosaic aneuploidy by interphase FISH with moderate scoring of 500 nuclei even with a low percentage of abnormal cells (<3% of chromosome gains). In contrast, the cases of low‐level XXY/XY mosaicism ascertained via FISH studying of around 100 metaphase spreads per case showed aneuploidy to involve the whole X chromosome. Although the frequency of additional chromosome X in metaphase spreads was lower in the interphase FISH study (not more than two 47,XXY metaphases per case), this allowed us to confirm these cases as low‐level XXY/XY mosaicism. The discrepancy of frequencies between interphase and metaphase surveys was probably produced either by different cell scoring numbers or by growth disadvantage of the aneuploid cell line during cell cultivation.
Discussion
Mosaicism refers to the presence of >1 genetically distinct cell line in an organism. Somatic mosaicism has emerged as important factor contributing to phenotypic variability and disease. It has been implicated in >30 monogenic disorders of variable expressivities. At the level of the whole organism, the mosaic phenotype or genotype depends on tissue to tissue variations that do not follow mendelian rules of inheritance.8 Mosaicism in an organism may not be apparent unless closely analysed. However, evaluation techniques of mosaic composition in an organism are scarce. Therefore, the role of genetic mosaicism appears to be underestimated. This seems to be of special importance for complex disorders, which are suggested to be the result of an interaction between genetic and environmental factors.
Somatic chromosomal mosaicism is commonly observed in humans. Chromosomal mosaic aneuploidy was documented in 50% of human preimplantation embryos, 25% of spontaneous miscarriages at 5–12 weeks' gestation and 1–2% of viable pregnancies.7,11 The gradual decrease in aneuploidy levels during the prenatal period suggests that there is substantial darwinian selection against aneuploid cells. Absence of selective pressure may lead to survival of mosaic cell populations. Intercellular chromosomal variations caused by stochastic errors in mitotic checkpoint machinery can be observed appearing as low‐level mosaic aneuploidy in virtually all mitotic and postmitotic cell populations.7,16 To date, however, the overall percentage of aneuploid cells (or the integral level of background aneuploidy) in human somatic tissues is unknown, owing to the absence of commonly approved approaches to identification of true low‐level mosaicism. Interphase FISH is considered one of the most efficient techniques of aneuploidy scoring at the cellular level, allowing aneuploidy detection in cells accounting for <1% of the total cell population.7,16 Chromosomal mosaicism can be analysed by other techniques, but probably with much lower efficiency than with interphase FISH. For instance, array CGH can detect chromosomal mosaicism with 15–30% of abnormal clone content.5 In this context, accurate delineation of a borderline between background and pathogenic levels of mosaic aneuploidy by alternative approaches is an essential condition for launching systematic studies of somatic mosaicism in health and disease.
The presence of mosaic chromosome abnormalities is documented in children with syndromic autism. The latter usually manifests as mosaic structural chromosome abnormalities and marker chromosomes affecting a significant proportion of cells studied.2,3,4 To our knowledge, cases of idiopathic autism associated with low‐level mosaicism presenting as chromosomal aneuploidy, such as those reported here, have not been previously described. The lack of data concerning low‐level mosaic aneuploidy in idiopathic autism can be explained by the fact that related studies are usually performed through classic cytogenetic evaluation or array CGH techniques, which are less efficient for mosaicism studies. Furthermore, no commonly accepted criteria for definition of chromosomal mosaicism in somatic human tissues are available.7 Therefore, to identify chromosomal mosaicism, the investigator must first define these criteria. Our data, showing for the first time that low‐level chromosomal mosaicism occurs at high frequency in children with unexplained autism, suggest that low‐level mosaicism does have clinical manifestations.
