Abstract
l‐2‐hydroxyglutaric aciduria (l‐2‐HGA) is a neurometabolic disorder that produces a variety of clinical neurological deficits, including psychomotor retardation, seizures and ataxia. The biochemical hallmark of l‐2‐HGA is the accumulation of l‐2‐hydroxyglutaric acid (l‐2‐HG) in cerebrospinal fluid, plasma and urine. Mutations within the gene L2HGDH (Entrez Gene ID 79944) on chromosome 14q22 encoding L‐2‐hydroxyglutaric acid dehydrogenase have recently been shown to cause l‐2‐HGA in humans. Using a candidate gene approach in an outbred pet dog population segregating l‐2‐HGA, the causal molecular defect was identified in the canine homologue of L2HGDH and characterised. DNA sequencing and pedigree analysis indicate a common founder effect in the canine model. The canine model shares many of the clinical and MRI features of the disease in humans and represents a valuable resource as a spontaneous model of l‐2‐HGA.
L‐2‐hydroxyglutaric aciduria (l‐2‐HGA) is a neurometabolic disorder that produces a variety of clinical neurological deficits in humans, including psychomotor retardation in the early years of life, progressive cerebellar dysfunction (ataxia and intention tremor), learning disability and in some cases seizures.1,2l‐2‐HGA was first recognised in 1980 and has the biochemical hallmark of increased levels of l‐2‐hydroxyglutaric acid (l‐2‐HG) in a variety of tissues and fluids, including cerebrospinal fluid, plasma and urine.3l‐2‐hydroxyglutarate is converted into 2‐oxoglutarate (a widely distributed compound formed through deamination of glutamate and formed within the Krebs cycle) by l‐2‐hydroxglutarate dehydrogenase, a flavin adenine dinucleotide‐dependent dehydrogenase linked to mitochondrial membranes.4,5 The recognition of a spontaneous canine model of l‐2‐HGA sharing many clinical similarities with humans who have l‐2‐HGA offers a valuable opportunity for better understanding of the pathophysiology and treatment options of l‐2‐HGA in humans.6,7 In dogs, as in humans, l‐2‐HGA seems to be inherited as an autosomal recessive condition.
Three separate studies have recently identified mutations responsible for the condition in humans within the gene L2HGDH (Entrez Gene ID 79944; previously known as chromosome 14 open reading frame 160 (C14orf160), duranin and FLJ12618) on chromosome 14q22.1. Topçu et al8 used homozygosity mapping whereas Rzem et al4 used a functional candidate gene approach to demonstrate the genetic mutation responsible for l‐2‐HGA within L2HGDH.9L2HGDH has been shown to encode l‐2‐HG dehydrogenase, with the demonstrated mutations within L2HGDH in patients with l‐2‐HGA resulting in a complete loss of l‐2‐HG dehydrogenase function and defective l‐2‐HG processing in an in vitro model.5,9
To address whether the underlying biochemical defect in the spontaneous canine model of l‐2‐HGA is the same as that in man, we (1) used homozygosity mapping to determine whether the region of canine chromosome 8 (CFA08) that is syntenic with human 14q22.1 (containing L2HGDH) was predominantly homozygotic in dogs with l‐2‐HGA and heterozygotic in obligate carriers, and (2) sequenced and compared the coding sequence of canine L2HGDH in affected, carrier and unaffected dogs. Significantly, in this study, the use of outbred pet dogs rather than a purpose‐bred colony offered a direct welfare benefit to the affected animal breed while providing the potential to recruit a large population of affected animals of relatively long life expectancy and large body size with which to study an uncommon and poorly understood neurometabolic disease in man.
Methods
Animals
Twenty‐one Staffordshire bull terriers with l‐2‐HGA and 127 closely related normal control Staffordshire bull terriers were enlisted from an outbred population of pet dogs. A presumptive diagnosis of l‐2‐HGA was made based on suggestive clinical signs and/or characteristic MRI features. Urine samples were collected from all dogs and stored at −20°C until analysis. Positive cases were confirmed by demonstrating l‐2‐HG accumulation in urine. Normal levels of 2‐HG were present in all control dogs. MRI studies of the brain were available in 12 l‐2‐HGA‐positive Staffordshire bull terriers. MRI was performed under general anaesthesia using a 1.5 T superconducting magnet (1.5 T Signa Echospeed system, General Electric Medical Systems, Waukesha, Wisconsin, USA) and an extremity coil. The following pulse sequences were used: T1‐weighted pre‐contrast and post‐contrast, T2‐weighted, T2*‐weighted (“gradient echo”) and fluid‐attenuated inversion recovery (FLAIR). Images were acquired in all three planes.
