Abstract
Background
Smith‐Magenis syndrome (SMS) is rare (prevalence 1 in 25 000) and is associated with psychomotor delay, a particular behavioural pattern and congenital anomalies. SMS is often due to a chromosomal deletion of <4 Mb at the 17p11.2 locus, leading to haploinsufficiency of numerous genes. Mutations of one of these gemes, RAI1, seems to be responsible for the main features found with heterozygous 17p11.2 deletions.
Methods
We studied DNA from 30 patients with SMS using a 300 bp amplimers comparative genome hybridisation array encompassing 75 loci from a 22 Mb section from the short arm of chromosome 17.
Results
Three patients had large deletions (10%). Genotype–phenotype correlation showed that two of them had cleft palate, which was not found in any of the other patients with SMS (p<0.007, Fisher's exact test). The smallest extra‐deleted region associated with cleft palate in SMS is 1.4 Mb, contains <16 genes and is located at 17p11.2‐17p12. Gene expression array data showed that the ubiquitin B precursor (UBB) is significantly expressed in the first branchial arch in the fourth and fifth weeks of human development.
Conclusion
These data support UBB as a good candidate gene for isolated cleft palate.
Keywords: Smith‐Magenis, cleft palate, CGH varray
Smith‐Magenis syndrome (SMS) [MIM 182290] is a rare (prevalence 1 in 25 000) syndrome exhibiting multiple congenital anomalies and mental retardation, with distinctive behavioural characteristics, sleep disturbance and dysmorphic features, associated with a heterozygous interstitial deletion of chromosome 17p11.2.1,2,3 Most patients have the same interstitial genomic deletion of around 4 Mb at chromosome 17p11.2, comprising 20 expressed genes.4,5,6 Heterozygous frameshift mutations of the RAI1 gene, leading to protein truncation, is likely to be responsible for the majority of the SMS features, but other deleted genes in the SMS region may modify the overall phenotype in the patients with 17p11.2 deletions.7,8,9 In this study, we report comparative genome hybridisation (CGH) analysis of the short arm of chromosome 17 in patients with SMS.
Methods
Patients
Patients with SMS were recruited by voluntary participation of members of the French Smith‐Magenis Association (www.smithmagenis.com). All patients with SMS were diagnosed using commercial fluorescence in situ hybridisation probes (Oncor Inc., Gaithersburg, Maryland, USA and Vysis, Inc., Downers Grove, Illinois, USA) encompassing FLII. After informed consent was given, blood samples were obtained from 30 patients.
DNA preparation
Genomic DNA for CGH arrays was isolated from leucocytes from peripheral blood samples of each patient using genomic DNA columns (Qiagen Inc., Valencia, California, USA). DNA concentration was evaluated using a spectrophotometer (Nanodrop, Wilmington, Delaware, USA). Reference DNA was obtained in the same condition from a pool of 15 healthy men and women without cytogenetic abnormalities.
Design of the probes
We have made the choice of an “in silico” design based directly on an extraction of DNA sequences in the region of interest directly obtained from the Human Genome Browser Gateway (University of California Santa Cruz, Santa Cruz, California, USA; 24 April 2007; http://genome.ucsc.edu/cgi‐bin/hgGateway?org = human) and a PCR‐based production of the probes. For each of these sequences, primers were designed to obtain a nested PCR DNA fragment of 250–400 bp. This PCR fragment was tested to verify lack of redundancy, homology, freedom form repeats and specificity for the hybridisation. This process was fully automated design using bespoke software (available on our website at http://genopole‐lille.fr) which combines BLAT results (http://www.genome.ucsc.edu/cgi‐bin/hgBlat), identification of non‐redundant sequences and absence of primer crosshybridisation.
All the primers had an identical melting point (mean (SD) 60 (1)°C), in order to have one GC clamp so that all primers could be used under the same PCR conditions. The 75 loci and the primers used are listed in a supplementary file available online at http://jmg.bmj.com/supplemental.
Array construction
PCR DNA products after purification were spotted at a concentration of 100 ng/µl in 3× SSC in triplicate onto Telechem Superamine glass slides by using a microarrayer (Eurogridderd; Eurogentec, Seraing, Belgium). The slides were heated at 80°C for 10 min, and DNA fragments were crosslinked by UV light (2 × 150 mJ). Finally, the slides were stored at room temperature after denaturation (2 minutes in boiling water).
