Abstract
Background
The AZFc region on the human Y chromosome has been found to be functionally important in spermatogenesis. Complete AZFc deletion is one of the most frequent causes of male infertility and the roles of partial AZFc deletions (gr/gr and b2/b3 deletions) in spermatogenesis are controversial.
Methods
To further study the roles of partial AZFc deletions in spermatogenic impairment and the relationship between complete and partial AZFc deletions, these deletions were typed and quantitative analysis of DAZ gene copies and Y chromosome haplogrouping were performed for seven pedigrees of complete AZFc deletion carriers, comprising 296 infertile and 280 healthy Chinese men.
Results
Neither the gr/gr nor the b2/b3 deletion was found to be associated with spermatogenic failure. In one pedigree, a complete AZFc deletion was observed to result from the gr/gr deletion, suggesting that complete deletions of AZFc can be preceded by partial deletions. In addition, a new gr/gr‐deleted Y haplogroup Q1 was identified and the reported fixation of the b2/b3 deletion in haplogroup N confirmed. The frequency of complete AZFc deletion in haplogroups Q1 and N was significantly higher than that in the other haplogroupsm with fewer partial deletions. Duplications of DAZ gene copies were also observed in this study.
Conclusions
To date, these observations comprise the first evidence showing that partial AZFc deletions can increase the risk of complete AZFc deletion. The susceptibility of partial AZFc deletions to complete AZFc deletion deserves further examination, especially in the populations or Y haplogroups abundant in partial AZFc deletions.
Keywords: Y chromosome; AZFc deletion, haplogroup; male infertility; spermatogenesis
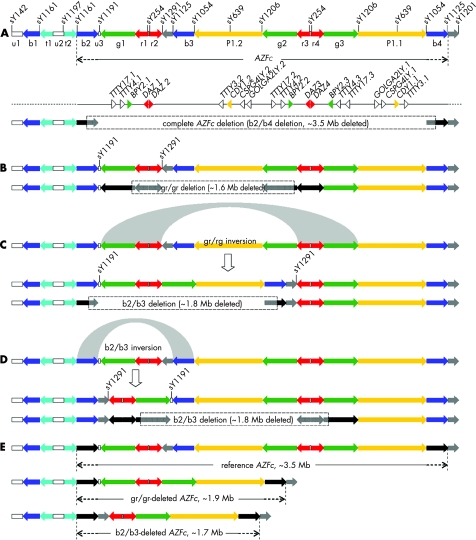
Deletions on the human Y chromosome are one of the main causes of male infertility.1,2 Three azoospermia factors (AZFa, AZFb and AZFc) have been mapped to Yq11,3 and of these, AZFc is the region most often involved in deletions.4,5 As AZFc is composed of several distinct families of long repeats (amplicons) (fig 1A), it is susceptible to non‐allelic homologous recombination between amplicons, which induces the recurrence of various deletions in the gene 6,11,12
Figure 1 Structures and rearrangements of amplicons in the AZFc region. (A) The amplicon structure, sequence‐tagged sites, three protein‐coding gene families (solid triangles) and five non‐coding gene families (open triangles) in AZFc of the GenBank reference sequence.6,7 The recombination between two b amplicons (shown in black) can lead to complete deletion of AZFc. (B) The gr/gr deletion removed the part of AZFc that included sY1291.8 (C) The b2/b3 deletion removed the part of the gr/rg‐inversed AZFc that included sY1191.9 (D) The b2/b3 deletion removed the part of the b2/b3‐inversed AZFc that included sY1191.9,10 (E) Comparison of the distances between the b amplicons (the recombination targets of complete AZFc deletion) of reference AZFc, gr/gr‐deleted AZFc and b2/b3‐deleted AZFc.
