Abstract
Background
Recently the genotype/phenotype map of Wolf‐Hirschhorn syndrome (WHS) has been refined, using small 4p deletions covering or flanking the critical region in patients showing only some of the WHS malformations. Accordingly, prenatal‐onset growth retardation and failure to thrive have been found to result from haploinsufficiency for a 4p gene located between 0.4 and 1.3 Mb, whereas microcephaly results from haploinsufficiency of at least two different 4p regions, one of 2.2–2.38 Mb and a second one of 1.9–1.28 Mb.
Methods and Results
We defined the deletion size of a ring chromosome (r(4)) in a girl with prenatal onset growth retardation, severe failure to thrive and true microcephaly but without the WHS facial gestalt and mental retardation. A high‐resolution comparative genome hybridisation array revealed a 760 kb 4p terminal deletion.
Conclusions
This case, together with a familial 4p deletion involving the distal 400 kb reported in normal women, may narrow the critical region for short stature on 4p to 360–760 kb. This region is also likely to contain a gene for microcephaly. “In silico” analysis of all genes within the critical region failed to reveal any strikingly suggestive expression pattern; all genes remain candidates for short stature and microcephaly.
Attempts are ongoing to better correlate specific symptoms or malformations in patients with 4p deletions to specific genes or, at least, molecularly defined regions. The most recent genotype–phenotype map1 confirmed that WHSC1 hemizygosity is essential to the development of the Wolf‐Hirschhorn syndrome (WHS) facial gestalt with the typical “Greek helmet” profile whereas the other key features (growth retardation, microcephaly, cleft palate, mental retardation and epilepsy) result from haploinsufficiency of more than one gene in that region. Particularly, the presence of a dose‐sensitive gene at 4p15–p16, involved in linear growth, was suggested several years ago because of the finding of prenatal and postnatal harmonic short stature in most patients with WHS.2 Patients with the typical WHS facial gestalt but normal stature and interstitial rather than terminal 4p deletions confirmed the presence on 4p of at least two genes with complete penetrance, one for linear growth and the other, more proximal, for distinctive facial features.
We have detected a 760 Kb terminal deletion of the short arm of chromosome 4 due to a ring chromosome in a 34‐month‐old girl examined for growth retardation with microcephaly and normal psychomotor development. Comparison of this case to other critical 4p cases seems to define a 360 kb region containing a gene(s) for which haploinsufficiency correlates with short stature and microcephaly.
Methods
Case report
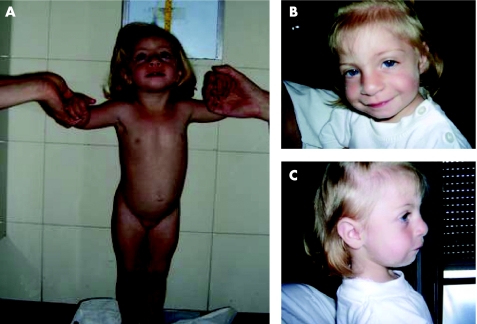
This girl was the second child of unrelated parents, with a negative family history for genetic diseases. Pregnancy was normal. The proband was delivered at 40 weeks of gestation. Perinatal events were normal; birth weight was 2000 g (<3rd centile), length 44.0 cm (−4.5 SD) and head circumference 30.0 cm (−3.7 SD). At the age of 1 month and 15 days her weight was 2900 g (<3rd centile), length 48.5 cm (−2.25 SD), head circumference 32.5 cm (−4.6 SD). Cranial and cardiac ultrasonography were normal; a renal ultrasound showed the absence of left kidney, confirmed by a renal scintigraphy. At the age of 12 months, she had persistent failure to thrive: weight 5100 g (<3rd centile), length 62.5 cm (−4.35 SD), head circumference 38.5 cm (−5.5 SD). Eye examination and EEG were normal. At the age of 13 months, she had age‐appropriate psychomotor development, evaluated by the Brunet‐Lezine scale, with a development quotient of 102. To investigate the severe weight and length failure, the following tests were performed: thyroid