Abstract
The clinical phenotypes of maternal and paternal uniparental disomy of chromosome 14 (UPD14) are attributed to dysregulation of imprinted genes. A large candidate locus exists within 14q32, under the regulation of a paternally methylated intergenic differentially methylated region (IG‐DMR). We present a patient with clinical features of maternal UPD14, including growth retardation, hypotonia, scoliosis, small hands and feet, and advanced puberty, who had loss of methylation of the IG‐DMR with no evidence of maternal UPD14. This case provides support for the hypothesis that the maternal UPD14 phenotype is due to aberrant gene expression within the imprinted domain at 14q32.
Keywords: UPD14, imprinting, DNA methylation, DLK1, GTL2
Since the first reports of Wang et al1 and Temple et al2 in 1991, a well‐characterised clinical phenotype has emerged for both paternal and maternal uniparental disomy of chromosome 14 (UPD14). Maternal UPD14, the inheritance of both chromosome homologues from the mother with no contribution from the father, is characterised by prenatal and postnatal growth retardation, hypotonia, joint laxity, motor delay, early onset of puberty, and minor dysmorphic features of the face, hands and feet.3 Paternal UPD14 has a more severe presentation, with polyhydramnios, thoracic and abdominal‐wall defects, growth retardation, severe developmental delay and characteristic dysmorphism.3 The constancy of features has pointed to aberrant imprinting as the likely cause of the phenotypes. Cases of segmental UPD14 have established distal 14q as the critical region for this phenotype.4,5
An imprinted locus exists at 14q326 under the control of a paternally methylated intergenic differentially methylated region (IG‐DMR).7 The imprinted genes in this region include the paternally expressed DLK1 (delta, Drosophila homologue‐like 1) a transmembrane signalling protein and growth regulator homologous to proteins in the Notch/delta pathway.8 Genes for RNA species are also found within the imprinted domain, including the maternally expressed GTL2 (gene trap locus 2), 15 kb distal to the IG‐DMR, and a large microRNA cluster.9
The functional hemizygosity of imprinted genes means that a single imprinting disorder can arise from multiple mechanisms, such as UPD, copy‐number change in imprinted genes, disruption of regulatory sequences, or mutation of the single active allele.10 We describe a patient referred to a joint clinical‐genetics community child‐health clinic with features overlapping those of maternal UPD14, and with isolated methylation deficit at the IG‐DMR.
Case report
The proband was the third child of non‐consanguineous parents, conceived normally and born after a normal pregnancy, with a birth weight of 2.04 kg. He has three sisters. The early neonatal period was complicated by poor feeding and hypotonia and at 1 week of age, a ventricular septal defect was diagnosed but did not require treatment. The patient's head was noted to be on the 75th centile with normal fontanelles and no clinical evidence of hydrocephalus. By 3 months, he had developed mild scoliosis, treated with a spinal brace. No vertebral abnormalities were detected on skeletal radiography.
All motor milestones were delayed; the child did not walk until 3 years of age, when fine motor delay was also noted. Other developmental skills were assessed as normal for age. Although nonverbal comprehension and communication were assessed as normal, he had delayed speech with poor intelligibility, which led to a referral for palatal movement assessment. He was noted to have a high palate but no movement abnormality. There was no submucous cleft. He presented clinically with features of oromotor dyspraxia because of his slow eating and poor pronunciation. He required supplements to maintain his weight, and a programme to improve speech and language. He was investigated for muscle disease, including a muscle biopsy, but power was assessed as normal and muscle histology was also normal. An abdominal ultrasound showed a large single cyst of his left kidney but kidney function was normal.
At the age of 4 years, the patient's height was on the second centile, weight was less than the 40th centile and head circumference on the 25th centile for age. He was noted to have frontal bossing, maxillary hypoplasia with malocclusion and crowded upper teeth. He had a low posterior hairline and a short neck with slight, bilateral webbing, more marked on the left. He had a prominent philtrum and a small mouth with full lips. His hands and feet were noted to be small, but were normal in shape except for fifth‐finger clinodactyly. The genitalia were normal. Neurological examination demonstrated hypotonia but no reduction in muscle power. A skull radiograph showed absence of the sphenoid bones.