Array hybridisation, DNA test and reference labelling
The PCR primers allowed us to selectively amplify the region of interest in the test and reference DNA before the labelling to obtain more efficient hybridisation kinetics and a low complexity target. Very low amounts of genomic DNA (15 ng), were used in each multiplex. Test and reference muliplex‐obtained DNA were fluorescently labelled by incorporation of CyTm3‐conjugated and CyTm5‐conjugated dUTP (Amersham Biosciences, Amersham, Buckinghamshire, UK) by random priming (Bioprime DNA labelling system; Invitrogen, Carlsbad, California, USA). Unincorporated nucleotides were removed using column centrifugation (YM30 Microcon membrane; Millipore, Billerica, Massachusetts, USA). For hybridisation, labelled test DNA and reference DNA in a ratio of 1:1 were coprecipitated and resolved in 200 µl of hybridisation buffer (Chipspread; Ventana, Tucson, Arizona, USA). After denaturation (85°C for 5 min), the mixture was hybridised for 8 h at 42°C then slides were washed automatically three times at 37, 50 and 55°C in washing buffer (Ventana). Finally, slides were dried by centrifugation at 100 g for 5 min at 2500 rpm.
Image and data processing
Arrays were scanned using an 428 GMS scanner (MWG, Affymetrix, Santa Clara, California, USA). Fluorescent intensities were extracted after subtraction of local background using Jaguar software V.2.0. In addition, probes that we had identified in previous control hybridisations as inducing consistently weak signals or irreproducible or aberrant ratios were excluded from the analysis. The data normalisation was performed using an automatic algorithm added in the R package MANOR (Micro‐Array NORmalisation; http://bioinfo‐out.curie.fr/projects/manor, which is available from http://www.bioconductor.org). To determine gain and loss segments we used a hidden Markov model (HMM)‐based method that assigns probes to different state (loss, normal or gain).10 The HMM outputs are state medians weighted by the estimated probability of being in each state.
Results
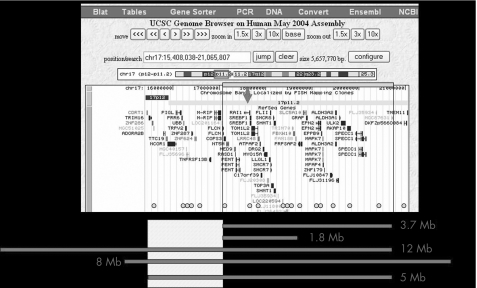
We studied deletion sizes from 30 patients with SMS using the 300 bp amplimer CGH array, encompassing 75 loci on a 22 Mb segment from the short arm of chromosome 17 (17 Mb). Medium resolution of this array was 300 kb. Three patients showed larger deletions (5, 8 and 12 Mb), and one also showed a smaller deletion (1.8 Mb) (fig 1). Genotype–phenotype correlation including mental retardation, craniofacial and skeletal anomalies, and behavioural abnormalities, showed that two of these three patients had cleft palate, which was not found in any other patient with SMS (p<0.007, Fisher's exact test).
Figure 1 Genes in the 17p11.2–17p12 region (Human Genome Browser, http://genome.ucsc.edu/cgi‐bin/hgGateway?org = human). Thick lines are the estimated locations of the deletions compared with the Genome Browser gene position. Spots are the estimated locations of the CGH array amplimer probes. The empty square is the estimated location of the smallest SMS extra‐deleted region.
The smallest extra‐deleted region associated with cleft palate in SMS is 1.4 Mb (chr17:15 637 376 to 17 052 919). The telomeric breakpoint of this deletion was studied using fluorescence in situ hybridisation (FISH) analysis of bacterial and P1‐derived artificial chromosomes and was estimated to be between chr17:15 700 000 and chr17: 15 800 000 (table 1). This region contains <16 genes and is located at the 17p11.2–17p12 junction. The first branchial/pharyngeal arch is a conserved embryonic structure that develops into the palate and jaw. Expression array data from the first branchial arch at the fourth and fifth weeks of human development allows the study of 7 of these 16 genes on an Affymetrix expression array (between ADORA2B and MRIP). These gene expression array data showed that only the ubiquitin B precursor (UBB) is significantly expressed in the first branchial arch at at the fourth and fifth weeks of human development (table 2). No such data were available for the nine other genes.
