Abstract
Background
Age‐related hearing impairment (ARHI) is the most common sensory impairment in older people, affecting 50% of those aged 80 years. The proportion of older people is increasing in the general population, and as a consequence, the number of people affected with ARHI is growing. ARHI is a complex disorder, with both environmental and genetic factors contributing to the disease. The first studies to elucidate these genetic factors were recently performed, resulting in the identification of the first two susceptibility genes for ARHI, NAT2 and KCNQ4.
Methods
In the present study, the association between ARHI and polymorphisms in genes that contribute to the defence against reactive oxygen species, including GSTT1, GSTM1 and NAT2, was tested. Samples originated from seven different countries and were combined into two test population samples, the general European population and the Finnish population. Two distinct phenotypes for ARHI were studied, Zlow and Zhigh, representing hearing in the low and high frequencies, respectively. Statistical analysis was performed for single polymorphisms (GSTM1, GSTT1, NAT2*5A, NAT2*6A, and NAT2*7A), haplotypes, and gene–environment and gene–gene interactions.
Results
We found an association between ARHI and GSTT1 and GSTM1 in the Finnish population sample, and with NAT2*6A in the general European population sample. The latter finding replicates previously published data.
Conclusion
As replication is considered the ultimate proof of true associations in the study of complex disorders, this study provides further support for the involvement of NAT2*6A in ARHI.
Keywords: N‐acetyltransferase, age related hearing impairment, association study, complex disorder, glutathione S‐transferase
Age‐related hearing impairment (ARHI) is the most frequent sensory impairment in older people. It is a complex disorder; both environmental and genetic factors contribute to the disease. The contribution of genetic factors to the development of ARHI has been clearly demonstrated by several heritability studies.1,2,3 The number of factors, their individual contribution, and their interaction with each other remain unknown. Much research effort has been put into elucidation of the environmental factors that are involved in ARHI, such as noise exposure,4,5,6,7 ototoxic medication,8,9,10 exposure to chemicals,11,12 medical conditions,13,14 malnutrition,15,16,17 tobacco smoking and alcohol abuse,14,18,19,20,21,22 although some of these factors remain controversial. In contrast, little is known about the contributory genetic factors.
Two recent genomewide linkage studies and a handful of association studies on candidate genes for ARHI were performed, which aimed to identify some of the genetic factors involved in ARHI. The genomewide linkage studies resulted in a total of seven candidate susceptibility regions for ARHI.23,24 Association studies on GSTM and GSTT25 and DFNA526 failed to detect an association with ARHI. Two other studies, one studying the involvement of NAT27 and one studying KCNQ4,28 resulted in the identification of the first susceptibility genes for ARHI.
Oxidative stress is considered to participate in the ageing process, and consequently, also in ARHI. It is caused by an imbalance between the production and the removal of reactive oxygen species (ROS).29 The exact mechanism by which ROS may cause ARHI remains unknown. A series of insults in the inner ear during the lifetime, such as the influence of drugs, exposure to noise, ear diseases and age‐dependent degeneration, all against a specific genetic background, might cause increased ROS levels, in turn leading to hair‐cell damage.30,31
Glutathione (GSH) and glutathione‐related antioxidant enzymes, such as glutathione S‐transferases (GST), are involved in the detoxification of cytotoxic and carcinogenic molecules, and ROS. GSTs are present in the inner ear,32,33 which may suggest that they may play a central role in the metabolism and inactivation of ototoxic compounds. Decreased GSH and GST activity levels cause increased susceptibility of cells to noxious insults and cell damage.31 In the outer hair cells from the basal end of the cochlea, levels of GSH are lower, rendering the basal end of the cochlea more vulnerable to damage by free radicals.34 Five different GST classes have been identified in humans. Some classes such as GSTM1 (mu, chromosome 1p13.3), GSTP1 (pi, chromosome 11q13), and GSTT1 (theta, chromosome 22q11.2) are polymorphic.35 The GSTT1 polymorphism is caused by a deletion of a substantial part of the gene, causing a reduction of GSTT1 activity in all tissues, whereas in people homozygous for the GSTM1 deletion, the enzyme is absent.35
The N‐acetyltransferase (NAT) enzymes are responsible for the detoxification of exogenic substrates by N‐acetylation or O‐acetylation, and are important for the balance of the oxidative status. Insufficiently acetylated drugs accumulate and may be converted to reactive drug metabolites by oxidative enzymes. It has been suggested that NAT reduces the possibility that these reactive metabolites are formed.36NAT1 and NAT2, two isoenzymes, are both highly polymorphic.37 Some of these polymorphisms result in the production of slow acetylating enzymes, while others will form rapid acetylators. Both slow and rapid acetylators have been linked to disease. Within this study, we focused on polymorphisms of NAT2 as putative causative factors for ARHI. The missense substitutions of the NAT2 alleles, G191A (NAT2*14A), T341C (NAT2*5A), G590A (NAT2*6A) and/or G857A (NAT2*7A), are associated with slow acetylator phenotypes38 with different functional characteristics. NAT2*5A and NAT2*6A lead to decreased protein expression, and NAT2*14A and NAT2*7A result in reduced protein stability.37 The NAT2*5 cluster (all different polymorphisms for NAT2*5) shows the greatest reduction in acetylation, followed by NAT2*14, NAT2*6 and the NAT2*7 cluster.37 In a previously published study, a Turkish Caucasian population was investigated by Ünal et al, who found an association of the NAT2*6A polymorphism and ARHI.27
For the current study, 2111 Caucasian subjects (age range of 53–67 years), from nine different centres and seven different countries, were genotyped. The subjects were divided into two population groups representing the general European population and the Finnish population. The aim of this study was to detect an association between genes related to oxidative stress and ARHI. Single gene polymorphisms (GSTM1, GSTT1, NAT2*5A, NAT*6A, and NAT2*7A), haplotypes, and gene–environment and gene–gene interactions were analysed.
Methods
Samples
Caucasian volunteers aged 53–67 years from nine different centres in seven countries were collected from population registries or through clinical consultations. When samples were collected through audiological consultations, the spouses of included subjects were also included. Table 1 gives an overview of the nine different centres and their general collection data. Air conduction was measured at 0.125, 0.25, 0.5, 1, 2, 3, 4, 6 and 8 kHz, and bone conduction at 0.5, 1, 2 and 4 kHz from all participating volunteers. The full inclusion and exclusion criteria have been described previously.28 Two distinct phenotypes of ARHI, Zlow and Zhigh, were tested within this study. Z‐scores were calculated as follows.39 Based on the ISO 7029 standard,40 frequency‐specific thresholds were converted to Z‐scores independent of sex and age,39 defined as the number of standard deviations that the hearing threshold differs from the median value at a specific frequency. People with better hearing than the age‐specific and sex‐specific median at a certain frequency have a negative Z‐score. For each subject, the better hearing ear was selected by averaging the Z‐scores at 0.25, 0.5 and 1 kHz (Zlow), and at 0.2, 4 and 8 kHz (Zhigh). All further calculations were performed using the respective Z‐scores of the better hearing ear. After calculating the Z‐scores and exclusion of phenotypic outliers for Zhigh, samples were selected for analysis by selecting the 34% best and 34% worst hearing subjects of both sexes in each separate sample set, based upon Zhigh. The selection of the samples was performed before data polishing. Analyses for Zlow were conducted after excluding the phenotypic outliers for Zlow as well.