At 8 years the patient's height was measured on the 0.4th centile, but growth velocity was normal. Weight continued on the 0.4th centile. Endocrine studies showed a normal level of insulin‐like growth factor 1 and normal thyroid function. There was no evidence of premature puberty.
By 10 years 7 months, progression of the scoliosis to 60° required continued spinal‐jacket usage and corrective surgery using spinal rods that require lengthening every 9 months. The patient had made considerable progress educationally and was at normal school, performing within the average range, according to his teachers and grades. He was generally healthy. He was reported as having ongoing difficulties with fine‐motor coordination, particularly with handwriting, using cutlery and tying shoelaces. He was diagnosed as having organisational motor dyspraxia. His head circumference was on the 10th centile and his height on the 0.4th centile. Hand measurements were 11 cm (palm and finger length) with a middle finger measurement of 3.8 cm (<0.4th centile). His feet were small (UK size 11/European size 30). He was in early puberty, assessed at Tanner stage 2/3. He had enlarged testes, pubic and upper‐lip hair but no axillary hair. His voice had lowered in character during the 2 months prior to the clinic appointment. He was diagnosed with malocclusion of the teeth, with the secondary dentition having erupted behind the lower set so that the maxilla was not free to grow forward. His facial features are shown in fig 1.
Figure 1 Photograph of the proband aged 10 years and 7 months, showing characteristic facial features. Parental consent was given for the publication of this figure.
Initial investigations found a normal male karyotype and normal inheritance of microsatellite markers on chromosome 14. However, the combination of facial features, particularly frontal bossing and prominent philtrum, marked hypotonia and scoliosis in the presence of normal muscle power, dyspraxia, normal intelligence and small hands and feet, indicated that further analysis at 14q32 was warranted.
Methods and results
Methylation‐specific PCR of the 14q32 IG‐DMR
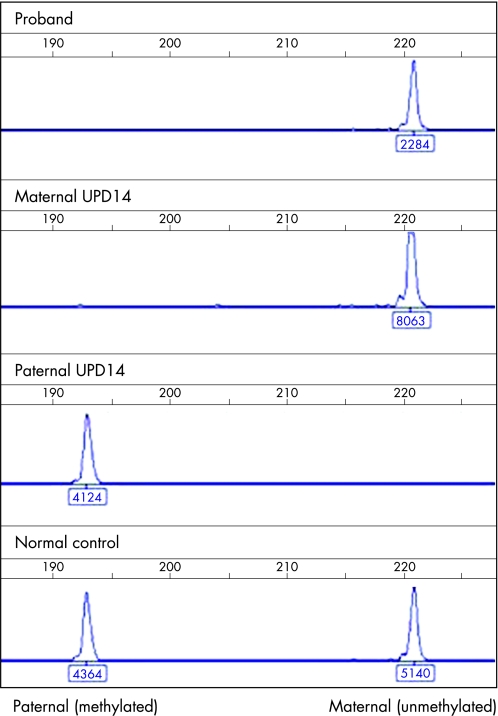
Methylation‐specific PCR (MS‐PCR) uses the divergent sequence changes deriving from bisulphite treatment of differentially methylated DNA, yielding differently sized products in a ratio reflecting that of the starting material. The reaction uses lymphocyte‐derived DNA purified by standard methods. The reaction contained a forward primer and divergent reverse primers, encompassing 5CpG dinucleotides, from the IG‐DMR within 14q32. The amplicon corresponds to chr14:100362206‐100362432 of the Human Genome Sequence (derived from primer sequences on http://genome.ucsc.edu; release March 2006): GTL2b‐fam CTCCAACAACAAAACCCAAAATCAAACAAACTCTC; GTL2b‐unmeth GTGTAGATGGTGGAGAGTAGAGAGGGAGTGTG; GTL2b‐meth CGCGTTTTGGTTCGTTGGTTTTGGCGGCG). The primer set was validated using 120 normal controls, for whom mean methylation ratios of 1.0 were obtained, and patient controls with maternal (n = 2) and paternal (n = 2), UPD14 in whom paternal and maternal amplicons, respectively, were not detected (fig 2 and data not shown).
