Abstract
Background
Pseudoxanthoma elasticum (PXE), an autosomal recessive disorder with considerable phenotypic variability, mainly affects the eyes, skin and cardiovascular system, characterised by dystrophic mineralization of connective tissues. It is caused by mutations in the ABCC6 (ATP binding cassette family C member 6) gene, which encodes MRP6 (multidrug resistance‐associated protein 6).
Objective
To investigate the mutation spectrum of ABCC6 and possible genotype–phenotype correlations.
Methods
Mutation data were collected on an international case series of 270 patients with PXE (239 probands, 31 affected family members). A denaturing high‐performance liquid chromatography‐based assay was developed to screen for mutations in all 31 exons, eliminating pseudogene coamplification. In 134 patients with a known phenotype and both mutations identified, genotype–phenotype correlations were assessed.
Results
In total, 316 mutant alleles in ABCC6, including 39 novel mutations, were identified in 239 probands. Mutations were found to cluster in exons 24 and 28, corresponding to the second nucleotide‐binding fold and the last intracellular domain of the protein. Together with the recurrent R1141X and del23–29 mutations, these mutations accounted for 71.5% of the total individual mutations identified. Genotype–phenotype analysis failed to reveal a significant correlation between the types of mutations identified or their predicted effect on the expression of the protein and the age of onset and severity of the disease.
Conclusions
This study emphasises the principal role of ABCC6 mutations in the pathogenesis of PXE, but the reasons for phenotypic variability remain to be explored.
Keywords: pseudoxanthoma elasticum, ABC transporters, genotype–phenotype correlations, heritable skin diseases
Pseudoxanthoma elasticum (PXE) is an inherited disorder in which mutations in the ABCC6 (ATP‐binding cassette family C member 6) gene lead to dystrophic mineralization and fragmentation of connective tissues.1,2,3,4 Multiple organs are affected, including the skin, eyes and cardiovascular system, and the pathogenic changes include lax and inelastic skin, angioid streaks in the retina and mineralization of the internal elastic lamina of midsized arteries, including the cerebral, coronary, gastrointestinal and peripheral vasculature. Onset is often in late childhood or adolescence, when yellowish cutaneous papules may be noted, most commonly on the neck, axillae and antecubital fossae. However, in many cases angioid streaks in the ocular fundus are the initial physical finding, which correspond to breaks in the elastin‐rich Bruch's membrane of the choroid. As the disease progresses, fragile new vessels may grow through the angioid streaks and hemorrhage, leading to central vision loss. The cardiovascular system may also be affected, leading to hypertension, intermittent claudication, gastrointestinal bleeding, ministrokes and rarely, myocardial infarction. Early identification of the disease and increased surveillance for its sequelae might improve the quality and length of life of those affected. Not only would early detection of mutations in the ABCC6 gene in at‐risk people facilitate diagnosis, but elucidation of possible genotype–phenotype correlations might enable the prediction of disease severity and development of early intervention strategies.
Inheritance of PXE is autosomal recessive; although a few families have been reported in which two generations are affected, pathogenetic mutations in both ABCC6 alleles of affected people were present indicating pseudodominance.2,5,6
Mineralization and fragmentation of elastic fibers, the hallmark of PXE, results from altered function of multidrug resistance‐associated protein 6 (MRP6), the gene product encoded by ABCC6. The ABCC6 gene is located at chromosome 16p13.17,8 along with two closely related but nonfunctional 5′ pseudogenes, ABCC6‐Ψ1 and ABCC6‐Ψ2, corresponding to exons 1–9 and 1–4, respectively.9 The close sequence similarity of the two pseudogenes to the actual coding gene complicates mutation detection and sequencing, although coding gene‐specific primer sets can be designed for each exon without interference from the pseudogene sequences.9 The ABCC6 gene is encoded in 31 exons spanning approximately 75 kb of the human genome and is transcribed into an mRNA of∼5 kb and translated into a 165 kDa protein of 1503 amino acids.7 It is expressed primarily in the liver and functions as a putative efflux transporter of currently unknown substrate specificity, although recent studies have shown that it can function as a transmembrane transporter of polyanionic, glutathione conjugated molecules in vitro.10,11,12,13 In addition to high levels of ABCC6 expression in the liver, clearly measurable levels of expression are detected in the proximal tubules of kidneys, and lower, barely detectable levels are found in the skin, retina and blood vessels, tissues most severely affected by PXE.14,15,16
MRP6 has three hydrophobic membrane‐spanning domains containing a total of 17 transmembrane helices and two intracellular nucleotide‐binding folds (NBFs), comprising 3 highly conserved Walker motifs, and is critical for the putative function of the protein in transmembrane transport driven by the energy from ATP hydrolysis. To date, >150 distinct mutations in the ABCC6 gene have been described in the literature including missense and nonsense mutations in 27 of the 31 exons and deletions spanning the entire coding region.2,15,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38 Clustering of missense mutations to exons corresponding to the nucleotide‐binding folds and a few intracellular segments connecting two transmembrane domains have been noted. Two recurrent mutations, R1141X and a large deletion of exons 23–29 (del23–29, p.A999_S1403del) have been described in a significant proportion of patients, leading to a mutation‐detection strategy that first identifies the recurrent mutations by restriction‐enzyme digestion, followed by sequencing of the remaining exons.39
In this study, we collected genotype data on 270 patients with PXE from 239 families and were able to characterise the phenotype in 198 of these patients to improve diagnosis, identify genotype–phenotype associations and facilitate genetic counselling of people at risk for PXE.
Methods
Informed consent was obtained from all patients and the study was approved by the institutional review board of Good Samaritan Medical Center, Brockton, Massachusetts and the Genetic Alliance BioBank Institutional Review Board.