Table 1 Overview of sample collections.
| Centre number | Country | City | Collection | Age range (years) | No of samples | No selected for analysis |
|---|---|---|---|---|---|---|
| 1 | Belgium | Antwerp | Population registries | 54–66 | 769 | 567 |
| 2 | Belgium | Ghent | Clinic + spouses | 55–65 | 237 | 203 |
| 3 | UK | Cardiff | Clinic + spouses | 53–67 | 374 | 252 |
| 4 | The Netherlands | Nijmegen | Clinic + spouses | 55–65 | 276 | 188 |
| 5 | Germany | Tübingen | Clinic + population registries | 53–67 | 395 | 276 |
| 6 | Denmark | Copenhagen | Clinic + spouses | 55–65 | 404 | 278 |
| 7 | Italy | Padova | Clinic + spouses | 53–67 | 359 | 246 |
| 8* | Finland | Oulu | Population registries | 53–67 | 504 | 346 |
| 9* | Finland | Tampere | Clinic + spouses | 55–65 | 256 | 184 |
For statistical analysis, the samples were grouped into two populations, a general European population containing all samples except those from Finland and the samples from centre 3, and the Finnish population, containing the two sample sets from Finland*.
Data polishing
Data polishing was based on a larger study that analysed 768 single nucleotide polymorphisms (SNPs) derived from 70 candidate susceptibility genes for ARHI (Illumina, San Diego, California; http://www.illumina.com). The first step in the data‐polishing process consisted of the removal of all samples that had ⩾10% missing genotypes. After removing these, SNP assays that led to >4% missing genotypes of all genotyped individuals were excluded. In the third step, Hardy‐Weinberg equilibrium was tested for every approved SNP on all approved samples by χ2 test. All the tests for the first three polishing steps were performed using an automated protocol with SAS software (SAS Institute Inc., North Carolina, USA; http://www.SAS.com). The threshold for significance for Hardy‐Weinberg equilibrium was set at 0.001. No SNPs were excluded on the basis of Hardy‐Weinberg equilibrium.
The fourth step of the polishing process consisted of the detection of genetic outliers, using two programs: CHECKHET41 and the Graphical Representation of Relationship errors program (GRR; http://bioinformatics.well.ox.ac.uk/GRR).42 As the homogeneous genetic background of each independent sample set enhances the power of a genetic association study, CHECKHET can be used to detect small numbers of subjects with a different genetic background compared with the genetic background of the majority of the tested sample set. The presence of related individuals in association studies using unrelated samples can lead to misleading conclusions about the significance of statistical tests. GRR detects putative relatives using the fact that related individuals share an excess of alleles identical by state. The cut‐off value to exclude samples was 1.75 identical by state. In addition, GRR is also able to detect sample duplications. Table 2 gives an overview of the number of samples that were excluded during the different data‐polishing steps. Apart from sample set 9, all centres contained samples with ⩾10% missing genotypes. Only in sample set 3 was an important fraction of the samples lost due to missing genotypes (6.3%). For the other centres, the number of failed samples was negligible (0–1.6%). CHECKHET and GRR were used to detect genetic outliers, which were found in all nine sample sets (1.5–3.7%) (table 2). Samples that were marked by CHECKHET and/or GRR as putative genetic outliers were excluded from further statistical analysis.
Table 2 Number of samples used for further analysis after various data‐polishing steps.
| Centre | No of selected samples | No of failed samples | CHECKHET | GRR | Total no of genetic outliers | Total no used for analysis | |
|---|---|---|---|---|---|---|---|
| 1 | 567 | 4 | 14 | 3 | 17 | 546 | |
| 2 | 203 | 2 | 2 | 1 | 3 | 198 | |
| 3 | 252 | 16 | 5 | 2 | 7 | 229 | |
| 4 | 188 | 1 | 3 | 0 | 3 | 184 | |
| 5 | 276 | 3 | 7 | 1 | 8 | 265 | |
| 6 | 278 | 1 | 7 | 1 | 8 | 269 | |
| 7 | 246 | 4 | 7 | 2 | 9 | 233 | |
| 8 | 346 | 2 | 5 | 4 | 9 | 335 | |
| 9 | 184 | 0 | 5 | 0 | 5 | 179 |
Correction for population stratification
Correction for population stratification was performed using the genomic control approach for quantitative traits as proposed by Devlin et al.43 In brief, 70 independent markers (SNPs) that were not associated with the phenotype were selected across the genome and genotyped. For each of these markers, the quantitative phenotype (Z‐score) was regressed on the genotype, coded linearly. In each regression, a test statistic was calculated as the squared ratio of the regression coefficient for genotype and its standard error. In the absence of stratification, this statistic has a χ2 distribution with one degree of freedom. In cases of population substructure, this test statistic is inflated by a factor λ. To estimate λ, the observed median of the test statistic was divided by 0.456, which is the median under the hypothesis of no substructure. Subsequently, all p values were multiplied by this estimate of λ.44 The λ values were calculated for the general European (λ = 1.29) and the Finnish population (λ = 1.09) groups.
Combining several sample sets
To visualise the relationships of the sample sets, a phylogram of all centres was constructed based upon the SNP markers genotyped by Illumina. For this purpose, we applied the unweighted pair group method with arithmetic mean (UPGMA) clustering method to the distance matrix, which was determined by the pairwise F‐statistics between the nine different sample sets.45 The calculations were performed using Powermarker V.3.0 software (http://statgen.ncsu.edu/powermarker/46) and the tree topology was plotted with Treeview V.1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The resulting phylogram showed that the samples fell into two population groups (figure 1). The general European population group (n = 1695) contained all sample sets, except for the two Finnish sample sets and the samples from centre 3 that were excluded from further analysis because of the low DNA quality. The Finnish population sample (n = 514) contained Finnish participants only. All further analyses were performed on the two population samples.
Figure 1 Phylogram based upon pairwise F statistics and the UPGMA method. From the nine original sample sets, the Finnish participants were combined together (Finnish population group, indicated in grey), and the remaining seven sample sets formed a second group (general European population group).