Urinary organic acid analysis
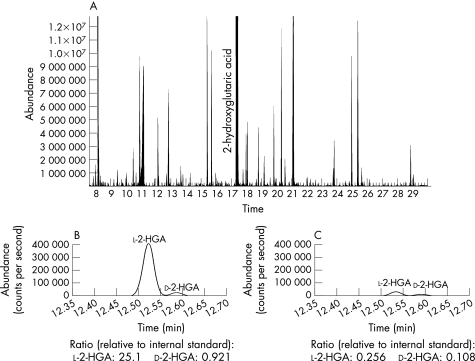
Accumulation of 2‐HG in urine was detected by gas chromatography (GC) mass spectrometric analysis. Urinary organic acids were extracted according to a modification of the protocol described by Tanaka et al.10 An aliquot of urine relative to creatinine was made up to 500 µl in non‐ionised water, to which 100 µl of internal standard was added (comprising 137.5 µmol/l heptadecanoic acid, heptanoylglycine and a deuterated analogue of methylmalonic acid). The resultant mixture was acidified with 50 µl of concentrated HCl to encourage the acidic species to favour the non‐ionised form and make them more soluble in organic solvents. Solid NaCl (1 g) was added and the organic acids were sequentially extracted with the organic solvents ethyl acetate and diethyl ether, and the extracts combined. The organic acid‐containing phase was separated and the organic solvents removed under a gentle stream of nitrogen at 60°C. The residue was reconstituted and derivatised at 80°C for 30 min in 75 µl of BSTFA (N,O‐bis(trimethylsilyl)trifluoroacetamide with trimethylchlorosilane; Sigma‐Aldrich, Gillingham, UK) and 20 µl pyridine (Sigma‐Aldrich). The derivatised organic acids were separated by GC and identified by mass spectrometry using an Agilent 6890 series GC system with a 5973 network mass‐selective detector (Agilent Technologies UK Ltd, Stockport, Cheshire, UK), operated in continuous scanning mode throughout the temperature programme. Urinary organic acids were identified based on the retention time, evaluation of their mass spectra and comparison of these mass spectra with computer library spectra. Differential diagnosis between l‐2‐HGA and D‐2‐HGA and quantification of l‐2‐HG and D‐2‐HG were performed in all dogs with evidence of 2‐HG accumulation by stable‐isotope‐dilution GC–mass spectrometry of O‐acetyl‐di‐(d)‐2‐butyl esters.11
L2HGDH microsatellite homozygosity mapping
To determine whether a mutation in L2HGDH was responsible for l‐2‐HGA in the canines, we downloaded DNA sequences corresponding to the canine homologue of L2HGDH (http://www.ensembl.org/Canis_familiaris/index.html) and approximately 500 000 nucleotides flanking either side of the gene. Within this region, we identified four closely spaced microsatellite markers and designed four sets of PCR primers to amplify these microsatellites in four separate PCR reactions, as follows: L2HGDH‐1 (C TTT repeat), forward primer: 5′‐CCAgggCCTgATAgAATCAA‐3′, reverse primer: 5′‐CCAgggCCTgATAgAATCAA‐3′; L2HGDH‐2 (CT and AT repeat), forward primer: 5′‐TATgTgCAgCCTTggCATAg‐3′, reverse primer: 5′‐TATgTgCAgCCTTggCATAg‐3′; L2HGDH‐3 (CTTT repeat), forward primer: 5′‐TgAgACTCCAgggATATgAACA‐3′, reverse primer: 5′‐TgAgACTCCAgggATATgAACA‐3′; and L2HGDH‐4 (CTTT and CT repeat), forward primer: 5′‐CTgCCACAgTgCCTTCAAT‐3′, reverse primer: 5′‐CTgCCACAgTgCCTTCAAT‐3′. The microsatellites were amplified using unlabelled forward and reverse primers, and a third fluorescently labelled primer, complementary to an 18‐nucleotide extension (TGA CCG GCA GCA AAA TTG), was added to the 5′‐end of each forward primer. PCR products were fractionated using ABI Prism 3100 genetic analysers (Applied Biosystems, Foster City, California, USA) and the resulting data analysed using GeneMapper software (Applied Biosystems). The markers were genotyped in the 19 clinically affected Staffordshire bull terriers, in 11 obligate carriers (assuming inheritance in a simple recessive manner) and in 12 clinically normal but closely related dogs. Clinical status was determined by the presence or absence of l‐2‐HG accumulation in urine. Obligate canine carriers demonstrated no l‐2‐HG accumulation, but were dogs that had produced affected offspring or were the offspring of affected animals.