panel, anti‐gliadin and anti‐endomisium antibodies, sweat test, ammonia, acid‐base status, urine and plasma determination of amino acids, radioimmunosorbent and radioallergosorbent tests, and toxoplasma–rubella–cytomegalovirus–herpes and other viruses (TORCH) test. All were found to be normal. At the age of 19 months, her bone age according to Greulich and Pyle3 was in the neonatal range. At the age of 2 years MRI of the brain and skeletal radiography were performed to exclude a skeletal dysplasia and found to be normal. Evaluation of the main growth regulator factors (growth hormone, insulin‐like growth factor 1, insulin‐like growth factor binding protein 3) was in the normal range. At the age of 34 months, the patient (figure 1A) had a weight of 8300 g (−3.6 SD), length of 80.7 cm (−3.2 SD), head circumference of 40.5 cm (−6.8 SD) and normal psychomotor development (100 on the Brunet‐Lezine scale). She had some dysmorphic features (figure 1B,C) such as a wide and prominent forehead, hyperthelorism, mild synophris, wide and prominent nasal bridge, broad nasal tip, downslanting palpebral fissures, mild micrognathia and posteriorly rotated ears. Bilateral fifth finger clinodactyly was also observed. Specific investigations excluded the presence of cranial asymmentry, posterior midline scalp defects, prominent glabella, preauricular tags and pits, hearing loss, strabismus, exophthalmus, ptosis, Rieger anomaly, nystagmus, iris coloboma, beaked nose, cleft lip/palate and downturned corners of the mouth (all signs that are present to varying extents in typical patients with WHS).
Figure 1 Patient at the age of 34 months. Parental informed consent was obtained for publication of this figure.
Cytogenetic, molecular analysis and results
Conventional chromosome analysis of peripheral blood lymphocytes from the proband and her parents showed a karyotype 46,XX,r(4)(pterqter) de novo. The ring chromosome was present in all 100 metaphases analysed. Fluorescence in situ hybridisation (FISH) analyses, using commercially available (Vysis, Downers Grove, Il, USA) 4p and 4q subtelomeric probes, showed a normal signal on 4q and a deletion of 4p on r(4). FISH with a WHS critical region probe (Vysis) was normal.
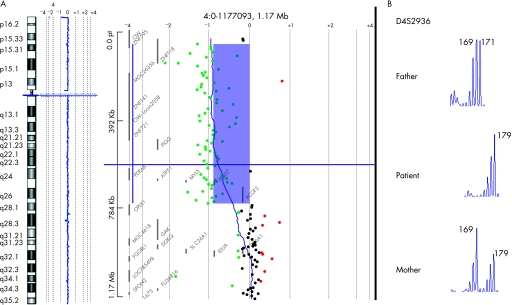
Array CGH was performed using a microarray kit (Human Genome CGH Microarray Kit 244A; Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer's instructions. This is an oligonucleotide‐based platform that allows genome‐wide survey and molecular profiling of genomic aberrations with an average spacing of 6.5 kb. Labelling and hybridisation were performed following the protocols provided by the manufacture. Briefly, 500 ng of purified DNA from a patient and a control (Promega Corporation, Madison, Wisconsin, USA) were double‐digested with RsaI and AluI for 2 hours at 37°C. After 20 minutes at 65°C, DNA of each digested sample was labelled by random priming (Invitrogen, Carlsbad, California, USA) for 2 hours using Cy5‐dUTP for the patient DNA and Cy3‐dUTP for the control DNA. Labelled products were column‐purified and prepared according to the manufacturer's protocol. After probe denaturation and pre‐annealing with 50 μg of Cot‐1 DNA, hybridisation was performed at 65°C with rotation for 40 hours. After two washing steps, the arrays were analysed with the Agilent scanner and Feature Extraction software (V.9.1.3.1). A graphical overview was obtained using CGH analytics software (V.3.4.27). Array CGH confirmed the 4p terminal deletion and localised the breakpoint to 4p16.3 between 759.758 Kb, the last oligomer deleted, and 769.090 Kb, the first oligomer present (genome assembly of May 2004) (figure 2A). Further FISH experiments, performed with the bacterial artificial chromosome RP11‐69L7, confirmed the breakpoint's location showing a weaker signal (not shown).