Genotyping
GSTM1 and GSTT1
All PCR reactions were conducted using 40 ng of DNA in 16 mmol/l (NH4)2SO4, 67 mmol/l Tris‐HCl (pH8.8 at 25°C), 0.1% Tween‐20 and 1.5 mmol/l MgCl2 (Eurogentec, Seraing, Belgium). Primer sequences are listed in table 3. To detect the homozygous null and the homozygous wild‐type genotype of GSTM1, the PCR reaction was performed with GSTM1 primers (0.1 μmol/l; length of PCR product 1030 bp)47 and HBB1 (haemoglobin β b1 subunit) primers (0.1 μmol/l; length of PCR product 1566 bp) (both Invitrogen, Carlsbad, California, USA), 250 μmol/l dNTPs (Amresco, Ohio, USA) and 0.04 U/μl Silverstar Taq polymerase (Eurogentec, Seraing, Belgium). The PCR reaction was initiated by a denaturation step at 94°C for 5 min. The amplification reaction consisted of 35 cycles of denaturation at 94°C for 45 s, annealing at 57.4°C for 45 s and extension at 72°C for 90 s. A final extension was performed at 72°C for 10 min. The GSTT1 genotyping assay consisted of a multiplex PCR reaction (0.5 μmol/l of each primer; length of PCR products 466 bp and 1460 bp) (Invitrogen),48 with 200 μmol/l dNTPs (Amresco) and 0.0072 U/μl Silverstar Taq polymerase (Eurogentec). After a denaturation step of 94°C for 5 min, the amplification reaction comprised 35 cycles of denaturation at 94°C for 1 min, annealing at 68°C for 1 min and extension at 72°C for 1 min. A final extension was performed at 72°C for 10 min.
Table 3 Primer and probe sequences for GSTM1, GSTT1 and NAT2 analysis.
| Sequence | |
|---|---|
| Primers | |
| GSTM1 forward | 5′‐AGACAGAAGAGGAGAAGATTC‐3′ |
| GSTM1 reverse | 5′‐TCCAAGTACTTTGGCTTCAGT‐3′ |
| HBB1 forward | 5′‐GACACAACTGTGTTCACTAGC‐3′ |
| HBB1 reverse | 5′‐CAGAATCCAGATGCTCAAGG‐3′ |
| GSTT1 (1) forward | 5′‐CAGTTGTGAGCCACCGTACCC‐3′ |
| GSTT1 (1) reverse | 5′‐CGATAGTTGCTGGCCCCCTC‐3′ |
| GSTT1 (2) forward | 5′‐CCAGCTCACCGGATCATGGCCAG‐3′ |
| GSTT1 (2) reverse | 5′‐CCTTCCTTACTGGTCCTCACATCTC‐3′ |
| NAT2*5A*6A*7A_forward | TGCATTTTCTGCTTGACA |
| NAT2*5A*6A*7A_reverse | GTTGGGTGATACATACACAA |
| NAT2*14A_forward | CATTGTGGGCAAGCCA |
| NAT2*14A_reverse | GTTGGGTGATACATACACAA |
| Probes | |
| NAT2*5A_FL | GCATTTTCTGCTTGACAGAAGAGAGAGGA‐FL |
| NAT2*5A_LC | LC‐TCTGGTACCTGGACCAAATCAGGA‐PH |
| NAT2*6A_FL | GACGTCTGCAGGTATGTATTCATAGACTCAAAAT‐FL |
| NAT2*6A_LC | LC‐TCAATTGTTCGAGGTTCAAGCGT‐PH |
| NAT2*7A_FL | TTCCTTGGGGAGAAATCTCGTGC‐FL |
| NAT2*7A_LC | LC‐CAAACCTGGTGATGGATCCCT‐PH |
| NAT2*14A_FL | CACCCACCCCGGTTTCTTC‐FL |
| NAT2*14A_LC | LC‐TACAATGTGATCAAAAATAGCCTCTAAGCCC‐PH |
FL, fluorescein label with a detection wave length of 533 nm; LC, LightCycler red label with a detection wavelength of 640 nm; PH, phosphate.
NAT2
Genotyping of the NAT2 polymorphisms (NAT2*5A, NAT2*6A, NAT2*7A and NAT2*14A) was performed using primers and probes (TIB MOLBIOL, Berlin, Germany) designed for genotyping analysis on a real‐time PCR system (LightCycler 480; Roche Diagnostics, Basel, Switzerland) (table 3). To perform the reaction, the system kit (LightCycler Genotyping Master Kit; Roche Diagnostics) was used in a final volume of 5 μl in 384 well plates with 20 ng of DNA. Final concentrations of PCR primers and probes were 0.5 μmol/l and 0.2 μmol/l, respectively. Protocols for genotyping of the four polymorphisms are given in table 4.
Table 4 LightCycler protocols for NAT2*5A, NAT2*6A, NAT2*7A and NAT2*14A.
| Programme | Cycles | Analysis mode | Temperature (°C) | Hold | Ramp (°C/s) | Acquisition |
|---|---|---|---|---|---|---|
| Denaturation | 1 | None | 95 | 10 min | 4.8 | None |
| Amplification | 45 | Quantification | 95 | 5 s | 4.8 | None |
| 52/55 | 10 s | 2.5 | Single | |||
| 72 | 20/35 s | 4.8 | None | |||
| Melting curve | 1 | Melting curve | 95 | 1 min | 4.8 | None |
| 50 | 1 min | 2 | None | |||
| 75 | 1 s | – | Continuous | |||
| Cooling | 1 | None | 40 | 30 s | 2 | None |
Numbers in bold are the target temperature and incubation time for NAT2*14A.
Statistical analysis
Genotype frequencies of GSTs and NAT2 in the investigated sample sets
The frequencies of the investigated polymorphisms (GSTM1, GSTT1, NAT2*5A, NAT2*6A and NAT2*7A) in the population samples were calculated using SPSS V.12.0 for Windows (SPSS Inc., Chicago, Illinois, USA). The differences in frequencies between the general European and the Finnish population samples were calculated using Fisher's exact test with R Console V.2.0.1 software (http://www.r‐project.org).