Figure 2 Electropherograms of methylation‐specific PCR of the DLK1/GTL2 intergenic DMR. The x axis represents product size (in bp), and the y axis the peak height (fluorescence units), as do the figures under each peak. The maternal, unmethylated product is 221 bp; the paternal, methylated product is 193 bp. Trace 1, proband; trace 2, maternal uniparental disomy (UPD) control; trace 3, paternal UPD control; trace 4, normal control.
The amplification reaction contained 1 μl DNA, 0.2 mmol/l dNTPs, 5 pmol of each primer, Taq polymerase (HotStar; Qiagen, Valencia, California, USA) and buffer containing 1.5 mmol/l Mg2+, in a final volume of 10 μl. Reaction conditions were 95°C for 15 min, followed by 28 cycles at 95°C for 20 seconds, 60°C for 20 seconds and 72°C for 20 seconds, then a final cycle at 72°C for 5 min. Methylated (paternal) and unmethylated (maternal) product sizes were 193 and 221 bp respectively. PCR products were visualised on a genetic analyser (ABI 3130×l; Applied Biosystems, Foster City, CA, USA). Peaks were inspected and peak heights <100 or > 8000 pixels were discarded, and the degree of methylation calculated as paternal/maternal peak heights and normalised against normal controls (n = 6/experiment).
MS‐PCR of the proband demonstrated a complete absence of the methylated (paternal) product, giving an epigenotype indistinguishable from maternal UPD14 (fig 2). An identical epigenotype was obtained with primer set GTL2a (amplifying hg18 chr14:100362206‐100362432;11 data not shown). This finding eliminated the possibility of a primer binding site mutation causing artefactual amplification failure of the paternal product.
Microsatellite analysis
To determine whether the absence of the paternal amplicon was due to a small region of segmental maternal UPD14, microsatellite analysis was carried out on proband and parental samples according to standard methods, using primer sets throughout chromosome 14q (table 1). The results indicated biparental inheritance of chromosome 14 (table 1 and supplementary fig 1; available at http://jmg.bmj.com/supplemental). Notably, the biparentally inherited markers D14S1006 and D14S985 closely flanked the IG‐DMR, so that segmental UPD of that region was unlikely and, if present, would be <117 kb in extent.
Table 1 Microsatellite analysis of chromosome 14q in the proband and mother.
| D14S | Cytogenetic position | Size (Mb) | Proband | Mother | Father | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 72 | q11.2 | 20.4 | 1.2 | 2.3 | 1.4 | |||||
| 277 | q24.2 | 72.1 | 2.3 | 3.3 | 1.2 | |||||
| 51 | q32.2 | 96.4 | 1.2 | 2.2 | 1.3 | |||||
| 267 | q32.2 | 98.3 | 1.4 | 2.4 | 1.3 | |||||
| 1006 | q32.2 | 100.3 | 1.2 | 1.1 | 1.2 | |||||
| 985 | q32.2 | 100.4 | 1.2 | 2.3 | 1.3 |
Long‐range PCR
Finally, it was possible that the paternal product was absent because of a microdeletion within the paternal allele of the IG‐DMR. Such microdeletions within the H19 DMR have been associated with familial Beckwith–Wiedemann syndrome.12 To test this possibility, long‐range PCR primers were designed, which would amplify a 4.3‐kb genomic DNA sequence spanning the IG‐DMR (hg18:chr14:100360151‐100364428: primer sequences LR‐DMR‐F: GACAGGAGAGACTGGACATTAGGTG and LR‐DMR‐R: GGGAGGGGGTAAGGATGATTTGAC). Amplification was performed using a commercial kit (Roche Expand PCR Kit; Roche, Basel, Switzerland) according to the manufacturer's instructions, and products were separated on 1% agarose gel (supplementary fig 2; available at http://jmg.bmj.com/supplemental). Comparing the proband with normal controls, there was no evidence of a microdeletion in the IG‐DMR.