Figure 2 (A) Array CGH of the patient showing deletion at distal 4p and enlargement of the deleted region; (B) microsatellite analysis (D4S2936) showed the paternal origin of the ring chromosome.
Genomic DNAs was extracted from the patient's and parents' venous blood using standard protocols. Genotyping of polymorphic loci in the deleted region was performed by amplification with primers labelled with fluorescent probes (ABI 5‐Fam, Hex and Tet) followed by analysis on an automated analyser (ABI 310 Genetic Analyzer Applied Biosystems, Foster City, CA, USA). Amplifications were performed with Taq Gold (Applied Biosystems) using standard protocols. Microsatellite analysis (D4S2936) showed that the origin of the ring chromosome was paternal (figure 2B).
Discussion
We have characterised, by array CGH and FISH, the 4p terminal deletion of a ring chromosome r(4) in a patient with normal psychomotor development, severe microcephaly and short harmonic stature without the typical WHS facial dysmorphisms. The deletion spans from the telomere up to 759.758 kb; the next oligomer on the array, at 769.090 Kb, is present.
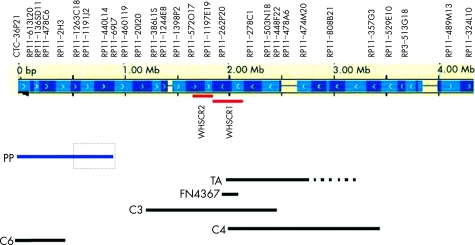
WHS can be considered a contiguous gene syndrome with an increasing number and severity of clinical manifestations positively correlated with increasing size of the deletion.4 No patients with terminal deletions <1.9 Mb have been reported to date, with the exception of a phenotypically normal woman with repeated spontaneous miscarriages and a deletion of around 0.4 Mb also present in her normal mother (patient C6 reported by Van Buggenhout et al1). The patient reported here confirms that 4p deletions involving the telomeric 760 kb are not associated with the typical facial features of WHS, but are associated with prenatal and postnatal short stature and severe microcephaly, increasing with age as in our patient, whose occipital frontal circumference was −3.7 SD at birth and −6.8 SD at the age of 34 months. Van Buggenhout1 described two patients with WHS with 4p interstitial deletions who did not present growth retardation. Rauch et al5 had reported a similar patient. The most distal breakpoint of the 4p interstitial deletions in these patients was at 1.3 Mb (patient C3 in figure 3). Based on the findings in their patients C3 and C6 (the latter a phenotypically normal woman with repeated spontaneous abortions, carrier of a 0.4 Mb deletion) Van Buggenhout et al1 hypothesised that growth retardation in WHS can be mapped to a region between 0.4 and 1.3 Mb from the 4p telomere. These conclusions are also in agreement with patient TA in the review of Bergemann,6 whereas they seem to be in contrast with patient LGL7447 in the same review, simply described in a short abstract as “with growth retardation” at the age of 1.7 years.7 Thus in the absence of a more precise clinical description, we may conclude that, altogether, the data from the literature indicate that patients with WHS with interstitial deletion proximal to 1.3 Mb of 4p do not have the severe growth retardation characteristic of the syndrome. Our findings further confirm these conclusions and narrow the region containing the gene(s) responsible for the short stature associated with 4p‐ to a region of around 400–760 kb (figure 3). Of course we cannot exclude that the responsible gene is located in the distal 400 kb deleted in C6 and that this woman has normal stature because of her genetic background compensating for her haploinsufficiency. Our proband has also severe true microcephaly. According to Zollino et al,4 microcephaly is present in 93% of patients with WHS having a deletion <3.5 Mb. The same authors proposed that 4p‐ microcephaly is caused by hemizygosity for genes proximal to 2.2 Mb whereas Van Buggenhout et al1 localised these genes in two regions, one between 2.2 and 2.39 Mb and the other between 1.28 and 1.9 Mb. Our findings suggest that the critical region we defined for short stature, between about 400 and 760 kb, also contains gene(s) involved in microcephaly. Here again, the distal 400 kb are excluded by proband C6 in Van Buggenhout et al1 and her mother.