Table 1 Results of the fluorescence in situ hybridisation of bacterial and P1‐derived artificial chromosomes (BACs/PACs) for the smallest cytogenetic spreads associated with cleft palate in patients with CMS.
| BAC/PAC | Location | Molecular position | FISH result | |
|---|---|---|---|---|
| RP11‐90G21 | 17p12 | 15 321 344 | 15 511 681 | Non‐deleted |
| RP11‐304M13 | 17p12 | 15 362 705 | 15 560 308 | Non‐deleted |
| RP11‐273K13 | 17p12 | 15 398 210 | 15 560 308 | Non‐deleted |
| RP1‐121M24 | 17p12 | 15 637 376 | 15 823 053 | Non‐deleted, asymmetrical signal |
| RP11‐459E6 | 17p12‐17p11.2 | 15 770 053 | 15 929 608 | Deleted |
| RP1‐188B3 | 17p11,2 | 15 995 972 | 16 077 445 | Deleted |
| RP11‐138I1 | 17p11.2 | 16 123 263 | 16 294 186 | Deleted |
| RP1‐77H15 | 17p11.2 | 16 289 474 | 16 438 473 | Deleted |
Table 2 Expression array data from the first branchial/pharyngeal arch at the fourth and fifth weeks of human development.
| Gene name | Probe set | 4th week, 1st pharyngeal arch | 5th week, 1st pharyngeal arch | ||
|---|---|---|---|---|---|
| Call | Log average * | Call | Log average* | ||
| ADORA2B | 36324_at | A/A | 234/−301 | A/A | 7/92 |
| TTC19 | ND | ND | ND | ND | ND |
| NCOR1 | ND | ND | ND | ND | ND |
| PIGL | 34496_at | A/A | 1691/−301 | A/A | 1968/296 |
| PRR6(p30) | ND | ND | ND | ND | ND |
| UBB | 1323_at | P/P | 151075/92691 | P/P | 45433/79534 |
| TRPV2 | ND | ND | ND | ND | ND |
| MGC40157 | 39077_at | A/A | 919/1372 | A/A | 316/676 |
| FLJ35696 | ND | ND | ND | ND | ND |
| LOC440403 | ND | ND | ND | ND | ND |
| ZNF287 | ND | ND | ND | ND | ND |
| ZNF624 | ND | ND | ND | ND | ND |
| KIAA0565 | 35535_f_at | A/A | 170/−39 | A/A | 60/−42 |
| LOC96597 | ND | ND | ND | ND | ND |
| TNFRSF13B | 31410_at | ND | 910/−2177 | A/A | 245/386 |
| MRIP | 38731_at | A/A | 191/−302 | A/A | 25/−101 |
ND, data not determined.
*Positive/negative.
Discussion
Smaller or larger deletions were seen in around 12% and around 10% of patients with SMS, respectively.11 Genotype–phenotype studies of the patients with SMS showed no phenotype difference between deletions of 1.5–9 Mb in size.12 However, because of less accurate cytogenetics techniques at the time, two of the nine patients with SMS and cleft palate reported by Smith and Magenis1 may have had a larger 17p11.2 deletion. Cleft palate occurs in about 10% of patients with SMS but to date, there has been no evidence of a link between this 10% and the 10% with larger deletions.
Two linking studies suggest the presence of one gene on 17p implicated, directly or as a cofactor, in cleft palate: 17p13.1, near D17S974 and D17S130313 and 17p11.2–p11, near D17S1843 and D17S953.14 These studies suggest that a gene at 17p11.2–11.1, together with the Van der Woude gene interferon regulatory factor 6 (IRF6) at 1q32, enhances the probability of cleft palate in an individual carrying these two risk genes.