Around 70% of children with autism have learning difficulties,3 but the genetic causes correlate poorly for unexplained autism and mental retardation, especially when autism cases associated with chromosome abnormalities are not taken into account. Cryptic chromosomal mosaicism in children with unexplained mental retardation has not been yet evaluated, mainly because of the difficulties associated with the identification. It is interesting to make an arbitrary parallel with other type of subtle chromosomal imbalances, which are also difficult to evaluate by conventional cytogenetics, namely, subtelomeric deletions and duplications. These abnormalities occur with a frequency of 6–7% among children with unexplained mental retardation.17 To date, >2500 subjects with mental retardation have been tested and the incidence of subtelomeric rearrangements appears to be around 5%.18 Recently, two independent screening studies of subtelomeric rearrangements in 104 and 72 individuals with autism, respectively, found a lack of subtelomeric anomalies, suggesting an insignificant association with autism.19,20 However, array CGH shows the presence of cryptic chromosomal rearrangements (but not subtelomeric or mosaic chromosome rearrangements) in syndromic autism spectrum disorders.6 Studies of low‐level mosaicism might be more successful, as intercellular genomic variations can produce exceedingly diverse phenotypes, such as the spectrum found in both autism and mental retardation. We consider that studies of cryptic mosaicism or tissue‐specific mosaicism in idiopathic autism and mental retardation offer a new perspective for medical genetics of childhood disorders. Thus, low‐level mosaic aneuploidy may be considered as a new diagnostic and therapeutic target for disorders of mental development in childhood.
Autism is recognised as a disorder of prenatal and postnatal brain development. The pathogenesis of the disease assumed to be related to dysfunction in the amygdala, hippocampus and related cortical structures. The cerebellum may be involved in a widely distributed neuronal network responsible for social cognition and communication.1,2,3 Large‐scale genomic alterations such as loss or gain of whole chromosomes (aneuploidy) have been described in cells of the normal and diseased human brain.7,12,21 Intercellular genomic (chromosomal) variations in the brain are likely to be an important mechanism underlying generation of neuronal diversity, complexity and brain disease susceptibility.7,22 Aneuploidy simultaneously involves hundreds or even thousands of genes, and therefore produces more dramatic alterations in neuronal physiology and brain circuitry than other types of genetic instabilities. The human brain is largely composed of postmitotic differentiated cells, including 95–100 billion neurons. A typical human mature neuron has approximately 5000–200 000 synapses.22 Aneuploid neurons may be integrated into the neuronal network and, theoretically, even low levels of chromosomally abnormal neurons should negatively affect the functional activity of the brain. This speculation is in accordance with pilot data suggesting that at least some cases of Alzheimer's disease and schizophrenia are probably associated with an increased level of chromosome imbalances in the brain. Mosaic aneuploidy in the brain, therefore, could be involved in the pathogenesis of mental disorders.7 Therefore, subsequent molecular cytogenetic studies of autism should investigate not only chromosomal mosaicism in tissues usually used for cytogenetic analysis (cultured peripheral blood lymphocytes), but also mosaic aneuploidy in postmitotic neuronal and glial cells in the brain.
Key points
Autism is a devastating psychiatric disorder in childhood, frequently associated with chromosomal abnormalities, but subtle chromosome imbalances (including low‐level mosaicism) have not been evaluated to date.
Chromosomal mosaicism was investigated in the peripheral blood cells of 60 unaffected children and 116 children with idiopathic autism, and low‐level mosaic aneuploidy was found to affect 16% (19 of 116) of children with idiopathic autism.
The findings propose low‐level aneuploidy as a new genetic risk factor for autism.
Acknowlegements
The first three authors contributed equally to this work. This work was supported by grant from INTAS (03‐51‐4060) and RGNF (060600639a, Russian Federation).
Abbreviations
ADI‐R - Autism Diagnostic Interview, revised
CGH - comparative genomic hybridisation
DSM‐IV - Diagnostic and statistical manual of mental disorders, 4th edition
FISH - fluorescence in situ hybridisation
Footnotes
Competing interests: None declared.