Association testing
All data were analysed using SPSS V.12.0 for Windows (SPSS Inc.). We tested for association between the Z‐scores for both high and low frequencies and the polymorphisms of GSTT1, GSTM1, and NAT2 respectively. A two‐way analysis of variance (ANOVA) was used to account for sex differences.49 Two‐way ANOVA models were constructed in a stepwise backward fashion, with a saturated model including main effects for sex and genotype, as well as the interaction term sex × genotype. If no significant interaction was found, the interaction term was omitted from the analysis and a new model, consisting solely of the main effects for sex and genotype, was used. The normality of the residuals was tested for each genotype of each tested marker. We did not detect major differences in variance between the genotype groups, which is in agreement with the assumptions of ANOVA. If the interaction term was significant, one‐way ANOVA was used to test men and women separately. The interaction between GSTM1 and GSTT1 and the haplotypes of NAT2 were also analysed. To test the interaction between GSTM1 and GSTT1, a similar two‐way ANOVA model was used, replacing the sex interaction term by the GSTM1 × GSTT1 interaction term. Haplotypes were built using FAMHAP (http://iuni‐bonn.de/˜umt7oe/becker.htm50) and were analysed in the same way as the single SNPs. Significance was set at 0.05.
Environment–gene interaction
All study subjects were asked to fill in a questionnaire regarding their medical history, smoking habits, exposure to noise and solvents, and some general features such as body height, body weight, and eye colour (questionnaire available on request). Some of the answers on the questionnaire (see supplementary table S1, available online at http://jmg.bmj.com/supplemental) had to be processed before statistical analysis could be initiated. For whiplash and diabetes, subjects were dichotomised into affected and non‐affected groups. The latter group also contained subjects who did not recall a history of whiplash or did not know whether they had diabetes. The body mass index for each subject was calculated by dividing weight (kg) by height square (m2). For the analysis of smoking habits, subjects were dichotomised into smokers and never smokers. Next, the number of pack years was estimated by multiplying the number of years a subject had been smoking, weighting for daily consumption of tobacco (weighting of 0.5 for <10 cigarettes/day, 1 for 10–20 cigarettes/day, 1.5 for >20 cigarettes/day). For non‐smokers the number of pack years was set to zero. Noise exposure was not analysed in this study because the information obtained on hearing protection and noise exposure from our questionnaire was limited.
We tested interactions between a single GST or NAT2 SNP and an environmental factor for both phenotypes (high and low frequency hearing loss) using SPSS V.12.0 for Windows (SPSS Inc). Because Zhigh shows sex differences, sex was included in the statistical model as a main effect to correct for these differences. Zlow does not show such sex differences. Hence, sex was not taken into account for the statistical analysis of Zlow.
A two‐way ANOVA was used to analyse the interaction of the GST or the NAT2 genotype and categorical environmental factors. To test for interactions for Zhigh, a saturated model was constructed including sex, environmental factor, and genotype as main effects, and an environmental × genotype interaction term. For Zlow, sex was omitted from the model. In the case of a continuous environmental factor, or if the size of the smallest test group for a categorical environmental factor was <10 (which violates the assumptions necessary to perform a two‐way ANOVA) linear regression was performed. In addition, linear regression was performed if an interaction was found using two‐way ANOVA on a categorical environmental factor. For linear regression, a two‐model based approach was pursued. The first model contained sex (only for Zhigh), genotype and environmental factor as main effects. In the second model, the main effect was the genotype × environment interaction term. If the interaction term remained significant, a one‐way ANOVA was used to test the effect of the particular environmental factor.
Gene–gene interactions
Analysis for gene–gene interactions was conducted with R software (http://www.r‐project.org). To test genetic interactions with GST or NAT2, we used a hypothesis‐driven based approach, selecting only SNPs from other candidate ARHI susceptibility genes that were related to oxidative stress and genotyped by Illumina. In total, 22 SNPs in the superoxide dismutase 1 (SOD1), glutathione peroxidase 1 (GXP1), CATALASE, glutathione reductase (GSR) and glutathione S transferase ρ 1 (GSTP1) genes were tested for interactions. For this analysis, phenotypic outliers for Zlow were also excluded in the Zhigh analysis. A two‐way ANOVA was performed to detect interactions. Because of sex differences in hearing, sex was introduced into the statistical analysis as a main effect (only for Zhigh), correcting for differences between men and women. Interaction was fitted by introducing sex (only for Zhigh), genotypes, and the genotype × genotype interaction term as main effects. Further strategies were identical to those for the single SNP and haplotype analyses.
Results
Genotype frequencies of GSTs and NAT2 in the investigated sample sets
Table 5 shows the frequencies for the null deletions of GSTM1 and GSTT1, and the slow acetylating genotypes of NAT2*5A, NAT2*6A and NAT2*7A in the eight different centres that we included in the final investigations. Because NAT2*14A was not polymorphic within our study sample, the genotyping analysis of NAT2*14A was limited to a subset of 920 samples. For this same reason, NAT2*14A was omitted from further analysis, and was not included in this table. We noticed significant differences in frequencies of the GSTM1 (p = 0.0005) and GSTT1 (p = 0.004) deletions and of NAT2*6A (p = 0.028) between the general European and the Finnish population samples, but the difference between the frequencies of the general European and the Finnish population groups for NAT2*7A was not significant (p = 0.15).
Table 5 Frequencies (%) of GST null polymorphisms and NAT2 slow acetylating genotypes in different European countries.
| Centre | Country | GSTM1 | GSTT1 | NAT2*5A | NAT2*6A | NAT2*7A |
|---|---|---|---|---|---|---|
| Antwerp | Belgium | 50 | 17 | 17 | 9 | 0 |
| Gent | Belgium | 60 | 11 | 20 | 6 | 0 |
| Copenhagen | Denmark | 50 | 13 | 21 | 10 | 0.4 |
| Tübingen | Germany | 48 | 12 | 17 | 8 | 0 |
| Padova | Italy | 46 | 14 | 19 | 9 | 0 |
| Nijmegen | The Netherlands | 51 | 15 | 17 | 10 | 0 |
| Oulu* | Finland | 42 | 10 | 16 | 7 | 0.3 |
| Tampere* | Finland | 38 | 6 | 19 | 4 | 0.5 |
| General European population | 50 | 15 | 18 | 9 | 0.06 | |
| Finnish population | 40 | 9 | 17 | 6 | 0.4 | |
*Finnish population.