Discussion
The patient has many clinical similarities to the maternal UPD 14 phenotype; a comparison of phenotypes is shown in table 2. His intrauterine growth retardation followed by subsequent poor growth is typical and was exacerbated by severe scoliosis secondary to marked hypotonia. His hands and feet were strikingly small. Onset of puberty, at approximately 10 years and 7 months (the interval between examinations made it impossible to ascertain the exact time of onset) was at the lower end of the normal range. Early puberty is a recognised feature of maternal UPD14, and the observations in this patient are in keeping with the syndrome. Interestingly, there was no excessive weight gain and he did not present with a Prader–Willi‐like phenotype, although this diagnosis had been considered at an earlier age because of severe hypotonia. His facial shape, broad forehead, prominent nose, fleshy nasal tip and prominent philtrum were consistent with the minor dysmorphic features of maternal UPD14. This is the first patient reported with neck webbing, but this may be secondary to his severe scoliosis and relative immobility. His intellect had tended to be underestimated because of his motor delay, delayed speech and difficulty with oromotor coordination. However, by 10 years he was functioning well in a normal school, with average ability.
Table 2 A comparison of clinical features between the proband and maternal UPD 14 cases.
| Features | Patients with maternal UPD 143 | Proband | ||
|---|---|---|---|---|
| Birth weight <5% | 10/13 | Yes | ||
| Weight at presentation <5% | 5/13 | No | ||
| Height at presentation <5% | 9/14 | Yes | ||
| Head circumference at presentation <5% | 4/12 | No | ||
| Frontal bossing/ broad forehead | 6/6 | Yes | ||
| Fleshy nasal tip | 5/6 | Yes | ||
| Short philtrum | 6/7 | Yes | ||
| High palate | 4/5 | Yes | ||
| Micrognathia | 3/4 | No | ||
| Small hands | 8/8 | Yes | ||
| Hyperextensible joints | 5/5 | Yes | ||
| Hypotonia | 10/10 | Yes | ||
| Scoliosis | 4/4 | Yes | ||
| Hydrocephalus | 3/6 | No* | ||
| Early puberty | 7/9 | Yes | ||
| Mental retardation/developmental delay | 11/14 | No |
*But occipitofrontal circumference disproportionately large (75th centile).
Molecular investigations showed the proband to have aberrant loss of paternal methylation at the 14q32 IG‐DMR. Maternal UPD14 was excluded by showing biparental inheritance of microsatellite markers throughout the chromosome; however, the possibility of a segmental UPD of <117 kb, the distance between the microsatellites tested, cannot be excluded. Apparent loss of paternal methylation could also have been caused by a microdeletion within the paternal IG‐DMR; however, the child had a normal male karyotype, and no IG‐DMR deletion was detected. The most likely explanation, therefore, is that he has a methylation mutation at the 14q32 IG‐DMR on the paternal allele. The consonance of clinical features between the proband and maternal UPD14 cases indicates that aberrations at the 14q32 imprinted locus are likely to be responsible for the phenotype of maternal UPD14, and suggests that no other imprinted regions on chromosome 14 are of major clinical relevance to this syndrome.
The changes in gene expression resulting from the proband's methylation mutation have not been determined. However, reduced expression of DLK1 is predicted. Murine studies indicate that Dlk1 is a regulator of somatic growth; Moon et al13 showed that Dlk1 null mice have poor postnatal growth and accelerated fat deposition, a phenotype more severe than, but consistent with, that seen in this individual.
It has recently been shown that maternal loss of methylation (LOM) at one imprinted locus may be associated with LOM at other loci and that an overarching mechanism may be responsible for generalised LOM in some patients with imprinting disorders.11,14 We examined the proband's DNA for evidence of LOM at both the paternally methylated H19 DMR and maternally methylated DMRs including TNDM and SNRPN but found no evidence of any methylation abnormality at these loci (data not shown). The cause of the LOM in the proband therefore remains unknown.
In conjunction with this case study, we analysed IG‐DMR methylation in 35 further patients referred to the Wessex Genetics Service with clinical features of UPD14 analysis but no molecular evidence of UPD14. We failed to identify other cases with paternal LOM (data not shown), and so at present must conclude that a methylation mutation is an uncommon cause of the phenotype. However, with methylation‐based diagnostic tests for maternal UPD14 now in routine use, it is likely that further cases will be recognised. This will enable more extensive clinical documentation of this disorder, a more precise comparison of genotype–phenotype correlation between this disorder and maternal UPD14, and potentially the dissection of the genetic causes of this syndrome.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
Acknowledgements
We thank Dr DO Robinson at WRGL Salisbury for helpful discussions.