References
- 1.Volkmar F R, Pauls D. Autism. Lancet 20033621133–1141. [DOI] [PubMed] [Google Scholar]
- 2.Castermans D, Willquet V, Steyert J, Van de Ven W, Fryns J P, Devriendt K. Chromosomal anomalies in individuals with autism: a strategy towards the identification of genes involved in autism. Autism 20048141–161. [DOI] [PubMed] [Google Scholar]
- 3.Vorstman J A S, Staal W G, van Daalen E, van Engeland H, Hochstenbach P F, Franke L. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry 20061118–28. [DOI] [PubMed] [Google Scholar]
- 4.Xu J, Zwaigenbaum L, Szatmari P, Scherer S W. Molecular cytogenetics of autism. Curr Genomics 20044347–368. [Google Scholar]
- 5.Ballif B C, Rorem E A, Sundin K, Lincicum M, Gaskin S, Coppinger J, Kashork C D, Shaffer L G, Bejjani B A. Detection of low‐level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet 20061402757–2767. [DOI] [PubMed] [Google Scholar]
- 6.Jacquemont M L, Sanlaville D, Redon R, Raoul O, Cormier‐Daire V, Lyonnet S, Amiel J, Le Merrer M, Heron D, de Blois M C, Prieur M, Vekemans M, Carter N P, Munnich A, Colleaux L, Philippe A. Array‐based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet 200643843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iourov I Y, Vorsanova S G, Yurov Y B. Chromosomal variations in mammalian neuronal cells: known facts and attractive hypotheses. Int Rev Cytol 2006249143–191. [DOI] [PubMed] [Google Scholar]
- 8.Youssoufian H, Pyeritz R. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet 20023748–758. [DOI] [PubMed] [Google Scholar]
- 9.Gottlieb B, Beitel L K, Trifiro M A. Somatic mosaicism and variable expressivity. Trends Genet 20011779–82. [DOI] [PubMed] [Google Scholar]
- 10.Yurov Y B, Soloviev I V, Vorsanova S G, Marcais B, Roizes G, Lewis R. High resolution fluorescence in situ hybridization using cyanine and fluorescein dyes: ultra‐rapid chromosome detection by directly fluorescently labeled alphoid DNA probes. Hum Genet 199697390–398. [DOI] [PubMed] [Google Scholar]
- 11.Vorsanova S G, Kolotii A D, Iourov I Y, Monakhov V V, Kirillova E A, Soloviev I V, Yurov Y B. Evidence for high frequency of chromosomal mosaicism in spontaneous abortions revealed by interphase FISH analysis. J Histochem Cytochem 200553375–380. [DOI] [PubMed] [Google Scholar]
- 12.Yurov Y B, Iourov I Y, Monakhov V V, Soloviev I V, Vostrikov V M, Vorsanova S G. The variation of aneuploidy frequency in the developing and adult human brain revealed by an interphase FISH study. J Histochem Cytochem 200553385–390. [DOI] [PubMed] [Google Scholar]
- 13.American Psychiatric Association Diagnostic and statistical manual of mental disorders: 4th ed. Washington DC: American Psychiatric Association, 2000
- 14.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview‐Revised: a revised version of a diagnostic interview for care givers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 199424659–685. [DOI] [PubMed] [Google Scholar]
- 15.Yurov Y B, Vorsanova S G, Soloviev I V, Demidova I A, Alexandrov I A, Sharonin V O, Beresheva A K. Original collection of DNA probes for preimplantational, fetal prenatal and postnatal diagnosis of chromosomal anomalies by FISH. In: Macek M, Bianchi D, Cucle H, eds. Early prenatal diagnosis, fetal cells and DNA in the mother. Present state and perspectives Prague: Charles University in Prague, 2002
- 16.Iourov I Y, Vorsanova S G, Yurov Y B. Intercellular genomic (chromosomal) variations resulting in somatic mosaicism: mechanisms and consequences. Curr Genomics 20067435–446. [Google Scholar]
- 17.Knight SJL Regan R, Nicod A, Horsley S W, Kearney L, Homfray T, Winter R M, Bolton P, Flint J. Subtle chromosomal rearrangement in children with unexplained mental retardation. Lancet 19993541676–1681. [DOI] [PubMed] [Google Scholar]
- 18.De Vries B B, Winter R, Schinzel A, van Ravenswaaij‐Arts C. Telomeres: a diagnosis at the end of the chromosomes. J Med Genet 200340385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassink T H, Losh M, Piven J, Sheffield V C, Ashley E, Westin E R, Patil S R. Systematic screening for subtelomeric anomalies in a clinical sample of autism. J Autism Dev Disord 200737703–708. [DOI] [PubMed] [Google Scholar]
- 20.Battaglia A, Bonaglia M C. The yield of subtelomeric FISH analysis in the evaluation of autistic spectrum disorders. Am J Med Genet 20061428–12. [DOI] [PubMed] [Google Scholar]
- 21.Iourov I Y, Liehr T, Vorsanova S G, Kolotii A, Yurov Y B. Visualization of interphase chromosomes of the human brain by multicolour banding (MCB). Chromosome Res 200614223–229. [DOI] [PubMed] [Google Scholar]
- 22.Muotri A R, Gage F H. Generation of neuronal variability and complexity. Nature 2006441903–910. [DOI] [PubMed] [Google Scholar]