Sequencing of canine L2HGDH
The 10 exons of canine L2HGDH, using Ensembl‐predicted exon/intron positions (Canis familiaris Ensembl Gene ID ENSCAFG00000014237) and including 120 base pairs of the flanking 5′‐intron and 3′‐intron regions, were amplified by PCR and sequenced in their entirety in one affected and two known carrier dogs, using the following PCR primers: exon 1, forward primer: 5′‐TCGATTGGCCCTTGAGTG‐3′, reverse primer: 5′‐CGGATCCTGACCTTCTCTCAC‐3′; exon 2, forward primer: 5′‐CCAGCAATGCCGTAATTCTT‐3′, reverse primer: 5′‐GCTCCTGCCTCCTAAACAAC‐3′; exon 3, forward primer: 5′‐CCAGTGCTCTGCCTGTAAGA‐3′, reverse primer: 5′‐CATGGAGTTAGTCATACCACCTTTT‐3′; exon 4, forward primer: 5′‐TGAGGTCATGAAGAAAGGAAGA‐3′, reverse primer: 5′‐TGAATGTGCCTTGTTGTTGTT‐3′; exon 5, forward primer: 5′‐GGCACTGAATTTCTGCCTGA‐3′, reverse primer: 5′‐CCCATATTCATCCCTTCTGC‐3′; exon 6, forward primer: 5′‐AAAGAGCATTAGAATGAAAAGTAGGA‐3′, reverse primer: 5′‐TTTGCTGGCACAATCACTTC‐3′; exon 7, forward primer: 5′‐CCTTGCATGCTGGAGAACTT‐3′, reverse primer: 5′‐ACCCAAAAAGCACTTCAAAA‐3′; exon 8, forward primer: 5′‐GTGTGGCACTTCTAGGCAGA‐3′, reverse primer: 5′‐TCCACAAAGCTCTTAATCTGGA‐3′; exon 9, forward primer: 5′‐CCTCCTTATATTGACCAAAAGTTGA‐3′, reverse primer: 5′‐AACAGCCATGAGACAGCAAG‐3′; and exon 10, forward primer: 5′‐TTTCCACATAATTGTGCATTTAGAA‐3′, reverse primer: 5′‐TTCACAAAGTAAAAGAGCATGGA‐3′. PCR products were purified and bidirectionally sequenced using ABI Prism 3100 genetic analysers (Applied Biosystems). Exons 8 and 10, including 120 base pairs of the flanking 5′‐intron and 3′‐intron regions, were additionally sequenced in 21 affected dogs, 11 known carriers and 4 normal related dogs (homozygotic for the wild‐type haplotype on homozygosity mapping). Pedigree analysis was performed using the software analysis program Cyrillic V.2.1 (CyrillicSoftware, Wallingford, UK).
Results
Affected dogs have dramatically increased levels of l‐2‐HG in urine and demonstrate characteristic changes in brain MRI
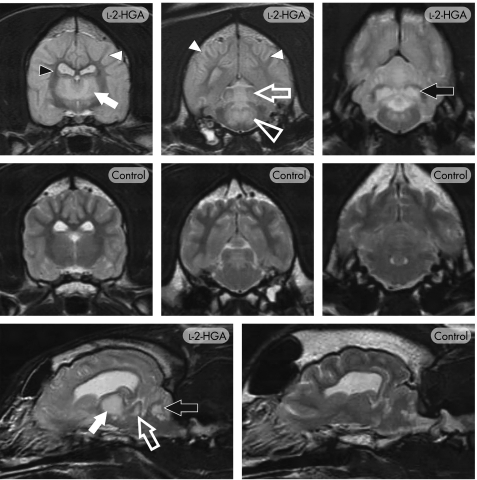
The biological hallmark of l‐2‐HGA is the finding of increased levels of l‐2‐HG in a variety of tissues and fluids, including urine. Organic acid profiles by GC–mass spectrometry represent one of the more sophisticated screening methods currently available, as a large number of metabolic disorders may be detected either directly or indirectly by this technique, including 2‐HG. GC–mass spectrometry demonstrated a dramatic increase of 2‐HG in the urine of all affected dogs, but in none of the unaffected dogs. Stable‐isotope‐dilution GC–mass spectrometry confirmed the presence of l‐2‐HG (vs d‐2‐HG) in all affected dogs (fig 1). Mean (SEM) l‐2‐HG concentrations in the affected (n = 8) and control (n = 25) dog population were determined as 2856 (262) and 3.12 (0.34) mmol/mol creatinine, respectively. MRI features in 12 affected dogs demonstrated a highly conserved pattern of bilaterally symmetrical regions of hyperintensity on T2‐weighted (fig 2) and FLAIR images similar to that observed in humans.1,12,13,14,15,16 Within the cerebral cortex there was evidence of white‐matter hyperintensity, with sparing of the central region of the internal capsule and the corpus callosum, and a zone of marked hyperintensity at the level of the grey–white matter interface. Consistent hyperintensity was evident in the caudal (inferior) colliculi, dorsomedian tegmentum, cerebellar nuclei (including the dentate nuclei) and thalamus. The basal nuclei, including the putamen and globus pallidus, were variably affected. On T1‐weighted images, the parenchyma demonstrating hyperintensity on T2‐weighted images seemed mildly hypointense. No abnormal contrast enhancement was evident.