Figure 3 4p deletions size in patients with normal stature (black lines) and in the present patient (blue line). The top part shows BAC clones from the telomeric 4 Mb together with the DNA contig representation from Ensembl (January 2004). The Wolf‐Hirschhorn critical regions WHSCR1 and WHSCR2 are indicated with red lines under the Ensembl contig representation. The bottom shows the 4p deletion size in cases reported by 1 (C3, C4, C6), 5 (FN4367) and 6 (TA). Our patient is indicated by a blue line; the dotted square indicates the critical region for prenatal and postnatal short stature.
Our patient also has only one kidney. Renal hypoplasia/agenesis has been reported in roughly half of the patients with simple distal 4p deletions4 and is among the prenatal sonographic signs that, together with the typical face and midline fusion defects, call for 4p cytogenetic investigations.8 No patients with renal abnormalities were detected by Zollino et al4 among those with deletions <3.5 Mb. Among 16 subjects with r(4)9, one had bilateral renal agenesis10 and another one, with a 4p deletion distal to the WHS critical region, had smaller than expected kidneys at renal ultrasound, with a diagnosis of oligomeganephronia based on electron microscopy studies.11 We might hypothesise that haploinsufficiency for a gene located in the telomeric 760 kb of 4p is responsible for abnormal kidney development, although with incomplete penetrance.
Human linear growth is a complex process controlled by multiple genetic, nutritional and environmental factors. Of the genes whose mutations are associated essentially with short stature in the absence of dysmorphic features or disproportionate stature are those in the growth hormone–insulin‐like growth factor‐1 axis.12 The short stature homeobox (SHOX) gene can also be numbered among these genes although its functional loss, originally correlated with “idiopathic” (or isolated) short stature, was subsequently also linked to the Leri‐Weill and Langer syndromes.13 It is not possible at the moment to discern which gene(s) within the critical 4p region we have identified is responsible for isolated short stature or for short stature and microcephaly.
The distal 760 kb of chromosome 4 harbours a limited number of genes. ZNF141 is a ubiquitously expressed zinc‐finger encoding gene.14GPI7 is involved in glycosylphosphatidylinositol biosynthesis.15PDEB6 codes for the β subunit of retinal rod photoreceptor cGMP phosphodiesterase; mutations in PDE6B cause autosomal recessive retinitis pigmentosa. ATP5I, a diet‐regulated gene, codes for subunit E of the mitochondrial F1F0‐ATP synthase. The F1F0‐ATP synthase carries out ATP synthesis from ADP and Pi. Mutations in ATP6, another component of ATP synthase, are responsible for several mitochondrial diseases, such as Leigh syndrome, neurogenic ataxia and retinitis pigmentosa and some cases of Leber hereditary optic neuropathy. MFSD7 codes for a membrane protein belonging to the major facilitator superfamily. MYL5 codes for a regulatory myosin light chain. PCGF3 codes for a RING finger protein of unknown function. Downregulation of the expression of ATP5I and MSFD7 mediated by RNA interference causes a phenotype of stunted/slow growth in Caenorhabditis elegans.
In conclusion, in the long and difficult path to dissect the complex puzzle of Wolf‐Hirschhorn contiguous gene syndrome into its single genetic components we can now assume that at least severe prenatal and postnatal short stature is due to 4p haploinsufficiency for one or more genes within a single region located between 400 and 760 kb. In contrast, no final conclusion can be drawn for microcephaly, which presently seems to be linked to three different regions, the one we defined for short stature and the other two already defined by Van Buggenhout et al.1
Acknowledgements
This work was supported by the Ministero dell'Università e della Ricerca (MIUR) (cofin05‐MIUR to O.Z. and cofin06‐MIUR to ER), the Mariani Foundation (R‐06‐55) and the Cariplo Foundation (both to O.Z.).
Abbreviations
CGH - comparative genomic hybridisations
FISH - fluorescence in situ hybridisation
WHS - Wolf‐Hirschhorn syndrome
Footnotes
Competing interests: None declared.
Parental informed consent was obtained for publication of figure 1.