Donors and samples
Blood samples were collected from 270 patients with PXE (239 probands and 31 affected family members) from the USA, Canada, Europe, Australia and South Africa. None of these families was previously used in other mutation analysis datasets. A detailed questionnaire was used, covering demographic information, family history, and dermatological, ophthalmological, gastrointestinal, cardiac, vascular, neurological, orthopaedic, gynaecological and nutritional histories and physical findings, which was used to ascertain the phenotype of the patients for genotype–phenotype correlations. The phenotypic characteristics were summarised according to organ system and severity (Phenodex PXE International; see table 1). In all index cases, the diagnosis of PXE was made according to the consensus criteria of Lebwohl et al3 and confirmed by skin biopsy specimens stained with H&E and/or von Kossa stain. People with a positive family history (ie confirmed PXE in a first‐degree relative and either eye or skin signs), were also considered to be affected.
Table 1 Phenodex assignment of patients with PXE to different phenotypic categories based on clinical findings in five organ systems.
| System | Findings |
|---|---|
| Skin | |
| S0 | No sign |
| S1 | Papules/bumps |
| S2 | Plaques of coalesced papules |
| S3 | Lax and redundant skin |
| Eye | |
| E0 | No sign |
| E1 | Peau d'orange |
| E2 | Angioid streaks |
| E3 | Bleeding and/or scarring |
| GI | |
| G0 | No sign |
| G1 | GI bleeding as related to PXE |
| Vascular | |
| V0 | No sign |
| V1 | Weak or absent pulses |
| V2 | Intermittent claudication |
| V3 | Vascular surgery |
| Cardiac | |
| C0 | No sign |
| C1 | Chest pain/angina/abnormal ECG or abnormal stress test with no symptoms |
| C2 | Heart attack |
ECG, electrocardiogram; GI, gastrointestinal.
Mutation detection
Genomic DNA was isolated from peripheral blood samples (Puregene DNA Isolation Kit; Gentra Systems, Minneapolis, Minnesota, USA). Control genomic DNA was obtained from the human lymphoblastoid cell line K562 (American Type Culture Collection, Manassas, Virginia, USA). All DNA samples were adjusted with water to a concentration of 10 ng/μL.
Our mutation‐detection strategy was based on: (1) identification of the recurrent mutations R1141X and del23–29 by restriction‐enzyme digestion; (2) optimised denaturing high‐performance liquid chromatography (dHPLC) scanning of PCR products corresponding to all exons in subjects in whom the two recurrent mutations were not identified on both alleles, followed by (3) sequencing of exons with altered dHPLC patterns; and (4) confirmation of novel mutations by restriction‐enzyme digestion or resequencing.
Screening for the recurrent mutations R1141X and del23–29 was performed as previously described.30,31 Conditions and primers for generating PCR products spanning all exons of the coding regions and flanking intronic sequences of the ABCC6 gene were identified for optimum dHPLC screening (supplementary table 1; available at http://jmg.bmj.com/supplemental). These primers were designed to exclude the pseudogenes homologous to exons 1–4 and 1–931 and to anneal within ∼50 bases of the 5′ and 3′ ends of the exon and to exclude known intronic polymorphisms where possible. In many cases, the primers originally designed for exons 10–31 were not suitable for sequencing, and new primers corresponding to sequences situated >50 bases from the intron–exon boundaries were designed to improve sequencing results. PCR for dHPLC analysis was performed using 1.5 U Taq polymerase (Qiagen Inc., Valencia, California, USA) mixed with 5 U Optimase Taq polymerase (Transgenomic, Gaithersburg, Maryland, USA) and Q buffer (Qiagen), according to the manufacturer' instructions. PCR reactions contained 200 ng DNA as template and 20 ng of each primer in a final volume of 50 μl. Cycling conditions for all primer pairs were 94°C for 5 min, followed by 41 cycles of 94°C for 1 min, annealing temperature for a particular primer pair (range 55–60°C) for 1 min and 72°C for 1 min, with a final step at 72°C for 5 min.
The PCR products generated using patients with PXE DNA as template were allowed to form heteroduplexes with an equal volume of a PCR product of the same exon amplified from template DNA of the lymphoblastoid control cell line K562. For this purpose, the PCR products were mixed in a 1:1 ratio and denatured at 94°C for 10 min, followed by reannealing at 65°C for 15 min and 37°C for 15 min. The PCR products were screened by dHPLC (WAVE; Transgenomic, Gaithersburg, Maryland, USA) using methods designed to enhance partial denaturation of the PCR products containing mismatched bases (supplementary table 1; available at http://jmg.bmj.com/supplemental). PCR products showing pattern shifts were sequenced in both directions in most cases. DNA sequencing was performed on an automated sequencer (ABI Prism 377 or ABI 3100; Perkin‐Elmer‐Cetus, Foster City, California, USA). Putative mutations were confirmed by restriction‐enzyme digestion followed by agarose‐gel electrophoresis or by resequencing of a new PCR product when a suitable restriction enzyme was not available. Novel amino acid substitution mutations that affected conserved residues and were not found in 200 control alleles were considered causal.40
Genotype–phenotype correlation analyses
Only patients who filled out the complete clinical questionnaire and in whom the mutations in both alleles had been characterised were included in the genotype–phenotype correlation analysis. Of an original cohort of 270 patients, 134 fulfilled both these criteria. These were 94% Caucasian (all of European descent: 80% USA, 8% Canada, 7% South Africa, 2% UK, 2% Australia, 1% other European countries), 4% Native Americans, 1% Hispanic, 0.5% African American and 0.5% Pacific Islander. Medical records for >10% of the respondents were compared with the questionnaire responses to determine whether the results were representative. Two epidemiologists (SH, MM) analysed the data, using SAS® version 9.1 (Cary, NC).
For the purposes of the genotype–phenotype correlation study, the symptoms and signs were scored as presented in table 1. The data gathered indicated that PXE primarily affected five main clinical areas: skin (S), eyes (E), gastrointestinal system (G), heart (C) and vasculature (V). The data also indicated that for each of these areas there were between two and four grades of severity.
The genotype–phenotype analysis consisted of two parts. First, subjects were grouped by the probable effect of their mutation on protein function: (1) no functional protein, including premature termination codon causing mutations and out‐of‐frame insertions and deletions; (2) some functional protein, such as in‐frame deletions or insertions and missense mutations; and (3) those where it was not possible to predict whether functional protein would be made, such as splicing mutations.