Statistical analysis of individual polymorphisms and haplotypes
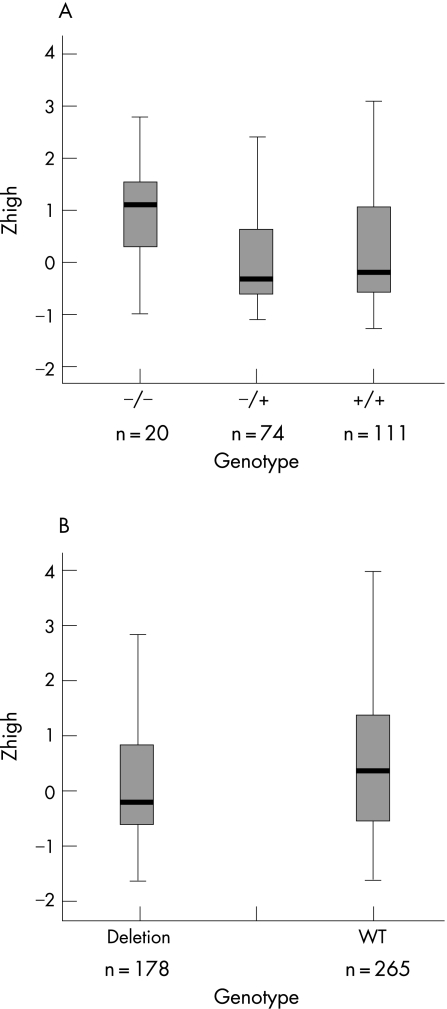
No significant associations for GSTM1, GSTT1 and ARHI could be detected in the general European population sample for Zlow and Zhigh. However, analysis of the Finnish population sample resulted in a nominally significant p value for GSTT1 in women only (Zhigh) (p = 0.035) (table 6). Women homozygous for the GSTT1 deletion had significantly worse hearing at high frequencies (figure 2A). Analysis for GSTM1 in the Finnish population sample resulted in a significant p value (0.027) for Zhigh (table 6). Subjects homozygous for the GSTM1 deletion had significantly better hearing in the high frequencies than those who were not homozygous for the deletion (figure 2B). Interactions of both deletions were tested, but no effect on hearing could be demonstrated (data not shown).
Table 6 Two way analysis of variance (ANOVA) analysis for GSTM1, GSTT1 and NAT2 polymorphisms.
| Zlow | Zhigh | ||
|---|---|---|---|
| General European population | |||
| GSTM1 | 1.00 | 1.00 | |
| GSTT1 | 0.37 | 0.80 | |
| NAT2*5A | 1.00 | 0.68 | |
| NAT2*6A | 0.21 | 0.013 | |
| NAT2*7A | 0.72 | 1.00 | |
| Finnish population | |||
| GSTM1 | 0.78 | 0.027 | |
| GSTT1 | 0.33 | F = 0.035, M = 0.32 (0.040*) | |
| NAT2*5A | 0.73 | 0.54 | |
| NAT2*6A | 0.64 | 0.74 | |
| NAT2*7A | 0.57 | 0.79 | |
All p values were multiplied by λ to correct for population stratification (λEurope = 1.29; λFinland = 1.09). If p was >1, these values were rounded down to 1.00.
Significant p values (p<0.05) are in bold.
Results of NAT2*14A are not included in this table because it was not polymorphic within our study populations.
*Two‐way ANOVA sex × genotype interaction.
Figure 2 Box plots of significantly associated GSTT1 and GSTM1 polymorphisms in the Finnish population: (A) GSTT1 for Zhigh in women; (B) GSTM1 for Zhigh in the overall Finnish population. The sample size for each genotype is indicated below each box. The upper flag represents the 90th percentile (P90), the upper border of the box P75, the bold line P50, the lower border of the box P25 and the lower flag P10.
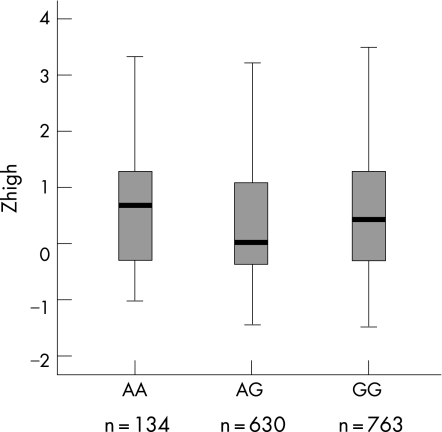
Analysis for Zlow and Zhigh in the general European population sample resulted in significant p values for NAT2*6A (Zhigh, p = 0.013) (table 6). Subjects with the AA genotype for NAT2*6A had worse hearing compared with heterozygous or homozygous GG subjects, indicating that in the general European population sample, an increased risk of developing ARHI existed for subjects with the AA genotype of NAT2*6A (figure 3). No significant p values were found for the NAT2 polymorphisms in the Finnish population sample.
Figure 3 Box plots of the significantly associated NAT2*6A polymorphism in the general European population for Zhigh. The sample size for each genotype is indicated below each box. The upper flag represents the 90th percentile (P90), the upper border of the box P75, the bold line P50, the lower border of the box P25 and the lower flag P10.
For both the European and the Finnish population samples, six different haplotypes were constructed with three NAT2 polymorphisms (NAT2*5A, NAT2*6A and NAT2*7A). For each population, three haplotypes were considered to be ‘rare' and were grouped together. Subsequent statistical haplotype analysis did not result in any significant p values (data not shown).
Gene–environment interaction
For every tested subject, data on 20 environmental factors were available. Statistical analysis for gene–environment interactions in the general European and the Finnish population samples resulted in several significant p values for the GST deletions and NAT2 in both samples. A false discovery rate (FDR) correction for multiple testing was calculated independently for GST deletions and NAT2 in combination with the other SNPs following the Benjamini and Hochberg method described by Sabatti et al.51 None of the significant p values survived this correction for multiple testing. All data on gene–environment interactions are available on request.
Gene–gene interaction
Gene–gene interactions were performed between GSTM1, GSTT1 and NAT2 polymorphisms and 22 additional SNPs originating from five candidate susceptibility genes for oxidative stress in ARHI. Statistical analysis resulted in many significant p values for the GSTT1, GSTM1 and NAT2 polymorphisms (range p = 0.05 to 0.0001). FDR was calculated using the method of Benjamini and Hochberg described by Sabatti et al.51 None of the interaction p values remained significant after correction for multiple testing, except for two interactions with NAT2*6A and GSR for both Zlow (6.5×10−6) and Zhigh (0.00061) in the Finnish population sample. The other gene–gene interaction results are not described within this paper. All data are available on request.