Ethics approval granted by Università “Magna Graecia” di Catanzaro, Catanzara, Italy.
References
- 1.Van Buggenhout G, Melotte C, Dutta B, Froyen G, Van Hummelen P, Marynen P, Matthijs G, de Ravel T, Devriendt K, Fryns J P, Vermeesch J R. Mild Wolf‐Hirschhorn syndrome: micro‐array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype‐phenotype map. J Med Genet 200441691–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battaglia A, Carey J C, Wright T J. Wolf‐Hirschhorn (4p‐) syndrome. Adv Pediatr 20014875–113. [PubMed] [Google Scholar]
- 3.Greulich W W, Pyle S I.Radiographic atlas of skeletal development of the hand and wrist, 2nd ed. Stanford, CA: Stanford University Press, 1959
- 4.Zollino M, Lecce R, Fischetto R, Murdolo M, Faravelli F, Selicorni A, Butte C, Memo L, Capovilla G, Neri G. Mapping the Wolf‐Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR‐2. Am J Hum Genet 200372590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch A, Schellmoser S, Kraus C, Dorr H G, Trautmann U, Altherr M R, Pfeiffer R A, Reis A. First known microdeletion within the Wolf‐Hirschhorn syndrome critical region refines genotype‐phenotype correlation. Am J Med Genet 200199338–342. [DOI] [PubMed] [Google Scholar]
- 6.Bergemann A D, Cole F, Hirschhorn K. The etiology of Wolf‐Hirschhorn syndrome. Trends Genet 200521188–195. [DOI] [PubMed] [Google Scholar]
- 7.Somer M, Peippo M, Keinanen M. Controversial findings in two patients with commercially available probe D4S96 for the Wolf–Hirschhorn syndrome. Am J Hum Genet 199557A127 [Google Scholar]
- 8.Boog G, Le Vaillant C, Collet M, Dupre P F, Parent P, Bongain A, Benoit B, Trastour C. Prenatal sonographic patterns in six cases of Wolf‐Hirschhorn (4p‐) syndrome. Fetal Diagn Ther 200419421–430. [DOI] [PubMed] [Google Scholar]
- 9.Balci S, Engiz O, Aktas D, Vargel I, Beksac M S, Mrasek K, Vermeesch J, Liehr T. Ring chromosome 4 and Wolf‐Hirschhorn syndrome (WHS) in a child with multiple anomalies. Am J Med Genet A 2006140628–632. [DOI] [PubMed] [Google Scholar]
- 10.Fryns J P, Kleczkowska A, Jaeken J, Van den Berghe H. Ring chromosome 4 mosaicism and Potter sequence. Ann Genet 198831120–122. [PubMed] [Google Scholar]
- 11.Anderson C E, Wallerstein R, Zamerowski S T, Witzleben C, Hoyer J R, Gibas L, Jackson L G. Ring chromosome 4 mosaicism coincidence of oligomeganephronia and signs of Seckel syndrome. Am J Med Genet 199772281–285. [PubMed] [Google Scholar]
- 12.Walenkamp M J, Wit J M. Genetic disorders in the growth hormone ‐ insulin‐like growth factor‐I axis. Horm Res 200666221–230. [DOI] [PubMed] [Google Scholar]
- 13.Blaschke R J, Rappold G. The pseudoautosomal regions, SHOX and disease. Curr Opin Genet Dev 200616233–239. [DOI] [PubMed] [Google Scholar]
- 14.Tommerup N, Aagaard L, Lund C L, Boel E, Baxendale S, Bates G P, Lehrach H, Vissing H. A zinc‐finger gene ZNF141 mapping at 4p16.3/D4S90 is a candidate gene for the Wolf‐Hirschhorn (4p‐) syndrome. Hum Molec Genet 199321571–1575. [DOI] [PubMed] [Google Scholar]
- 15.Shishioh N, Hong Y, Ohishi K, Ashida H, Maeda Y, Kinoshita T. GPI7 is the second partner of PIG‐F and involved in modification of glycosylphosphatidylinositol. J Biol Chem 20052809728–9734. [DOI] [PubMed] [Google Scholar]