Figure 1 Identification and quantitation of l‐2‐hydroxyglutaric acid (HG) in canine urine. (A) Total ion chromatogram following organic acid extraction demonstrating accumulation of 2‐HG in an affected dog. Control dogs demonstrated minimal or no detectable 2‐HG. Stable‐isotope‐dilution gas chromatography‐mass spectrometry of an affected dog (B) and control dog (C), with the ratio of the l‐2‐HG versus d‐2‐HG expressed as a ratio relative to an internal standard. Marked increase of l‐2‐HG is evident in the affected dog.
Figure 2 T2‐weighted MRI of canine l‐2‐hydroxyglutaric aciduria (l‐2‐HGA). Transverse images through the level of the thalamus, midbrain and cerebellum (top and middle panels) and para‐sagittal images (bottom panel) in an affected (l‐2‐HGA) and normal (control) Staffordshire bull terrier. Extensive, bilaterally symmetrical increased signal is present, particularly affecting the cerebral cortex, cerebral white matter (white arrowheads), thalamus (closed white arrow), caudal colliculi (open white arrow), dorsomedian tegmentum (open white arrowhead) and the cerebellar nuclei (black arrow), including the dentate nucleus. The central region of the internal capsule (small black arrowhead) and the corpus callosum retained the normal white‐matter signal.
Microsatellite homozygosity mapping indicates a mutation within L2HGDH is responsible for l‐2‐HGA in the spontaneous canine model
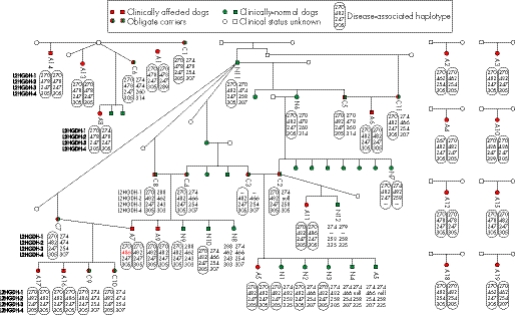
Four microsatellite markers were genotyped in 42 Staffordshire bull terriers (including 19 clinically affected dogs, 11 obligate carriers and 12 clinically normal but closely related dogs). The results demonstrated that clinically affected dogs were largely homozygotic for microsatellites closely flanking the L2HGDH gene, with a recognisable disease‐associated haplotype, and obligate carriers were heterozygous (fig 3). Within the clinically normal but related group, the microsatellite data indicated that eight of the dogs were heterozygotic, with one disease‐associated haplotype and one wild‐type haplotype. The remaining four clinically normal but related dogs were either heterozygotic for different wild‐type haplotypes or homozygotic for identical wild‐type haplotypes. These findings support the hypothesis that l‐2‐HGA is a recessive condition in the Staffordshire bull terrier and that a mutation within the canine homologue of L2HGDH is responsible for l‐2‐HGA in affected animals.
Figure 3 Genotyping data for microsatellites flanking canine L2HGDH in 19 clinically affected (A), 11 obligate carrier (C) and 12 clinically normal but related (N) Staffordshire bull terriers. Clinical status was determined by the presence or absence of l‐2‐hydroxyglutaric acid (l‐2‐HG) accumulation in urine. Clinically affected dogs demonstrated dramatic accumulation and clinically normal dogs demonstrated no to trace accumulation. Obligate carriers demonstrated no l‐2‐HG accumulation, but were dogs that had produced affected offspring or were the offspring of affected animals. The data demonstrate that clinically affected dogs are homozygotic for the disease‐associated haplotype and obligate carriers are heterozygotic, with one disease‐associated haplotype and one wild‐type haplotype. This is consistent with the hypothesis that a mutation in L2HGDH is the cause of l‐2‐HGA in dogs. Clinically normal but related Staffordshire bull terriers had either one or no disease‐associated haplotype. Clinically normal dogs N4 and N5 seemed to be homozygotic for allele 466 of L2HGDH‐2, which is discrepant with the genotypes of their parents at this marker. It is therefore assumed that parent C2 is heterozygotic for allele 482 and for a null allele that fails to amplify, and that N4 and N5 are therefore heterozygotic for 466 and for the null allele. The high mutation rate of canine tetranucleotide microsatellite markers has resulted in an obligate mutation in L2HGDH‐2, from sire and dam C8 and C4, to clinically affected offspring A7 (red allele).