Discussion
To date, only a handful of association studies on candidate susceptibility genes for ARHI have been published. A few of these studies concentrated on genes involved in oxidative stress.25,27 Ates et al studied the contribution of variations in GSTs (GSTM1, GSTP1, GSTT1) to ARHI.25 Individuals homozygous for the deletion of GSTM1 or GSTT1 cannot conjugate metabolites or toxins specific for these enzymes.31,35 This might cause an increased susceptibility to ototoxic compounds and oxidative stress, with cell damage in the inner ear as a consequence. Starting from this working hypothesis, Ates et al performed a case–control study with 68 subjects and 69 healthy controls, but could not detect an association. The relatively small sample size and the consequent low power of this study might have been responsible for this negative result. Within the current study, a large set of 2111 independent samples was divided into two population groups, a general European and a Finnish population samples. The null mutation frequencies that we found for GSTM1 and GSTT1 in our study population are in agreement with previously published findings.25,52 We found a significant difference in the frequencies of the GSTM1 and GSTT1 deletions between the general European and the Finnish population samples, perhaps because the Finnish population is genetically distinct from the general European population.53
In the general European population sample, we could not detect an association between GSTM1 or GSTT1 and ARHI. However, in the Finnish population sample, we found significant associations for both genes. In addition to the genetic distinctness observed in the Finnish population sample,53 it has been demonstrated that the distribution of the GSTM1 and GSTT1 deletions differ between ethnic groups, and even between subpopulations of the same ethnicity,54,55 thus it is very plausible that there is an increased risk for ARHI due to GSTM1 and GSTT1 polymorphisms in Finnish inhabitants only. This might also partly explain why Ates et al did not detect an association, as they were studying a Turkish population.25
A striking, but unexpected finding was that Finnish subjects homozygous for the GSTM1 deletion had better hearing abilities than those without the deletion. This might indicate that this finding represents a false positive result. Bekris et al, however, had suggested that deletions of GSTs might have a protective effect on the development of diabetes.56 In addition, an unexplainable protective effect against coronary artery disease and acute myocardial infarction of the GSTM1 null genotype has been found.57
The fact that subjects lacking GSTT1 had worse hearing is in agreement with our working hypothesis. The fact that this could only be found in women requires explanation. In another study, GST enzyme activity was found to be higher in women than in men.58 If we extend this to our study, this might suggest that women are more vulnerable to develop ARHI due to oxidative stress and GSTT1 deletions.
Ünal et al investigated the effect of NAT2 polymorphisms on ARHI,27 and found a significant association with NAT2*6A. In the current study we were able to replicate this finding, as we observed significant associations between ARHI and NAT2*6A for Zhigh in the general European population sample. This is a strong indication of the contribution of NAT2*6A to the development of hearing impairment in older people. The NAT2*6A AA genotype encodes a slow acetylator, which slows down the detoxification mechanisms.36 This might lead to a higher concentration of xenobiotics in the inner ear, which in turn might increase the number of acquired mitochondrial mutations, eventually leading to cell damage and hearing loss. Ünal et al observed a prevalence of 1–7% for slow acetylators in the controls of their tested population samples. Within our study, 9% slow acetylating genotypes (AA) were present for NAT2*6A in the general European population sample, and 6% in the Finnish population sample. This is in agreement with Ünal's findings. In addition, all slow acetylating genotype frequencies of NAT2*5A, NAT2*6A and NAT2*7A in the eight different sample sets were similar to previously published data.27,52
In addition to single SNP analysis, a gene–gene interaction analysis between NAT2 and the GSTM and GSTT genes and five other genes that are involved in oxidative stress was performed. Two of these interactions, between NAT2*6A and the GSR gene, survived the FDR correction. This may indicate that some genes of the oxidative stress defence mechanism, specifically GSR, do contribute with NAT2 to the development of ARHI.
Performing association studies for common disorders with many contributing factors with small effects will probably result in only marginally significant p values,59 and there is always a question as to whether these are true findings or false positives. Usually, a correction for multiple testing is suggested. However, exactly how to correct remains an unresolved issue among genetic epidemiologists. The Bonferroni correction is usually considered too strict, and might lead to false negative results. Other, less conservative methods, such as FDR, have been suggested;51 however, if we would perform an FDR correction for the single SNP analysis, none of the significant p values would survive. For single gene analysis, some authors have expressed doubt as to whether a correction for multiple testing is necessary, and stress the importance of replication rather than detecting very low p values.60 For gene–environment and gene–gene interactions, we opted to rely on FDR to correct for multiple testing, as we performed many tests for these analyses.
In conclusion, the current study provides evidence that GST genes may be involved in ARHI, buth additional research is required to prove our recent findings. In addition, we present here the first replication of a previously described association, providing further support for the hypothesis that NAT2 is one of the genes involved in the development of ARHI.
The supplementary table 1 can be viewed on the JMG website at http://jmg.bmj.com/supplemental
Acknowledgements
E.V.E. holds a predoctoral research position with the Instituut voor aanmoediging van Innovatie voor Wetenschap en Technologie van Vlaanderen (IWT‐Vlaanderen). This work was supported by grants of the British Royal National Institute for Deaf individuals (RNID), the EU Fifth Framework Quality of Life programme (QLK6‐CT‐2002‐00331), the FWO‐Vlaanderen (G.0131.04) and the University of Antwerp (TOP grant).
Abbreviations
ARHI - age‐related hearing impairment
FDR - false discovery rate
GSH - glutathione
GSR - glutathione reductase
GST - glutathione S‐transferase
GSTP1 - glutathione S transferase ρ
GXP1 - glutathione peroxidase 1
ROS - reactive oxygen species
SOD1 - superoxide dismutase 1
SNP - single nucleotide polymorphism
Footnotes
Competing interests: None declared.
The supplementary table 1 can be viewed on the JMG website at http://jmg.bmj.com/supplemental
References
- 1.Karlsson K K, Harris J R, Svartengren M. Description and primary results from an audiometric study of male twins. Ear Hear 199718114–120. [DOI] [PubMed] [Google Scholar]
- 2.Gates G A, Couropmitree N N, Myers R H. Genetic associations in age‐related hearing thresholds. Arch Otolaryngol Head Neck Surg 1999125654–659. [DOI] [PubMed] [Google Scholar]
- 3.Christensen K, Frederiksen H, Hoffman H J. Genetic and environmental influences on self‐reported reduced hearing in the old and oldest old. J Am Geriatr Soc 2001491512–1517. [DOI] [PubMed] [Google Scholar]
- 4.Mulroy M J, Henry W R, McNeil P L. Noise‐induced transient microlesions in the cell membranes of auditory hair cells. Hear Res 199811593–100. [DOI] [PubMed] [Google Scholar]
- 5.Flock A, Flock B, Fridberger A, Scarfone E, Ulfendahl M. Supporting cells contribute to control of hearing sensitivity. J Neurosci 1999194498–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamasoba T, Nuttall A L, Harris C, Raphael Y, Miller J M. Role of glutathione in protection against noise‐induced hearing loss. Brain Res 199878482–90. [DOI] [PubMed] [Google Scholar]
- 7.Pujol R, Puel J L. Excitotoxicity, synaptic repair, and functional recovery in the mammalian cochlea: a review of recent findings. Ann N Y Acad Sci 1999884249–254. [DOI] [PubMed] [Google Scholar]
- 8.Aran J M, Hiel H, Hayashida T. Noise, aminoglycosides, diuretics. In: Dancer A, Henderson D, Salvi R, Hamernik R, eds. Noise induced hearing loss. St. Louis: Mosby 1992188–195.