In our study, the smallestextra‐deleted region associated with cleft palate in SMS of 1.4 Mb contains <16 genes and is located at 17p11.2–17p12. Taking together the gene expression array data and previously published data, one gene, UBB, seems to be the best candidate to be implicated in cleft palate. This gene encodes ubiquitin, one of the most conserved proteins known. Ubiquitin is required for ATP‐dependent, nonlysosomal intracellular protein degradation of abnormal proteins and normal proteins with a rapid turnover.15 It is covalently bound to proteins to be degraded, and presumably labels these proteins for degradation (OMIM 191339, Genatlas). UBB is implicated in anomalies of the ubiquitin–proteasome system, particularly in neurodegenerative diseases.16
Several genes (UFD1L, MID1 and SUMO1) implicated in the ubiquitin‐mediated proteolysis pathways, are involved in syndromic or non‐syndromic cleft palate. In Opitz G/BBB syndrome, a genetic disorder characterised by developmental midline abnormalities, MID1 encodes a TRIM/RBCC protein that is anchored to the microtubules. The association of Mid1 with the cytoskeleton is regulated by dynamic phosphorylation through the interaction with the alpha4 subunit of phosphatase 2A (PP2A). MID1 acts as an E3 ubiquitin ligase, regulating PP2A degradation on microtubules.17,18
In DiGeorge syndrome (DGS), UFD1L encodes the human homolog of the yeast ubiquitin fusion degradation 1 protein (UFD1p), involved in the degradation of ubiquitin fusion proteins. Even though its mechanism remains unclear, UFD1L, expressed in embryonic branchial arches and in the conotruncus, appears to play a prominent role in the pathogenesis of the 22q11.2 deletion syndrome.19,20,21
Another gene implicated in cleft palate, the small ubiquitin‐related modifier SUMO1, reversibly modifies many proteins, including promoter‐specific transcription factors. Msx1 is conjugated to SUMO1, and studies in both humans and mice indicate that the Msx1 transcription factor is associated with specific disorders, including cleft palate.22,23
Conclusion
Cleft palate in SMS is a rare event (10%) in a rare syndrome (prevalence 1 25 000). In our study, larger deletions were associated with cleft palate in SMS. Constitutional hemizygosity for UBB has a role in ubiquitin‐mediated proteolysis and may act as a cofactor for cleft palate in patients with SMS. It may be a good candidate gene for non‐syndromic cleft palate, especially Van der Woude syndrome modifier (OMIM 604547).
Electronic database information
Human Genome Browser, http://genome.ucsc.edu/cgi‐bin/hgGateway?org = human
Expression array data from the first branchial arch: http://hg.wustl.edu/COGENE/
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = OMIM
Genatlas, http://www.genatlas.org
Supplementary file available on the JMG website — http://jmg.bmj.com/supplemental
Acknowledgements
We thank the patients and their families, the Smith‐Magenis Association, France, and its president Mrs Bommé. We also thank Mr Buffat for helping with the statistical analysis.
Abbreviations
CGH - comparative genome hybridisation
FISH - fluorescence hybridisation
HMM - hidden Markov model
UBB - ubiquitin B
Footnotes
Competing interests: None declared.
Supplementary file available on the JMG website — http://jmg.bmj.com/supplemental
References
- 1.Smith A C, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, Reiss J, Lahr M, Allen L, Magenis E. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet 198624393–414. [DOI] [PubMed] [Google Scholar]
- 2.Moncla A, Livet M O, Auger M, Mattei J F, Mattei M G, Giraud F. Smith‐Magenis syndrome: a new contiguous gene syndrome. Report of three new cases. J Med Genet 199128627–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith A C, Magenis R E, Elsea S H. Overview of Smith‐Magenis syndrome. J Assoc Genet Technol 200531163–167. [PubMed] [Google Scholar]
- 4.Elsea S H, Purandare S M, Adell R A, Juyal R C, Davis J G, Finucane B, Magenis R E, Patel P I. Definition of the critical interval for Smith‐Magenis syndrome. Cytogenet Cell Genet 199779276–281. [DOI] [PubMed] [Google Scholar]