In the second part of the analysis, subjects with PXE were grouped according to the location of their mutations along the putative MRP6 protein (ie, intracellular domains, transmembrane domains, or other).
For each approach, groups were compared using the Fisher exact test.
Results
Mutation detection
Mutation detection was performed on 270 samples from 239 families. The mean (SD) age of the patients was 45 (15.9) years ( range 3–81) and 72% were female (n = 173). Of the 239 index cases, 31 (31/240, 12.9%) had no identifiable mutations. Collectively, 316 mutant alleles were identified, yielding an overall detection rate of 66% (316/478) (all mutations and combinations are listed in supplementary table 2; available at http://jmg.bmj.com/supplemental).
Table 2 Distinct mutations identified in the international case series of 271 patients with PXE.
| Nucleotide change*† | Predicted consequence† | Frequency (alleles) | Exon–intron location | Domain affected‡ | Mutant alleles (%) | References§ |
|---|---|---|---|---|---|---|
| c.105delA | p.S37fsX80 | 2 | 2 | 0.6 | 28 | |
| c.177–185del9 | p.R60_Y62del | 1 | 2 | 0.3 | 9, 28 | |
| c.179del12ins3 | p. R60_W64del L60_R61ins | 1 | 2 | 0.3 | ||
| c.220‐1g→c | SJ | 1 | IVS 2 | 0.3 | ||
| c.724g→t | p.E242X | 1 | 7 | 0.3 | ||
| c.938insT | FS | 1 | 8 | 0.3 | 25 | |
| c.998+2delT | SJ | 1 | IVS 8 | 0.3 | 2, 21 | |
| c.998+2del2 | SJ | 1 | IVS 8 | 0.3 | 18 | |
| c.951c→g | p.S317R | 2 | 9 | TM6 | 0.6 | 28 |
| c.1087c→t | p.Q363X | 1 | 9 | 0.3 | ||
| c.1091g→a | p.T364R | 1 | 9 | TM7 | 0.3 | 9, 19, 21, 28 |
| c.1132c→t | p.Q378X | 4 | 9 | 1.2 | 9, 17–19, 28, 37 | |
| c.1144c→t | p.R382W | 2 | 9 | IC4 | 0.6 | |
| c.1171a→g | p.R391G | 3 | 9 | IC4 | 0.9 | 9, 18, 28, 37 |
| c.1176g→c | p.K392N | 1 | 9 | IC4 | 0.3 | |
| c.1388t→a | p.L463H | 1 | 11 | TM9 | 0.3 | |
| c.1484t→a | p.L495H | 1 | 12 | IC5 | 0.3 | 28 |
| c.1552c→t | p.R518X | 2 | 12 | 0.6 | 18, 19, 27, 28, 37 | |
| c.1553g→a | p.R518Q | 4 | 12 | IC5 | 1.2 | 18, 19, 24, 28, 31 |
| c.1603t→c | p.S535P | 1 | 12 | TM10 | 0.3 | |
| c.1703t→c | p.F568S | 1 | 13 | TM11 | 0.3 | 24 |
| c.1798c→t | p.R600C | 1 | 14 | TM11 | 0.3 | |
| c.1857insC | FS | 1 | 14 | 0.3 | ||
| c.1987g→t | p.G663C | 1 | 16 | NBF1 | 0.3 | |
| c.1999delG | FS | 1 | 16 | 0.3 | ||
| c.2070+5G→A | SJ | 2 | IVS 16 | 0.6 | ||
| c.2093a→c | p.Q698P | 2 | 17 | NBF1 | 0.6 | |
| c.2097g→t | p.E699D | 1 | 17 | NBF1 | 0.3 | |
| c.2177t→c | p.L726P | 1 | 17 | NBF1 | 0.3 | |
| c.2237ins10 | FS | 2 | 17 | 0.6 | ||
| c.2252t→a | p.M751K | 1 | 18 | NBF1 | 0.3 | 20, 37 |
| c.2263g→a | p.G755R | 2 | 18 | NBF1 | 0.6 | |
| c.2278c→t | p.R760W | 3 | 18 | NBF1 | 0.9 | 20, 28, 32, 37 |
| c.2294g→a | p.R765Q | 2 | 18 | NBF1 | 0.6 | 20‐22, 25, 28, 32, 37 |
| c.2329g→a | p.D777N | 1 | 18 | NBF1 | 0.3 | |
| c.2359g→t | p.V787I | 1 | 18 | NBF1 | 0.3 | |
| c.2432c→t | p.T811M | 1 | 19 | IC6 | 0.3 | 6 |
| c.2643g→t | p.R881S | 1 | 20 | IC6 | 0.3 | |
| c.2787+1G→T | SJ | 9 | IVS 21 | 2.8 | 17, 20, 24, 28, 31, 37 | |
| c.2814c→g | p.Y938X | 1 | 22 | 0.3 | ||
| c.2820insC | FS | 1 | 22 | 0.3 | ||
| c.2831c→t | p.T944I | 1 | 22 | TM12 | 0.3 | |
| c.2848g→a | p.A950T | 1 | 22 | TM12 | 0.3 | |
| c.2974g→c | p.G992R | 1 | 22 | TM13 | 0.3 | 2, 42 |
| c.3340c→t | p.R1114C | 2 | 24 | IC8 | 0.6 | 19, 28, 32, 37, 41 |
| c.3389c→t | p.T1130M | 3 | 24 | IC8 | 0.9 | 18, 19, 21, 22, 28, 30, 32, 37, 41 |
| c.3398g→c | p.G1133A | 1 | 24 | IC8 | 0.3 | |
| c.3412g→a | p.R1138W | 7 | 24 | IC8 | 2.2 | 28, 30, 37 |
| c.3413c→t | p.R1138Q | 7 | 24 | IC8 | 2.2 | 18, 19, 24, 25, 28, 30, 32, 37, 41 |
| c.3415g→a | p.A1139T | 2 | 24 | IC8 | 0.6 | |
| c.3415g→a & c.2070+5G→A* | p.A1139T & SJ | 1 | 24, IVS 16 | IC8 | 0.3 | |
| c.3415g→a & c.4335delG* | p.A1139T & FS | 1 | 24, 30 | IC8 | 0.3 | |
| c.3421c→t | p.R1141X | 92 | 24 | 29.3 | 5, 9, 15,18, 19, 21, 22, 24, 28, 30–32, 33, 37, 41 | |