Sequencing of canine L2HGDH demonstrates a substitution of two base pairs in exon 10 of the canine L2HGDH gene in affected dogs, resulting in a significant change in the encoded amino acid sequence
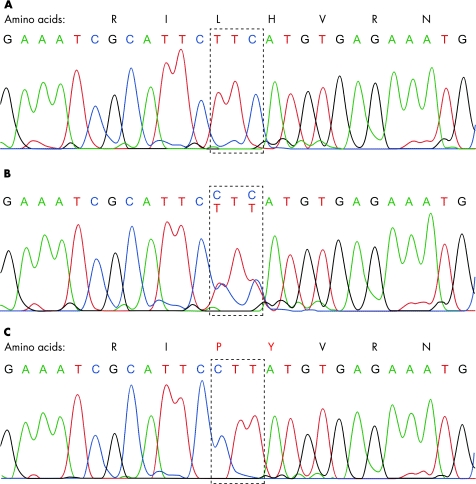
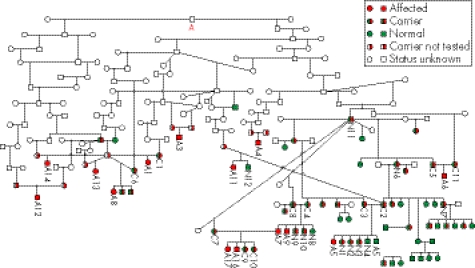
Comparison of the sequencing data from all 10 canine L2HGDH exons (with flanking intron regions) from the affected dogs and two carrier dogs with that of the published canine genome (Canis familiaris Ensembl Gene ID ENSCAFG00000014237) demonstrated the presence of two mutations within the genes: (1) two single‐nucleotide substitutions separated by a single invariant T nucleotide in exon 10 (c[1297T→C; 1299c→t]; p[Leu433Pro; His434Tyr]) and (2) a single nucleotide substitution in intron 8_9 (c[1064+8C→T]). All nucleotide numbering is in accordance with the current full Ensembl gene build for Canis familiaris (CanFam 1.0, http://www.ensembl.org/Canis_familiaris/). Sequencing of both mutations in additional dogs demonstrated that all 21 affected dogs were homozygotic and all carrier dogs were heterozygotic for the exon 10 double amino acid substitution. The mutation was present neither in any of the normal related dogs (determined by homozygosity mapping data to be homozygotic for the wild‐type haplotype), nor in a control population of unrelated clinically normal Staffordshire bull terriers. The mutation causes the substitution of two amino acids from leucine and histidine to proline and tyrosine, respectively (fig 4). The presence of an identical mutation in all affected dogs suggests a common founder effect within the Staffordshire bull terrier breed. The mutation within the 5′‐ flanking intron region of exon 8 did not modify an encoded amino acid and was homozygotic in some of the normal carriers, indicating that this is likely to be a polymorphism with no pathological significance. Five generation pedigrees were available for 14 affected dogs and 34 carrier dogs, and additional extension pedigrees were supplied by the Kennel Club. Although these dogs originated from various regions in the UK, all dogs could be traced back to a common ancestry, supportive of a common founder effect (fig 5).
Figure 4 Raw sequence data and resultant amino acid sequence from a normal control Staffordshire bull terrier (A), a Staffordshire bull terrier known to carry l‐2‐hydroxyglutaric aciduria (l‐2‐HGA) (B) and a Staffordshire bull terrier affected by l‐2‐HGA (C). A double amino acid substitution is present in exon 10 (amino acids substitution in red), resulting in a significant alteration in the amino acid sequence.
Figure 5 Extended pedigree analysis from all normal, carrier and affected dogs for which pedigree information was available. All of the 14 affected and 34 carrier dogs for which pedigree information was available could be traced back to a common ancestry (red letter A), supportive of a common founder effect. The numbering on the pedigree corresponds to the dog numbering in fig 3 (A, clinically affected; C, obligate carrier; N, clinically normal but related).
Discussion
l‐2‐HGA is a rare neurometabolic disease in man, and despite being recognised since 1980 only around 100 patients have been reported in the literature worldwide.1,3 This limited number of patients has considerably complicated the task of assessing potential treatments and providing an explanation for the clinical heterogeneity, despite a highly conserved biochemical and MRI phenotype.17,18,19 The MRI features of l‐2‐HGA in humans are suggestive of a white‐matter spongiform encephalopathy. This is supported by the neuropathological finding of spongiform changes with cystic cavitation, which predominate in the subcortical white matter but also include other regions of the brain.15,20,21 The exact cause of the pathology in l‐2‐HGA is unknown, but may represent a toxic effect of the l‐2‐HG accumulation. Some evidence suggests that this toxic effect may be mediated through induction of oxidative stress or mitochondrial dysfunction, as demonstrated by in vitro inhibition by l‐2‐HG of complex V of the mitochondrial respiratory chain and of the mitochondrial creatine kinase isoform in rat cerebellum.22,23,24 The demonstration that l‐2‐HG significantly increases intracellular calcium levels and induces cell death through stimulation of N‐methyl d‐aspartate glutamate receptors in chick embryo neuronal cultures has been used as supportive evidence to propose l‐2‐HG as an endogenous excitatory organic acid.25 Despite demonstrating that both l‐2‐HG and d‐2‐HG affect mitochondrial respiratory parameters in mitochondrial preparations of the rat brain, an inhibitory effect on mitochondrial energy metabolism has only been shown for d‐2‐HG.26 Differences in endogenous enzymatic function may provide an explanation for some of the variable effects in experimental models, with D‐2‐HG dehydrogenase, but not l‐2‐HG dehydrogenase, requiring Zn2+ or Co2+ as cofactors. Management of other organic acidurias where the exact enzyme defect has been characterised is by dietary protein restriction with appropriate amino acid, carnitine, mineral and vitamin supplementation. Currently, no specific treatment is available for l‐2‐HGA in affected humans.