- 9.Boettcher F A, Gratton M A, Bancroft B R, Spongr V. Interaction of noise and other agents: recent advances. In: Dancer A, Henderson D, Salvi R, Hamernik R, eds, Noise induced hearing loss. St. Louis: Mosby 1992175–187.
- 10.Stypulkowski P H. Mechanisms of salicylate ototoxicity. Hear Res 199046113–145. [DOI] [PubMed] [Google Scholar]
- 11.Johnson A C, Nylen P R. Effect of industrial solvents on hearing. Occup Med 199510623–640. [PubMed] [Google Scholar]
- 12.Rybak L P. Hearing: the effect of chemicals. Otolaryngol Head Neck Surg 1992106677–686. [DOI] [PubMed] [Google Scholar]
- 13.Kurien M, Thomas K, Bhanu T S. Hearing threshold in patients with diabetes mellitus. J Laryngol 1989103164–168. [DOI] [PubMed] [Google Scholar]
- 14.Gates G A, Cobb J L, D'Agostino R B, Wolf P A. The relation of hearing in the elderly to the presence of cardiovascular disease and cardiovascular risk factors. Arch Otolaryngol Head Neck Surg 1993119156–161. [DOI] [PubMed] [Google Scholar]
- 15.Willott J F, Erway L C, Archer J R, Harrison D E. Genetics of age‐related hearing loss in mice. II. Strain differences and effects of caloric restriction on cochlear pathology and evoked response thresholds. Hear Res 199588143–155. [DOI] [PubMed] [Google Scholar]
- 16.Houston D K, Johnson M A, Nozza R J, Gunter E W, Shea K J, Cutler G M, Edmonds J T. Age‐related hearing loss, vitamin B‐12, and folate in elderly women. Am J Clin Nutr 199969564–571. [DOI] [PubMed] [Google Scholar]
- 17.Seidman M D. Effects of dietary restriction and antioxidants on presbyacusis. Laryngoscope 2000110727–738. [DOI] [PubMed] [Google Scholar]
- 18.Cruickshanks K J, Klein R, Klein B E, Wiley T L, Nondahl D M, Tweed T S. Cigarette smoking and hearing loss: the epidemiology of hearing loss study. JAMA 19982791715–1719. [DOI] [PubMed] [Google Scholar]
- 19.Mellstrom D, Rundgren A, Jagenburg R, Steen B, Svanborg A. Tobacco smoking, ageing and health among the elderly: a longitudinal population study of 70‐year‐old men and an age cohort comparison. Age Ageing 19821145–58. [DOI] [PubMed] [Google Scholar]
- 20.Rosenhall U, Sixt E, Sundh V, Svanborg A. Correlations between presbyacusis and extrinsic noxious factors. Audiology 199332234–243. [DOI] [PubMed] [Google Scholar]
- 21.Fuortes L J, Tang S, Pomrehn P, Anderson C. Prospective evaluation of associations between hearing sensitivity and selected cardiovascular risk factors. Am J Ind Med 199528275–280. [DOI] [PubMed] [Google Scholar]
- 22.Brant L J, Gordon‐Salant S, Pearson J D, Klein L L, Morrell C H, Metter E J, Fozard J L. Risk factors related to age‐associated hearing loss in speech frequencies. J Am Acad Audiol 19967152–160. [PubMed] [Google Scholar]
- 23.DeStefano A L, Gates G A, Heard‐Costa N, Myers R H, Baldwin C T. Genomewide linkage analysis to presbycusis in the Framingham Heart Study. Arch Otolaryngol Head Neck Surg 2003129285–289. [DOI] [PubMed] [Google Scholar]
- 24.Garringer H J, Pankratz N D, Nichols W C, Reed T. Hearing impairment susceptibility in elderly men and the DFNA18 locus. Arch Otolaryngol Head Neck Surg 2006132506–510. [DOI] [PubMed] [Google Scholar]
- 25.Ates N A, Unal M, tamer L, Derici E, Karakas S, Ercan B, Pata Y S, Akbas Y, Vayisoglu Y, Camdeviren H. Glutathione S‐transferase gene polymorphisms in presbyacusis. Otol Neurotol 200526392–397. [DOI] [PubMed] [Google Scholar]
- 26.Van Laer L, DeStefano A L, Myers R H, Flothmann K, Thys S, Fransen E, Gates G A, Van Camp G, Baldwin C T. Is DFNA5 a susceptibility gene for age‐related hearing impairment? Eur J Hum Genet 200210883–886. [DOI] [PubMed] [Google Scholar]
- 27.Unal M, Tamer L, Dogruer Z N, Yildirim H, Vayisoglu Y, Camdeviren H. N‐acetyltransferase 2 gene polymorphism and presbycusis. Laryngoscope 20051152238–2241. [DOI] [PubMed] [Google Scholar]
- 28.Van Eyken E, Van Laer L, Fransen E, Topsakal V, Lemkens N, Laureys W, Nelissen N, Vandevelde A, Wienker T, Van De Heyning P, Van Camp G. KCNQ4: a gene for age‐related hearing impairment? Hum Mutat 2006271007–1016. [DOI] [PubMed] [Google Scholar]
- 29.Evans P, Halliwell B. Free radicals and hearing. Cause, consequence, and criteria. Ann N Y Acad Sci 199988419–40. [DOI] [PubMed] [Google Scholar]
- 30.Seidman M D, Ahmad N, Bai U. Molecular mechanisms of age‐related hearing loss. Ageing Res Rev 20021331–343. [DOI] [PubMed] [Google Scholar]
- 31.Lautermann J, Crann S A, McLaren J, Schacht J. Glutathione‐dependent antioxidant systems in the mammalian inner ear: effects of aging, ototoxic drugs and noise. Hear Res 199711475–82. [DOI] [PubMed] [Google Scholar]
- 32.Takumi Y, Matsubara A, Tsuchida S, Ottersen O P, Shinkawa H, Usami S. Various glutathione S‐transferase isoforms in the rat cochlea. Neuroreport 2001121513–1516. [DOI] [PubMed] [Google Scholar]
- 33.el Barbary A, Altschuler R A, Schacht J. Glutathione S‐transferases in the organ of Corti of the rat: enzymatic activity, subunit composition and immunohistochemical localization. Hear Res 19937180–90. [DOI] [PubMed] [Google Scholar]
- 34.Sha S H, Taylor R, Forge A, Schacht J. Differential vulnerability of basal and apical hair cells is based on intrinsic susceptibility to free radicals. Hear Res 20011551–8. [DOI] [PubMed] [Google Scholar]
- 35.Eaton D L, Bammler T K. Concise review of the glutathione S‐transferases and their significance to toxicology. Toxicol Sci 199949156–164. [DOI] [PubMed] [Google Scholar]
- 36.Dupret J M, Rodrigues‐Lima F. Structure and regulation of the drug‐metabolizing enzymes arylamine N‐acetyltransferases. Curr Med Chem 200512311–318. [DOI] [PubMed] [Google Scholar]