| c.3427c→t | p.Q1143X | 1 | 24 | 0.3 | ||
| c.3490c→t | p.R1164X | 15 | 24 | 4.7 | 18, 27, 28, 31, 33 | |
| c.3491g→a | p.R1164Q | 1 | 24 | IC8 | 0.3 | 28 |
| c.3661c→t | p.R1221C | 1 | 26 | IC9 | 0.3 | 21, 22, 28, 29 |
| c.3662g→a | p.R1221H | 2 | 26 | IC9 | 0.6 | 40 |
| c.3676c→a | p.L1226I | 1 | 26 | IC9 | 0.3 | |
| c.3722g→a | p.W1241X | 2 | 26 | 0.6 | ||
| c.3774insC | FS | 2 | 27 | 0.6 | ||
| c.3775delT | p.G1259fsX1272 | 3 | 27 | 0.9 | 15, 25, 28, 41 | |
| c.3880‐3882del | p.K1294del | 1 | 27 | 0.3 | ||
| c.3883‐5G→A | SJ | 1 | IVS 27 | 0.3 | ||
| c.3892g→t | p.V1298F | 1 | 28 | NBF2 | 0.3 | 25 |
| c.3904g→a | p.G1302R | 7 | 28 | NBF2 | 2.2 | 21, 22, 25, 28 |
| c.3907g→c | p.A1303P | 1 | 28 | NBF2 | 0.3 | 21, 22, 25, 28 |
| c.3912delG | FS | 1 | 28 | 0.3 | 28 | |
| c.3940c→t | p.R1314W | 4 | 28 | NBF2 | 1.2 | 24, 25, 32, 36 |
| c.3941g→a | p.R1314Q | 1 | 28 | NBF2 | 0.3 | 25, 28, 32, 36, 41 |
| c.4004t→a | p.L1335Q | 1 | 28 | NBF2 | 0.3 | |
| c.4015c→t | p.R1339C | 16 | 28 | NBF2 | 5.0 | 19, 25, 28, 33 |
| c.4016g→a | p.R1339H | 2 | 28 | NBF2 | 0.6 | |
| c.4025t→c | p.I1342T | 1 | 28 | NBF2 | 0.3 | |
| c.4041g→c | p.Q1347H | 1 | 28 | NBF2 | 0.3 | 25 |
| c.4104delC | FS | 1 | 29 | 0.3 | 25 | |
| c.4192c→t | p.R1398X | 2 | 29 | 0.6 | 25 | |
| c.4335delG | FS | 1 | 30 | 0.3 | ||
| c.4441g→a | p.G1481S | 1 | 31 | NBF2 | 0.3 | |
| c.4501g→a | p.G1501S | 1 | 31 | NBF2 | 0.3 | |
| Ex23_29del | p.A999_S1403del | 57 | 23–29 | 18.0 | 15, 18, 21, 25, 27, 28, 31, 32, 37, 44, 45 |
FS, frameshift; IC, intracellular domain; IVS, intron; NBF, nucleotide binding fold; SJ, splice junction; TM, transmembrane domain.
*The numbering system corresponds to ABCC6 cDNA (GenBank accession number NM00171.2), the adenosine residue in translation initiation codon ATG being +1. The combination of mutations indicated by an asterisk were identified on one allele of the same patient.
†Mutations in bold are novel.
‡In cases of missense mutations, the affected protein domain is indicated.
§In cases of previously published mutations, the corresponding references are listed.
In total, 82 distinct mutations were identified in this case series (table 2). The two most common mutations were R1141X (29.3%, 92/316) and del23–29 (18%, 57/316). In all, 23 subjects (9.6%) were homozygous for either the R1141X (n = 11) or del23–29 (n = 2) or compound heterozygous (n = 10) for these two mutations. All other mutations were found in <10 subjects (table 2). A total of 51 mutations occurred only once. Two subjects carried three mutations, with two PXE mutations on one allele, although haplotype phase could not be determined because of lack of parental samples. No mutations were identified in 161 alleles and there was an insufficient quantity of one patient's DNA for complete genotyping.
Further dHPLC screening of the ABCC6 gene resulted in the identification of recurrent but less common mutations. Of these, R1339C, R1164X and 2787+1g→c represented 5.0%, 4.7% and 2.8%, respectively, of the 316 alleles identified. R1339C was common in the South African Afrikaner population (15 of 40 alleles, 37.5%) and rare in the European/American case series (1/238 alleles). The high incidence of R1339C in the South African population is probably due to a founder effect.26 Conversely, R1164X (0/40 alleles) and 2787+1g→c (0 of 40 alleles) were absent in the South African case series but were prevalent in the European and American patient populations.
Novel mutations
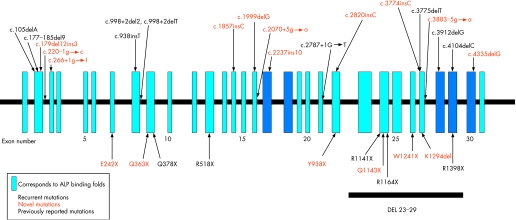
In all, 38 novel, previously unreported mutations were identified in 61 patients in this study (table 2, figs 1 and 2). Most were missense mutations (23 of 39), but there were also four splicing mutations, four insertions, three deletions and five nonsense mutations that had not been previously reported.