Abbreviations
DLK1 - delta, Drosophila homologue‐like1
GTL2 - gene trap locus 2
IG‐DMR - intergenic differentially methylated region
LOM - loss of methylation
MS‐PCR - methylation‐specific PCR
UPD14 - uniparental disomy of chromosome 14
Footnotes
Competing interests: None declared.
Funding: DJGM was funded by Diabetes UK.
Parental informed consent was obtained for the publication of this case report.
Supplementary material is available on the JMG website at http://jmg.bmj.com/supplemental
References
- 1.Wang J C C, Passage M B, Yen P H, Shapiro L J, Mohandas T K. Uniparental heterodisomy for chromosome‐14 in a phenotypically abnormal familial balanced 13/14 robertsonian translocation carrier. Am J Hum Genet 1991481069–1074. [PMC free article] [PubMed] [Google Scholar]
- 2.Temple I K, Cockwell A, Hassold T, Pettay D, Jacobs P. Maternal uniparental disomy for chromosome 14. J Med Genet 199128511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton V R, Shaffer L G. Search for imprinted regions on chromosome 14: Comparison of maternal and paternal UPD cases with cases of chromosome 14 deletion. Am J Med Genet 200093A381–387. [DOI] [PubMed] [Google Scholar]
- 4.Coveler K J, Yang S P, Sutton V R, Milstein J M, Wu Y Q, Knox‐Du Bois C, Beischel L S, Johnson J P, Shaffer L G. A case of segmental paternal isodisomy of chromosome 14. Hum Genet 2002110251–256. [DOI] [PubMed] [Google Scholar]
- 5.Kagami M, Nishimura G, Okuyama T, Hayashidani M, Takeuchi T, Tanaka S, Ishino F, Kurosawa K, Ogata T. Segmental and full paternal isodisomy for chromosome 14 in three patients: Narrowing the critical region and implication for the clinical features. Am J Med Genet 2005138A127–132. [DOI] [PubMed] [Google Scholar]
- 6.Wylie A A, Murphy S K, Orton T C, Jirtle R L. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res 2000101711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geuns E, De T N, Hilven P, Van S A, Liebaers I, De R M. Methylation analysis of the intergenic differentially methylated region of DLK1‐GTL2 in human. Eur J Hum Genet 200715352–361. [DOI] [PubMed] [Google Scholar]
- 8.Baladron V, Ruiz‐Hidalgo M J, Nueda M L, az‐Guerra M J, Garcia‐Ramirez J J, Bonvini E, Gubina E, Laborda J. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF‐like repeats. Exp Cell Res 2005303343–359. [DOI] [PubMed] [Google Scholar]
- 9.Seitz H, Royo H, Bortolin M L, Lin S P, Ferguson‐Smith A C, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1‐Gtl2 domain. Genome Res 2004141741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev 200414188–195. [DOI] [PubMed] [Google Scholar]
- 11.Mackay D J, Boonen S E, Clayton‐Smith J, Goodship J, Hahnemann J M, Kant S G, Njolstad P R, Robin N H, Robinson D O, Siebert R, Shield J P, White H E, Temple I K. A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum Genet 2006120262–269. [DOI] [PubMed] [Google Scholar]
- 12.Sparago A, Cerrato F, Vernucci M, Ferrero G B, Silengo M C, Riccio A. Microdeletions in the human H19 DMR result in loss of IGF2 imprinting and Beckwith‐Wiedemann Syndrome. Nat Genet 200436958–960. [DOI] [PubMed] [Google Scholar]
- 13.Moon Y S, Smas C M, Lee K, Villena J A, Kim K H, Yun E J, Sul H S. Mice lacking paternally expressed Pref‐1/Dlk1 display growth retardation and accelerated adiposity. Mol Cell Biol 2002225585–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossignol S, Steunou V, Chalas C, Kerjean A, Rigolet M, Viegas‐Pequignot E, Jouannet P, Le B Y, Gicquel C. The epigenetic imprinting defect of patients with Beckwith‐Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet 200643902–907. [DOI] [PMC free article] [PubMed] [Google Scholar]