This study demonstrated that a spontaneous canine model of l‐2‐HGA is caused by a mutation in canine L2HGDH and is thus a true homologue of human l‐2‐HGA, which offers an invaluable opportunity to further the understanding of the disease pathogenesis and treatment.4,8,9 We exploited the newly available canine whole‐genome sequence to identify microsatellites closely flanking canine L2HGDH. Subsequent genotyping and sequence analysis of affected and carrier dogs enabled the rapid confirmation that a mutation in canine L2HGDH is associated with l‐2‐HGA in this spontaneous canine model. The markers chosen for microsatellite homozygosity mapping were predominantly tetranucleotides that have a high mutation frequency.27 This resulted in one demonstrated obligate mutation in the microsatellite amplified by L2HGDH‐2, from sire and dam C8 and C4, to clinically affected offspring A7. The sequencing data demonstrated that the mutation in canine L2HGDH is a double amino acid substitution in exon 10, comprising two single‐nucleotide substitutions separated by a single invariant T nucleotide (c[1297T→C; 1299c→t]; p[Leu433Pro; His434Tyr]). The presence of a double nucleotide substitution raises the possibility that instead of occurring simultaneously, the nucleotide substitutions occurred in two steps. If the mutation did occur in two steps, it is likely that the first nucleotide substitution either did not confer any disease symptoms or only caused mild symptoms. In this situation, either the two mutations together confer disease or the second mutation is sufficient to confer disease on its own.
Key points
Mutations within the gene L2HGDH (Entrez Gene ID 79944) encoding l‐2‐hydroxyglutaric dehydrogenase have been demonstrated to cause l‐2‐hydroxyglutaric aciduria (l‐2‐HGA), producing a variety of clinical neurological deficits in affected patients. The causal molecular defect was identified and characterised in the canine homologue of L2HGDH in an outbred pet dog population segregating l‐2‐HGA.
Sequencing data demonstrated the mutation in canine L2HGDH as a double amino acid substitution in exon 10, comprising two single‐nucleotide substitutions separated by a single invariant T nucleotide (c[1297T→C; 1299c→t]; p[Leu433Pro; His434Tyr]).
The canine model shares many of the clinical and MRI features of the human disease. The relatively long life expectancy and larger body size of the dog makes it an appropriate comparative model, with the ethical advantage that it occurs spontaneously within an outbred population of pet dogs.
In all the dogs we tested, only the double amino acid substitution was identified and no dogs were identified with just one amino acid substitution, more evidence for the two mutations to have occurred simultaneously. Although the effect of the canine L2HGDH mutation was not determined through functional studies or predictive modelling in this study, the mutation was homozygotic in all affected animals and heterozygotic in all obligate carriers. The mutation resulted in the substitution of two amino acids in the protein sequence with, significantly, the introduction of a proline.
Our study of this spontaneous canine model of l‐2‐HGA, with its high degree of disease homology with human l‐2‐HGA, offers a particular ethical advantage over previous canine genetic studies because it used an outbred population of pet dogs instead of purpose‐bred laboratory dogs.28,29 The fact that all the outbred affected dogs in the study were homozygotic for the identical mutation strengthens our conclusion that the double amino acid substitution we identified is the cause of the disease in these animals. This is as opposed to being an innocent polymorphism in linkage disequilibrium with the true disease locus, which could be a criticism if the study had relied solely on results from an inbred colony of animals. The domestic dog has more naturally arising inherited diseases than any other species with the exception of man. Many canine diseases are known to have human equivalents, making the dog an excellent species with which to advance human medicine through comparative genetics. Until recently, however, the dog was man's poor relation, in terms of knowledge about its genome, but the recent availability of the canine genome sequence has changed that.30 The development of a mouse model to study l‐2‐HGA would hold significant advantages with regard to cost and speed of generation of an experimental model. However, the relatively long life expectancy and larger body size of the dog makes it a more appropriate comparative model, with the ethical advantage that it occurs spontaneously within the canine population. This study represents an example of how spontaneous disease models in the domestic dog can be successfully identified and used to study disease pathogenesis and evaluate clinical treatments.
Acknowledgements
This work was funded by Petsavers, the Animal Health Trust, the Metabolic Unit of the Department of Clinical Chemistry and Pediatrics, VU University Medical Centre and the University of Glasgow.
Abbreviations
GC - gas chromatography
l‐2‐HG - l‐2‐hydroxyglutaric acid
l‐2‐HGA - l‐2‐hydroxyglutaric aciduria
Footnotes
Competing interests: None declared.