Figure 1 The positions of nonsense, splice junction, insertion and deletion mutations identified in the ABCC6 gene in patients with pseudoxanthoma elasticum (PXE). Vertical blue boxes represent the 31 exons, and every fifth exon is numbered. Splicing, small insertion and deletion mutations are shown above the line, and nonsense mutations below, with the position of the recurrent del23–29 mutation. Dark blue boxes, nucleotide‐binding fold domains; bold, recurrent mutations; red, novel mutations.
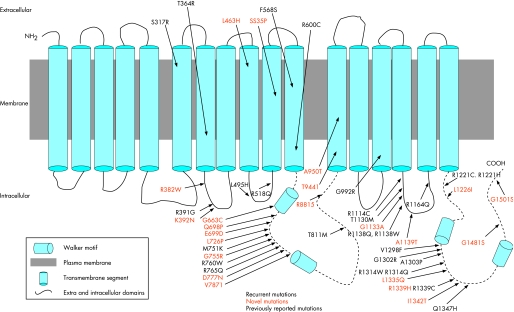
Figure 2 Schematic representation of the MRP6 protein and the positions of missense mutations identified in patients with pseudoxanthoma elasticum (PXE). Vertical blocks are the 17 transmembrane domains of the MRP6 protein; dotted lines, nucleotide‐binding fold; bold, recurrent missense mutations; red, novel mutations.
Prevalence of mutations by exon
Assignment of all of the mutations in this multi‐national case series by exon showed that most point mutations occurred in exons 24 (133/316, 42%) and 28 (36/316, 11%) with a smaller number in exons 9 (14/316, 4.4%) and 18 (10/316, 3%) of the ABCC6 gene (table 2, fig 1). Collectively, the mutations in exons 24 and 28, including the common mutations R1141X and del 23–29, accounted for 71.5% of all the 316 mutations identified in this study (table 2), and the 11 most prevalent mutations (R1141X, del23–29, R1339C, R1164X, 2787+1G→T, G1302R, R1138Q, R1138W, Q378X, R1314W, R518Q) accounted for 70% (223 of 316) of the mutant alleles identified (table 2).
Missense mutations cluster in certain domains
Of the 82 distinct mutations detected in this study, 48 (48/82, 58.5%) were missense mutations and were found to cluster in certain domains of the MRP6 protein (table 2, fig 2). The NBFs were found to harbour a large proportion of missense mutations (NBF1 10/49, 20.4%, NBF2 10/49, 20.4%), apparently reflecting the biological importance of these regions in the binding and hydrolysis of ATP and their critical role in the function of the MRP6 protein. Similarly, other cytoplasmic, intracellular domains (IC) were found to harbour a relatively large proportion of missense mutations (19/49, 38.7%) but these clustered in a number of domains, with the majority found in IC8 (7/49, 14.2%), again reflecting the potential importance of this domain in terms of the function of the protein. Notably, certain transmembrane domains (TM) were completely devoid of missense mutations (TM 3–5, 8, 14–17).
Phenotyping of index cases
Of the 197 patients with full phenotypic data, the characteristics were distributed as follows: 193 people (98%) had some skin signs, 176 (89%) had eye signs, 136 (69%) had vascular signs or symptoms, 50 (25%) had cardiac signs or symptoms and 16 (8%) had gastrointestinal signs (supplementary table 2; available at http://jmg.bmj.com/supplemental). To examine these distributions by age, these patients were categorised into three age categories: (1) <40, (2) 40–54 and (3) >55 years. The mean skin score was 2.2 in all three age categories, the mean scores for eye, vascular and cardiac symptoms increased with age, and the mean gastrointestinal score was nearly 0 in those aged <40 and was 0.1 in the other two age categories.
Genotype–phenotype correlations
The comparison of subjects whose mutations would probably have resulted in no functional protein with those whose mutations would probably have resulted in some functional protein did not yield significant differences. Secondly, subjects with PXE and with missense mutations on one or both alleles were grouped according to the location of their mutation (NBF, IC, TM). Each group was compared with the other three groups but no significant differences were obtained (data not shown). Subjects who had mutations in two different locations (n = 8) were assigned initially to the first location, and the analysis then repeated with these subjects assigned to the second mutation location. Again, no significant differences were found in these analyses.
Although there is no evidence to support the assumption that protein function can be revealed by type of mutation in the absence of functional data, we attempted to confirm the finding of Schulz et al,32 that the age of PXE diagnosis and the number of organ systems affected is significantly different between patients with predicted non‐functional protein compared with those with some potentially functional protein. The classifications were made according to predicted consequences to the protein; missense mutations were predicted to result in some protein that may be functional, whereas nonsense, insertion and deletion mutations were predicted to result in non‐functional protein. The consequence of splicing mutations could not be predicted. Unfortunately, the number of patients in whom a definite age of diagnosis was known was limited to 50 patients. The mean age at diagnosis of patients with a non‐functional protein in this study was 28.5 (n = 19), and that of patients with some protein function was 28.8 (n = 31). These differences were not significant (p = 0.95). When the analysis was repeated for reported age of onset of symptoms regardless of age of diagnosis, presuming that age at onset is younger than age at diagnosis, again no significant differences were found.
Discussion
This study identified a number of novel and recurrent mutations in the ABCC6 gene in a large multinational case series of patients with PXE. In all, 82 distinct mutations, of which 39 were novel, previously unreported, were identified in 239 probands. The detection rate of 66% is less than that reported by other groups, and potentially reflects (1) the lower detection rate when using dHPLC for screening,41,42 (2) the inability of the methods used to detect large deletions including heterozygous loss of entire exons or the entire ABCC6 gene, and (3) presence of mutations in regions such as the 5′ regulatory elements, 3′ untranslated region and central intronic sequences of ABCC6, which were not analysed in this study. Finally, there is the possibility that mutations in genes other than ABCC6 can result in a PXE phenotype, although no concrete evidence for this possibility currently exists. The recurrent mutations R1141X and del23–29 were the most prevalent, consistent with previous reports. In our international case series, mutations in exons 24 and 28 together with del23–29 accounted for the majority of the mutations detected (71.5%) in the ABCC6 gene. Several other recurrent mutations were also identified and comprised around 20% of the total number of mutations.