- 37.Hein D W. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res 2002506–50765–77. [DOI] [PubMed] [Google Scholar]
- 38.Hein D W, Doll M A, Fretland A J, Leff M A, Webb S J, Xiao G H, Devanaboyina U S, Nangju N A, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev 2000929–42. [PubMed] [Google Scholar]
- 39.Fransen E, Van Laer L, Lemkens N, Caethoven G, Flothmann K, Govaerts P, Van de Heyning P, Van Camp G. A novel Z‐score‐based method to analyze candidate genes for age‐related hearing impairment. Ear Hear 200425133–141. [DOI] [PubMed] [Google Scholar]
- 40.International Organization of Standardization Acoustic‐threshold of hearing by air conduction as a function of age and sex for otologically normal persons. Geneva: ISO 7029, 2000
- 41.Curtis D, North B V, Gurling H M, Blaveri E, Sham P C. A quick and simple method for detecting subjects with abnormal genetic background in case‐control samples. Ann Hum Genet 200266235–244. [DOI] [PubMed] [Google Scholar]
- 42.Abecasis G R, Cherny S S, Cookson W O, Cardon L R. GRR: graphical representation of relationship errors. Bioinformatics 200117742–743. [DOI] [PubMed] [Google Scholar]
- 43.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic‐based association studies. Theor Popul Biol 200160155–166. [DOI] [PubMed] [Google Scholar]
- 44.Bacanu S A, Devlin B, Roeder K. Association studies for quantitative traits in structured populations. Genet Epidemiol 20022278–93. [DOI] [PubMed] [Google Scholar]
- 45.Penny D, Hendy M. Chapter 13: Phylogenetics: Parisomy and distance methods. In: Balding D, Bischop M, Cannings C, eds. Handbook of statistical genetics, Vol 1. West Sussex: John Wiley and Sons, Ltd, 2000;348–88.
- 46.Liu K, Muse S V. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 2005212128–2129. [DOI] [PubMed] [Google Scholar]
- 47.Harada S, Misawa S, Nakamura T, Tanaka N, Ueno E, Nozoe M. Detection of GST1 gene deletion by the polymerase chain reaction and its possible correlation with stomach cancer in Japanese. Hum Genet 19929062–64. [DOI] [PubMed] [Google Scholar]
- 48.Sprenger R, Schlagenhaufer R, Kerb R, Bruhn C, Brockmoller J, Roots I, Brinkmann U. Characterization of the glutathione S‐transferase GSTT1 deletion: discrimination of all genotypes by polymerase chain reaction indicates a trimodular genotype‐phenotype correlation. Pharmacogenetics 200010557–565. [DOI] [PubMed] [Google Scholar]
- 49.Page G P, Amos C. I. Comparison of linkage‐disequilibrium methods for localization of genes influencing quantitative traits in humans. Am J Hum Genet 1999641194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becker T, Knapp M. Maximum‐likelihood estimation of haplotype frequencies in nuclear families. Genet Epidemiol 20042721–32. [DOI] [PubMed] [Google Scholar]
- 51.Sabatti C, Service S, Freimer N. False discovery rate in linkage and association genome screens for complex disorders. Genetics 2003164829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garte S, Gaspari L, Alexandrie A K, Ambrosone C, Autrup H, Autrup J L, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmoller J, Cascorbi I, Clapper M L, Coutelle C, Daly A, Dell'Omo M, Dolzan V, Dresler C M, Fryer A, Haugen A, Hein D W, Hildesheim A, Hirvonen A, Hsieh L L, Ingelman‐Sundberg M, Kalina I, Kang D, Kihara M, Kiyohara C, Kremers P, Lazarus P, Le Marchand L, Lechner M C, van Lieshout E M, London S, Manni J J, Maugard C M, Morita S, Nazar‐Stewart V, Noda K, Oda Y, Parl F F, Pastorelli R, Persson I, Peters W H, Rannug A, Rebbeck T, Risch A, Roelandt L, Romkes M, Ryberg D, Salagovic J, Schoket B, Seidegard J, Shields P G, Sim E, Sinnet D, Strange R C, Stucker I, Sugimura H, To‐Figueras J, Vineis P, Yu M C, Taioli E. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev 2001101239–1248. [PubMed] [Google Scholar]
- 53.Peltonen L, Jalanko A, Varilo T. Molecular genetics of the Finnish disease heritage. Hum Mol Genet 199981913–1923. [DOI] [PubMed] [Google Scholar]
- 54.Nelson H H, Wiencke J K, Christiani D C, Cheng T J, Zuo Z F, Schwartz B S, Lee B K, Spitz M R, Wang M, Xu X, Kelsey K T. Ethnic differences in the prevalence of the homozygous deleted genotype of glutathione S‐transferase theta. Carcinogenesis 1995161243–1245. [DOI] [PubMed] [Google Scholar]
- 55.Adams C H, Werely C J, Victor T C, Hoal E G, Rossouw G, van Helden P D. Allele frequencies for glutathione S‐transferase and N‐acetyltransferase 2 differ in African population groups and may be associated with oesophageal cancer or tuberculosis incidence. Clin Chem Lab Med 200341600–605. [DOI] [PubMed] [Google Scholar]
- 56.Bekris L M, Shephard C, Peterson M, Hoehna J, Van Yserloo B, Rutledge E, Farin F, Kavanagh T J, Lernmark A. Glutathione‐S‐transferase M1 and T1 polymorphisms and associations with type 1 diabetes age‐at‐onset. Autoimmunity 200538567–575. [DOI] [PubMed] [Google Scholar]
- 57.Wilson M H, Grant P J, Kain K, Warner D P, Wild C P. Association between the risk of coronary artery disease in South Asians and a deletion polymorphism in glutathione S‐transferase M1. Biomarkers 2003843–50. [DOI] [PubMed] [Google Scholar]
- 58.Orhan H, Evelo C T, Sahin G. Erythrocyte antioxidant defense response against cigarette smoking in humans‐‐the glutathione S‐transferase vulnerability. J Biochem Mol Toxicol 200519226–233. [DOI] [PubMed] [Google Scholar]
- 59.Todd J A. Statistical false positive or true disease pathway? Nat Genet 200638731–733. [DOI] [PubMed] [Google Scholar]
- 60.Neale B M, Sham P C. The future of association studies: gene‐based analysis and replication. Am J Hum Genet 200475353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]