References
- 1.Topcu M, Aydin O F, Yalcinkaya C, Haliloglu G, Aysun S, Anlar B, Topaloglu H, Turanli G, Yalnizoglu D, Kesimer M, Coskun T. L‐2‐Hydroxyglutaric aciduria: a report of 29 patients. Turk J Pediatr 2005471–7. [PubMed] [Google Scholar]
- 2.Barth P G, Hoffmann G F, Jaeken J, Lehnert W, Hanefeld F, van Gennip A H, Duran M, Valk J, Schutgens R B H, Trefz F K, Reimann G, Hartung H‐P. L‐2‐hydroxyglutaric acidemia: a novel inherited neurometabolic disease. Ann Neurol 19923266–71. [DOI] [PubMed] [Google Scholar]
- 3.Duran M, Kamerling J P, Bakker H D, van Gennip A H, Wadman S K. L‐2‐Hydroxyglutaric aciduria: an inborn error of metabolism? J Inherit Metab Dis 19803109–112. [DOI] [PubMed] [Google Scholar]
- 4.Rzem R, Veiga‐da‐Cunha M, Noel G, Goffette S, Nassogne M C, Tabarki B, Scholler C, Marquardt T, Vikkula M, Van Schaftingen E. A gene encoding a putative FAD‐dependent L‐2‐hydroxyglutarate dehydrogenase is mutated in L‐2‐hydroxyglutaric aciduria. Proc Natl Acad Sci USA 200410116849–16854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rzem R, Van Schaftingen E, Veiga‐da‐Cunha M. The gene mutated in L‐2‐hydroxyglutaric aciduria encodes L‐2‐hydroxyglutarate dehydrogenase. Biochimie 200688113–116. [DOI] [PubMed] [Google Scholar]
- 6.Abramson C, Garosi L, Platt S, Penderis J. Metabolic defect in Staffordshire bull terriers. Vet Rec 2001149532. [PubMed] [Google Scholar]
- 7.Abramson C J, Platt S R, Jakobs C, Verhoeven N M, Dennis R, Garosi L, Shelton G D. L‐2‐Hydroxyglutaric aciduria in Staffordshire Bull Terriers. J Vet Intern Med 200317551–556. [DOI] [PubMed] [Google Scholar]
- 8.Topçu M, Jobard F, Halliez S, Coskun T, Yalcinkayal C, Gerceker F O, Wanders R J, Prud'homme J F, Lathrop M, Ozguc M, Fischer J. L‐2‐Hydroxyglutaric aciduria: identification of a mutant gene C14orf160, localized on chromosome 14q22.1. Hum Mol Genet 2004132803–2811. [DOI] [PubMed] [Google Scholar]
- 9.Vilarinho L, Cardoso M L, Gaspar P, Barbot C, Azevedo L, Diogo L, Santos M, Carrilho I, Fineza I, Kok F, Chorao R, Alegria P, Martins E, Teixeira J, Cabral F H, Verhoeven N M, Salomons G S, Santorelli F M, Cabral P, Amorim A, Jakobs C. Novel L2HGDH mutations in 21 patients with L‐2‐hydroxyglutaric aciduria of Portuguese origin. Hum Mutat 200526395–396. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka K, West‐Dull A, Hine D G, Lynn T B, Lowe T. Gas‐chromatographic method of analysis for urinary organic acids. II. Description of the procedure, and its application to diagnosis of patients with organic acidurias. Clin Chem 1980261847–1853. [PubMed] [Google Scholar]
- 11.Gibson K M, ten Brink H J, Schor D S, Kok R M, Bootsma A H, Hoffmann G F, Jakobs C. Stable‐isotope dilution analysis of D‐ and L‐2‐hydroxyglutaric acid: application to the detection and prenatal diagnosis of D‐ and L‐2‐hydroxyglutaric acidemias. Pediatr Res 199334277–280. [DOI] [PubMed] [Google Scholar]
- 12.Barbot C, Fineza I, Diogo L, Maia M, Melo J, Guimaraes A, Pires M M, Cardoso M L, Vilarinho L. L‐2‐Hydroxyglutaric aciduria: clinical, biochemical and magnetic resonance imaging in six Portuguese pediatric patients. Brain Dev 199719268–273. [DOI] [PubMed] [Google Scholar]
- 13.D'Incerti L, Farina L, Moroni I, Uziel G, Savoiardo M. L‐2‐Hydroxyglutaric aciduria: MRI in seven cases. Neuroradiology 199840727–733. [DOI] [PubMed] [Google Scholar]
- 14.Moroni I, D'Incerti L, Farina L, Rimoldi M, Uziel G. Clinical, biochemical and neuroradiological findings in L‐2‐hydroxyglutaric aciduria. Neurol Sci 200021103–108. [DOI] [PubMed] [Google Scholar]
- 15.Seijo‐Martinez M, Navarro C, Castro dR, Vila O, Puig M, Ribes A, Butron M. L‐2‐hydroxyglutaric aciduria: clinical, neuroimaging, and neuropathological findings. Arch Neurol 200562666–670. [DOI] [PubMed] [Google Scholar]
- 16.Sener R N. L‐2 hydroxyglutaric aciduria: proton magnetic resonance spectroscopy and diffusion magnetic resonance imaging findings. J Comput Assist Tomogr 20032738–43. [DOI] [PubMed] [Google Scholar]