The NBF domains of the MRP6 protein harboured a large number of missense mutations (22/49), reflecting the strong amino acid sequence conservation in this region, necessary to maintain the function of the protein (fig 2). In addition, intracellular domain 8 harboured several missense mutations (8/49), suggesting the importance of sequence conservation in this region of the protein and perhaps a function that has yet to be identified.
Genotype–phenotype analysis of the 134 patients for whom both ABCC6 mutations and full phenotypic data were available failed to reveal any definitive correlations. The correlation of age of PXE diagnosis with the predicted consequence of the mutation (protein or no protein), as reported by Schulz et al,32 could not be confirmed. As there is no empirical confirmation of whether there is functional MRP6 protein as a result of each of the mutations, the accuracy of this conclusion rests on the validity of the presumption that the mutation either does or does not lead to functional MRP6 protein. Molecular studies of three different MRP6 missense mutations (V1298F, G1302R and G1321S) performed in vitro found that none of these three mutations resulted in ATP‐dependent substrate transport although ATP binding was normal.11 Although these in vitro data do not address whether the proteins bearing these missense mutations are actually formed in the cell and inserted properly into the membrane, it confirms the hypothesis that a missense mutation in a critical portion of the molecule can completely ablate the function of the protein and result in a phenotype as severe as if no protein was present at all.
Finding no genotype–phenotype correlations in complex mendelian disorders, such as metabolic diseases, is not uncommon.43,44 This lack of association suggests that no simple relationship exists between the type and position of mutations, the mutations themselves and the severity of disease. Thus, it is not possible from these data to predict the type or severity of phenotypic manifestations from mutation studies themselves. Because the type of phenotypic outcome and its severity cannot be predicted from genotype data, all patients with PXE should be screened regularly for potential serious and possibly life‐threatening complications regardless of their genotype.
An interesting observation was the absence of macroscopic skin lesions in four patients (9, 58, 131 and 171), although skin biopsy revealed typical histological characteristics of PXE. Patient 9 had significant ophthalmological complications, suggesting that this patient was not a carrier. In three of these patients, a complete genotype was determined, confirming the clinical diagnosis and emphasising that skin features, although present in the majority of patients with PXE, are not always mandatory for the diagnosis.
This study is limited by possible selection bias: participants volunteered and were contacted through the support group. Thus, it is skewed towards people who seek support, which might select for people with a more severe phenotype. In addition, more women than men responded, although that gender ratio is reflective of the PXE population in general. Furthermore, the phenotypic data were obtained through self‐reports, making their accuracy somewhat questionable, even though matching with medical records in a subgroup yielded a high correlation. Nonetheless, the results of this study, along with results of all previous genotype–phenotype correlation studies, indicate strongly that a straightforward and clinically usable correlation does not exist, as could be expected from the nature of this disorder. However, because of the significant clinical relevance, further work is needed to determine if there are subcellular phenotypes that correlate with genotypes in PXE.42 Although mutation detection in PXE has not been shown to have prognostic value, presymptomatic testing in families with history of PXE can provide early diagnosis and may be of value in surveillance for development of disease. In addition, it will probably prove to be of value in genetic counselling as well as in diagnosis of atypical cases or apparent phenocopies.
Acknowledgements
We thank the patients with pseudoxanthoma elasticum and their families for participation in this study; without their participation, this study would not have been possible. We also thank Muin Khoury, Christine Vocke, John McCloskey, Carol Kelly and P Van Acker for their valuable assistance. These studies were supported by the NIH/NIAMS, DHHS grants R01AR28450 (to EGP and JU) and R01AR52627 (to JU), and by an unrestricted grant from Transgenomic to PXE International.
Abbreviations
ABCC6 - ATP‐binding cassette family C member 6
dHPLC - denaturing high‐performance liquid chromatography
IC - intracellular domain
MRP6 - multidrug resistance‐associated protein 6
NBF - nucleotide‐binding fold
PTC - premature termination codon
PXE - pseudoxanthoma elasticum
TM - transmembrane domain
Footnotes
Competing interests: None declared.
References
- 1.Bercovitch L, Terry P. Pseudoxanthoma elasticum. J Am Acad Dermatol 200451S13–S14. [DOI] [PubMed] [Google Scholar]