- 5.Bi W, Yan J, Stankiewicz P, Park S S, Walz K, Boerkoel C F, Potocki L, Shaffer L G, Devriendt K, Nowaczyk M J, Inoue K, Lupski J R. Genes in a refined Smith‐Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 200212713–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoumans J, Staaf J, Jonsson G, Rantala J, Zimmer K S, Borg A, Nordenskjold M, Anderlid B M. Detection and delineation of an unusual 17p11.2 deletion by array‐CGH and refinement of the Smith‐Magenis syndrome minimum deletion to around 650 kb. Eur J Med Genet 200548290–300. [DOI] [PubMed] [Google Scholar]
- 7.Slager R E, Newton T L, Vlangos C N, Finucane B, Elsea S H. Mutations in RAI1 associated with Smith‐Magenis syndrome. Nat Genet 200333466–468. [DOI] [PubMed] [Google Scholar]
- 8.Bi W, Saifi G M, Shaw C J, Walz K, Fonseca P, Wilson M, Potocki L, Lupski J R. Mutations of RAI1, a PHD‐containing protein, in nondeletion patients with Smith‐Magenis syndrome. Hum Genet 2004115515–524. [DOI] [PubMed] [Google Scholar]
- 9.Girirajan S, Elsas L J, 2nd, Devriendt K, Elsea S H. RAI1 variations in Smith‐Magenis syndrome patients without 17p11.2 deletions. J Med Genet 200542820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridlyand J, Snijders A, Pinkel D, Albertson D, Jain A. Hidden Markov models approach to the analysis of array CGH data. J Multivar Anal 200490132–153. [Google Scholar]
- 11.Potocki L, Shaw C J, Stankiewicz P, Lupski J R. Variability in clinical phenotype despite common chromosomal deletion in Smith‐Magenis syndrome [del(17)(p11.2p11.2)]. Genet Med 20035430–434. [DOI] [PubMed] [Google Scholar]
- 12.Girirajan S, Vlangos C N, Szomju B B, Edelman E, Trevors C D, Dupuis L, Nezarati M, Bunyan D J, Elsea S H. Genotype‐phenotype correlation in Smith‐Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet Med 20068417–427. [DOI] [PubMed] [Google Scholar]
- 13.Wyszynski D F, Albacha‐Hejazi H, Aldirani M, Hammod M, Shkair H, Karam A, Alashkar J, Holmes T N, Pugh E W, Doheny K F, McIntosh I, Beaty T H, Bailey‐Wilson J E. A genome‐wide scan for loci predisposing to non‐syndromic cleft lip with or without cleft palate in two large Syrian families. Am J Med Genet A 2003123140–147. [DOI] [PubMed] [Google Scholar]
- 14.Sertie A L, Sousa A V, Steman S, Pavanello R C, Passos‐Bueno M R. Linkage analysis in a large Brazilian family with van der Woude syndrome suggests the existence of a susceptibility locus for cleft palate at 17p11.2–11.1. Am J Hum Genet 199965433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roos‐Mattjus P, Sistonen L. The ubiquitin‐proteasome pathway. Ann Med 200436285–295. [DOI] [PubMed] [Google Scholar]
- 16.Song S, Jung Y K. Alzheimer's disease meets the ubiquitin‐proteasome system. Trends Mol Med 200410565–570. [DOI] [PubMed] [Google Scholar]
- 17.Trockenbacher A, Suckow V, Foerster J, Winter J, Krauss S, Ropers H H, Schneider R, Schweiger S. MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat Genet 200129287–294. [DOI] [PubMed] [Google Scholar]
- 18.Berti C, Fontanella B, Ferrentino R, Meroni G. Mig12, a novel Opitz syndrome gene product partner, is expressed in the embryonic ventral midline and co‐operates with Mid1 to bundle and stabilize microtubules. BMC Cell Biol 200459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pizzuti A, Novelli G, Ratti A, Amati F, Mari A, Calabrese G, Nicolis S, Silani V, Marino B, Scarlato G, Ottolenghi S, Dallapiccola B. UFD1L, a developmentally expressed ubiquitination gene, is deleted in CATCH 22 syndrome. Hum Mol Genet 19976259–265. [DOI] [PubMed] [Google Scholar]
- 20.Botta A, Tandoi C, Fini G, Calabrese G, Dallapiccola B, Novelli G. Cloning and characterization of the gene encoding human NPL4, a protein interacting with the ubiquitin fusion‐degradation protein (UFD1L). Gene 200127539–46. [DOI] [PubMed] [Google Scholar]
- 21.Cuneo B F. 22q11. 2 deletion syndrome: DiGeorge, velocardiofacial, and conotruncal anomaly face syndromes, Curr Opin Pediatr 200113465–472. [DOI] [PubMed] [Google Scholar]
- 22.Alkuraya F S, Saadi I, Lund J J, Turbe‐Doan A, Morton C C, Maas R L. SUMO1 haploinsufficiency leads to cleft lip and palate. Science 20063131751. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V, Bei M. Modification of Msx1 by SUMO‐1. Biochem Biophys Res Commun 200634574–77. [DOI] [PubMed] [Google Scholar]