- 17.de Klerk J B, Huijmans J G, Stroink H, Robben S G, Jakobs C, Duran M. L‐2‐hydroxyglutaric aciduria: clinical heterogeneity versus biochemical homogeneity in a sibship. Neuropediatrics 199728314–317. [DOI] [PubMed] [Google Scholar]
- 18.Fujitake J, Ishikawa Y, Fujii H, Nishimura K, Hayakawa K, Inoue F, Terada N, Okochi M, Tatsuoka Y. L‐2‐hydroxyglutaric aciduria: two Japanese adult cases in one family. J Neurol 1999246378–382. [DOI] [PubMed] [Google Scholar]
- 19.Sztriha L, Gururaj A, Vreken P, Nork M, Lestringant G. L‐2‐hydroxyglutaric aciduria in two siblings. Pediatr Neurol 200227141–144. [DOI] [PubMed] [Google Scholar]
- 20.Larnaout A, Hentati F, Belal S, Ben Hamida C, Kaabachi N, Ben Hamida M. Clinical and pathological study of three Tunisian siblings with L‐2‐hydroxyglutaric aciduria. Acta Neuropathol (Berl) 199488367–370. [DOI] [PubMed] [Google Scholar]
- 21.Wilcken B, Pitt J, Heath D, Walsh P, Wilson G, Buchanan N. L‐2‐hydroxyglutaric aciduria: three Australian cases. J Inherit Metab Dis 199316501–504. [DOI] [PubMed] [Google Scholar]
- 22.da Silva C G, Bueno A R, Schuck P F, Leipnitz G, Ribeiro C A, Wannmacher C M, Wyse A T, Wajner M. L‐2‐hydroxyglutaric acid inhibits mitochondrial creatine kinase activity from cerebellum of developing rats. Int J Dev Neurosci 200321217–224. [DOI] [PubMed] [Google Scholar]
- 23.Kolker S, Okun J G, Ahlemeyer B, Mayatepek E, Krieglstein S, Hoffmann G F. D‐2 and L‐2‐hydroxyglutarate: characterisation of two novel endogenous excitotoxic organic acids. J Inherit Metab Dis 200124S1–140. [Google Scholar]
- 24.Latini A, Scussiato K, Rosa R B, Leipnitz G, Llesuy S, Bello‐Klein A, Dutra‐Filho C S, Wajner M. Induction of oxidative stress by L‐2‐hydroxyglutaric acid in rat brain. J Neurosci Res 200374103–110. [DOI] [PubMed] [Google Scholar]
- 25.Kolker S, Pawlak V, Ahlemeyer B, Okun J G, Horster F, Mayatepek E, Krieglstein J, Hoffmann G F, Kohr G. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d‐2‐hydroxyglutaric aciduria. Eur J Neurosci 20021621–28. [DOI] [PubMed] [Google Scholar]
- 26.Latini A, da Silva C G, Ferreira G C, Schuck P F, Scussiato K, Sarkis J J, Dutra Filho C S, Wyse A T, Wannmacher C M, Wajner M. Mitochondrial energy metabolism is markedly impaired by D‐2‐hydroxyglutaric acid in rat tissues. Mol Genet Metab 200586188–199. [DOI] [PubMed] [Google Scholar]
- 27.Francisco L V, Langston A A, Mellersh C S, Neal C L, Ostrander E A. A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome 19967359–362. [DOI] [PubMed] [Google Scholar]
- 28.Acland G M, Ray K, Mellersh C S, Langston A A, Rine J, Ostrander E A, Aguirre G D. A novel retinal degeneration locus identified by linkage and comparative mapping of canine early retinal degeneration. Genomics 199959134–142. [DOI] [PubMed] [Google Scholar]
- 29.Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong P J, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 199998365–376. [DOI] [PubMed] [Google Scholar]
- 30.Lindblad‐Toh K, Wade C M, Mikkelsen T S, Karlsson E K, Jaffe D B, Kamal M, Clamp M, Chang J L, Kulbokas EJ I I I, Zody M C, Mauceli E, Xie X, Breen M, Wayne R K, Ostrander E A, Ponting C P, Galibert F, Smith D R, DeJong P J, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin C W, Cook A, Cuff J, Daly M J, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli K P, Parker H G, Pollinger J P, Searle S M, Sutter N B, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait‐Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu A C, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger J P, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O'Neill B, O'Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange‐Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander E S. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005438803–819. [DOI] [PubMed] [Google Scholar]