- 2.Chassaing N, Martin L, Calvas P, Le Bert M, Hovnanian A. Pseudoxanthoma elasticum: a clinical, pathophysiological and genetic update including 11 novel ABCC6 mutations. J Med Genet 200542881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lebwohl M, Neldner K, Pope M F, DePaepe A, Christiano A M, Boyd C D, Uitto J, McKusick V A. Classification of pseudoxanthoma elasticum: report of a consensus conference. J Am Acad Dermatol 199430103–107. [DOI] [PubMed] [Google Scholar]
- 4.Uitto J. The gene family of ABC transporters‐‐novel mutations, new phenotypes. Trends Mol Med 200511341–343. [DOI] [PubMed] [Google Scholar]
- 5.Plomp A S, Hu X, de Jong P T, Bergen A A. Does autosomal dominant pseudoxanthoma elasticum exist? Am J Med Genet 2004A126403–412. [DOI] [PubMed] [Google Scholar]
- 6.Ringpfeil F, McGuigan K, Fuchsel L, Kozic H, Larralde M, Lebwohl M, Uitto J. Pseudoxanthoma elasticum is a recessive disease characterized by compound heterozygosity. J Invest Dermatol 2006126782–786. [DOI] [PubMed] [Google Scholar]
- 7.Cai L, Struk B, Adams M D, Ji W, Haaf T, Kang H L, Dho S H, Xu X, Ringpfeil F, Nancarrow J, Zach S, Schaen L, Stumm M, Niu T, Chung J, Lunze K, Verrecchia B, Goldsmith L A, Viljoen D, Figuera L E, Fuchs W, Lebwohl M, Uitto J, Richards R, Hohl D, Ramesar R. A 500‐kb region on chromosome 16p13.1 contains the pseudoxanthoma elasticum locus: high‐resolution mapping and genomic structure. J Mol Med 20007836–46. [DOI] [PubMed] [Google Scholar]
- 8.Le Saux O, Urban Z, Goring H H, Csiszar K, Pope F M, Richards A, Pasquali‐Ronchetti I, Terry S, Bercovitch L, Lebwohl M G, Breuning M, van den Berg P, Kornet L, Doggett N, Ott J, de Jong P T, Bergen A A, Boyd C D. Pseudoxanthoma elasticum maps to an 820‐kb region of the p13.1 region of chromosome 16. Genomics 1999621–10. [DOI] [PubMed] [Google Scholar]
- 9.Pulkkinen L, Nakano A, Ringpfeil F, Uitto J. Identification of ABCC6 pseudogenes on human chromosome 16p: implications for mutation detection in pseudoxanthoma elasticum. Hum Genet 2001109356–365. [DOI] [PubMed] [Google Scholar]
- 10.Belinsky M G, Chen Z S, Shchaveleva I, Zeng H, Kruh G D. Characterization of the drug resistance and transport properties of multidrug resistance protein 6 (MRP6, ABCC6). Cancer Res 2002626172–6177. [PubMed] [Google Scholar]
- 11.Ilias A, Urban Z, Seidl T L, Le Saux O, Sinko E, Boyd C D, Sarkadi B, Varadi A. Loss of ATP‐dependent transport activity in pseudoxanthoma elasticum‐associated mutants of human ABCC6 (MRP6). J Biol Chem 200227716860–16867. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Q, Uitto J. Pseudoxanthoma elasticum: A metabolic disease? J Invest Dermatol 20061261440–1441. [DOI] [PubMed] [Google Scholar]
- 13.Uitto J, Pulkkinen L, Ringpfeil F. Molecular genetics of pseudoxanthoma elasticum – a metabolic disorder at the environment‐genome interface? Trends Mol Med 2001713–17. [DOI] [PubMed] [Google Scholar]
- 14.Belinsky M G, Kruh G D. MOAT‐E (ARA) is a full length MRP/cMOAT subfamily transporter expressed in kidney and liver. Br J Cancer 1999801342–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergen A A, Plomp A S, Schuurman E J, Terry S, Breuning M, Dauwerse H, Swart J Kool M, van Soest S, Baas F, ten Brink J B, de Jong P T. Mutations in ABCC6 cause pseudoxanthoma elasticum. Nat Genet 200025228–231. [DOI] [PubMed] [Google Scholar]
- 16.Kool M, van der Linden M, de Haas M, Baas F, Boorst P. Expression of human MRP6, a homologue of the multidrug resistance protein MRP1, in tissues and cancer cells. Cancer Res 199959175–182. [PubMed] [Google Scholar]
- 17.Cai L, Lumsden A, Guenther U P, Neldner S A, Zach S, Knoblauch H, Ramesar R, Hohl D, Callen D F, Neldner K H, Lindpaintner K, Richards R I, Struk B. A novel Q378X mutation exists in the transmembrane transporter protein ABCC6 and its pseudogene: implications for mutation analysis in pseudoxanthoma elasticum. J Mol Med 200179536–546. [DOI] [PubMed] [Google Scholar]
- 18.Chassaing N, Martin L, Mazereeuw J, Barrie L, Nizard S, Bonafe J L, Calvas P, Hovnanian A. Novel ABCC6 mutations in pseudoxanthoma elasticum. J Invest Dermatol 2004122608–613. [DOI] [PubMed] [Google Scholar]
- 19.Gheduzzi D, Guidetti R, Anzivino C, Tarugi P, Di Leo E, Quaglino D, Ronchetti I P. ABCC6 mutations in Italian families affected by pseudoxanthoma elasticum (PXE). Hum Mutat 200424438–439. [DOI] [PubMed] [Google Scholar]
- 20.Hendig D, Schulz V, Eichgrun J, Szliska C, Gotting C, Kleesiek K. New ABCC6 gene mutations in German pseudoxanthoma elasticum patients. J Mol Med 200583140–147. [DOI] [PubMed] [Google Scholar]
- 21.Hu X, Plomp A, Wijnholds J, Ten Brink J, van Soest S, van den Born L I, Leys A, Peek R, de Jong P T, Bergen A A. ABCC6/MRP6 mutations: further insight into the molecular pathology of pseudoxanthoma elasticum. Eur J Hum Genet 200311215–224. [DOI] [PubMed] [Google Scholar]
- 22.Hu X, Plomp A, Gorgels T, Brink J T, Loves W, Mannens M, de Jong P T, Bergen A A. Efficient molecular diagnostic strategy for ABCC6 in pseudoxanthoma elasticum. Genet Test 20048292–300. [DOI] [PubMed] [Google Scholar]
- 23.Katona E, Aslanidis C, Remenyik E, Csikos M, Karpati S, Paragh G, Schmitz G. Identification of a novel deletion in the ABCC6 gene leading to pseudoxanthoma elasticum. J Dermatol Sci 200540115–121. [DOI] [PubMed] [Google Scholar]
- 24.Le Saux O, Urban Z, Tschuch C, Csiszar K, Bacchelli B, Quaglino D, Pasquali‐Ronchetti I, Pope F M, Richards A, Terry S, Bercovitch L, de Paepe A, Boyd C D. Mutations in a gene encoding an ABC transporter cause pseudoxanthoma elasticum. Nat Genet 200025223–227. [DOI] [PubMed] [Google Scholar]
- 25.Le Saux O, Beck K, Sachsinger C, Silvestri C, Treiber C, Goring H H, Johnson E W, De Paepe A, Pope F M, Pasquali‐Ronchetti I, Bercovitch L, Marais A S, Viljoen D L, Terry S F, Boyd C D. A spectrum of ABCC6 mutations is responsible for pseudoxanthoma elasticum. Am J Hum Genet 200169749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Saux O, Beck K, Sachsinger C, Treiber C, Goring H H, Curry K, Johnson E W, Bercovitch L, Marais A S, Terry S F, Viljoen D L, Boyd C D. Evidence for a founder effect for pseudoxanthoma elasticum in the Afrikaner population of South Africa. Hum Genet 2002111331–338. [DOI] [PubMed] [Google Scholar]
- 27.Meloni I, Rubegni P, De Aloe G, Bruttini M, Pianigiani E, Cusano R, Seri M, Mondillo S, Federico A, Bardelli AM andreassi L, Fimiani M, Renieri A. Pseudoxanthoma elasticum: Point mutations in the ABCC6 gene and a large deletion including also ABCC1 and MYH11. Hum Mutat 20011885. [DOI] [PubMed] [Google Scholar]
- 28.Miksch S, Lumsden A, Guenther U P, Foernzler D, Christen‐Zach S, Daugherty C, Ramesar R K, Lebwohl M, Hohl D, Neldner K H, Lindpaintner K, Richards R I, Struk B. Molecular genetics of pseudoxanthoma elasticum: type and frequency of mutations in ABCC6. Hum Mutat 200526235–248. [DOI] [PubMed] [Google Scholar]
- 29.Noji Y, Inazu A, Higashikata T, Nohara A, Kawashiri M A, Yu W, Todo Y, Nozue T, Uno Y, Hifumi S, Mabuchi H. Identification of two novel missense mutations (p.R1221C and p.R1357W) in the ABCC6 (MRP6) gene in a Japanese patient with pseudoxanthoma elasticum (PXE). Intern Med 2004431171–1176. [DOI] [PubMed] [Google Scholar]
- 30.Ringpfeil F, Lebwohl M G, Christiano A M, Uitto J. Pseudoxanthoma elasticum: mutations in the MRP6 gene encoding a transmembrane ATP‐binding cassette (ABC) transporter. Proc Natl Acad Sci USA 2000976001–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringpfeil F, Nakano A, Uitto J, Pulkkinen L. Compound heterozygosity for a recurrent 16.5‐kb Alu‐mediated deletion mutation and single‐base‐pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum. Am J Hum Genet 200168642–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz V, Hendig D, Szliska C, Gotting C, Kleesiek K. Novel mutations in the ABCC6 gene of German patients with pseudoxanthoma elasticum. Hum Biol 200577367–384. [DOI] [PubMed] [Google Scholar]
- 33.Struk B, Cai L, Zach S, Ji W, Chung J, Lumsden A, Stumm M, Huber M, Schaen L, Kim C A, Goldsmith L A, Viljoen D, Figuera L E, Fuchs W, Munier F, Ramesar R, Hohl D, Richards R, Neldner K H, Lindpaintner K. Mutations of the gene encoding the transmembrane transporter protein ABC‐C6 cause pseudoxanthoma elasticum. J Mol Med 200078282–286. [DOI] [PubMed] [Google Scholar]
- 34.Kiec‐Wilk B, Surdacki A, Dembinska‐Kiec A, Michalowska J, Stachura‐Deren M, Dubiel J S, Dudek D, Rakowski T, Szastak G, Bodzioch M, Aslanidis C, Schmitz G. Acute myocardial infarction and a new ABCC6 mutation in a 16‐year‐old boy with pseudoxanthoma elasticum. Int J Cardiol 2007116261–262. [DOI] [PubMed] [Google Scholar]
- 35.Schulz V, Hendig D, Schillinger M, Exner M, Domanovits H, Raith M, Szliska C, Kleesiek K, Götting C. Analysis of sequence variations in the ABCC6 gene among patients with abdominal aortic aneurysm and pseudoxanthoma elasticum. J Vasc Res 200542424–432. [DOI] [PubMed] [Google Scholar]
- 36.Bergen A A, Plomp A S, Hu X, de Jong P T, Gorgels T G. ABCC6 and pseudoxanthoma elasticum. Pflugers Arch 2007453685–691. [DOI] [PubMed] [Google Scholar]
- 37.Gotting C, Schulz V, Hendig D, Grundt A, Dreier J, Szliska C, Brinkmann T, Kleesiek K. Assessment of a rapid‐cycle PCR assay for the identification of the recurrent c.3421C>T mutation in the ABCC6 gene in pseudoxanthoma elasticum patients. Lab Invest 200484122–130. [DOI] [PubMed] [Google Scholar]
- 38.Gotting C, Hendig D, Adam A, Schon S, Schulz V, Szliska C, Kuhn J, Kleesiek K. Elevated xylosyltransferase I activities in pseudoxanthoma elasticum (PXE) patients as a marker of stimulated proteoglycan biosynthesis. J Mol Med 200583984–992. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Terry S F, Terry P F, Bercovitch L G, Gerard G F. Development of a rapid, reliable genetic test for pseudoxanthoma elasticum. J Mol Diagn 20079105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cotton R G H, Scriver C R. Proof of “disease causing” mutation. Hum Mutat 1998121–3. [DOI] [PubMed] [Google Scholar]
- 41.Klein B, Weinrich G, Brauch H. DHPLC‐based germline mutation screening in the analysis of the VHL tumor suppressor gene: usefulness and limitations. Human Genet 2001108376–384. [DOI] [PubMed] [Google Scholar]
- 42.Schulz V, Hendig D, Henjakovic M, Szliska C, Kleesiek K, Götting C. Mutational analysis of the ABCC6 gene promoter in German patients with Pseudoxanthoma elasticum (PXE). Human Mutat 200627831. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez‐Salguero P M, Sapone A, Wei X, Holt J R, Jones S, Idle J R, Gonzalez F J. Lack of correlation between phenotype and genotype for the polymorphically expressed dihydropyrimidine dehydrogenase in a family of Pakistani origin. Pharmacogenetics 19977161–163. [DOI] [PubMed] [Google Scholar]
- 44.Sidransky E. Gaucher disease: complexity in a “simple” disorder. Mol Genet Metab 2004836–15. [DOI] [PubMed] [Google Scholar]