Recently, two types of partial AZFc deletions were identified. One partial deletion is the gr/gr deletion, which is caused by homologous recombination between two g or two r amplicons (fig 1B).8 In particular, a 1.6 Mb DNA segment was excised from the AZFc region, which was considered as a significant risk factor for spermatogenic failure in Dutch, Spanish, Italian and Australian studies.8,13,14,15,16 However, the association of this deletion with spermatogenic failure was not confirmed in French, German, Brazilian, Japanese or Sri Lankan men, or in our previous study in Chinese men.17,18,19,20,21,22,23,24 Furthermore, this association was also contradicted by the fact that all the tested men of a common Y haplogroup, D2b, presumably fertile, carry the gr/gr deletion.8
The other partial deletion is the b2/b3 deletion (also known as the g1/g3 deletion or u3‐gr/gr deletion), which removes a 1.8 Mb DNA segment from the b2/b3‐inversed or gr/rg‐inversed AZFc (fig 1C, D).9,10,17 This deletion is found to be fixed (100%) in N, a haplogroup widely distributed in northern Eurasia.25,26 The association of the b2/b3 deletion with male infertility was recently reported in Chinese men, whereas no predisposition was detected in other populations.9,10,16,17,18,24
In contrast, complete AZFc deletion (referred to as the b2/b4 deletion), which removes all eight testis‐specific expressed gene families in AZFc (fig 1A), has long been known to cause azoospermia or oligozoospermia with few exceptions.6,27 In fact, it accounts for over half of the Y chromosome deletions causing male infertility.4,5
In this study, we typed complete and partial AZFc deletions in 296 patients with spermatogenic impairment and 280 healthy donors (controls) in a Chinese population to study the roles of these deletions in spermatogenesis and male infertility. We also typed 19 binary markers on the Y chromosome and subdivided patients and controls into different haplogroups. Studying the deletion distribution in Y haplogroups could help uncover the effect of the genetic background of each Y haplogroup in determining the incidence of deletions, which may account for the diverse frequency of partial AZFc deletions and their inconsistent association with spermatogenic impairment across populations. In addition, we performed quantitative analysis of DAZ gene copy in AZFc to further characterise partial AZFc deletions in this study.
METHODS
Subjects
This study was approved by the institutional ethics committees of Nanjing Medical University and Renji Hospital, and informed consent was obtained from all participants. In total, 296 unrelated patients with non‐obstructive azoospermia (216 men) or oligozoospermia (<20 million sperm per mL; 80 men) were sampled from the infertility clinic at the First Affiliated Hospital with Nanjing Medical University at Jiangsu and Renji Hospital, Shanghai. As controls, 280 healthy donors, who had normal sperm concentration, motility and morphology (90 men), or had fathered one or more healthy children (190 men), were also sampled from these two hospitals. In addition, seven pedigrees with infertile patients carrying complete AZFc deletion were sampled at Renji Hospital. The semen analysis for sperm concentration, motility and morphology was performed following World Health Organization criteria.28 These patients and controls are were all Han Chinese from East China.
Deletion typing
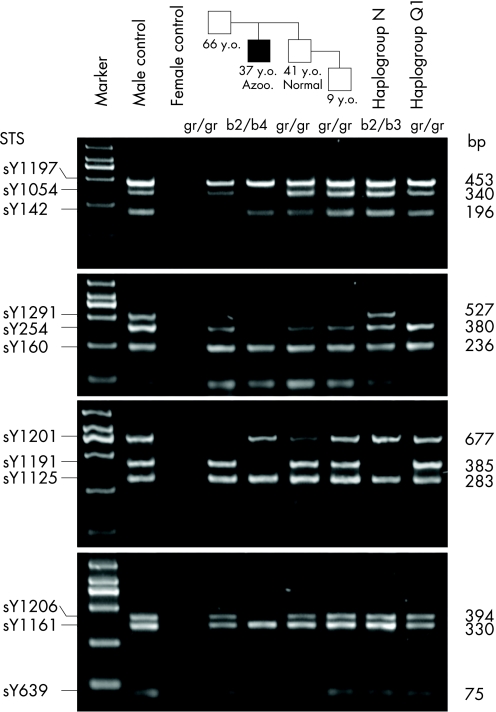
Details of the procedure were described in our previous study.23 Briefly, genomic DNA was extracted from peripheral blood samples. Partial AZFc deletions were typed by the following sequence‐tagged site (STS) pattern: gr/gr deletion (sY1161, sY1191, sY1201 and sY1206 are positive; sY1291 is negative) and b2/b3 deletion (sY1161, sY1201, sY1206 and sY1291 are positive; sY1191 is negative) (fig 2).8,9 Complete AZFc deletions were typed by STS analysis as described previously6 (fig 2). The locations of the STSs adopted are shown in fig 1A. Quantitative analyses of DAZ gene copies were performed to further characterise partial AZFc deletions. Using the method of Machev et al,17 we typed the sequence family variant (SFV) at sY587—that is, the single nucleotide variant (SNV) II of DAZ (DAZ‐SNV II), which can distinguish DAZ1/2 from DAZ3/4. Quantitative real‐time PCR of the DAZ gene was also performed to measure DAZ copy number. M159, a Y chromosomal locus outside AZFc, was used as the reference locus to calibrate measurement. DAZ‐SNV 1 and DAZ‐SNV 5, which are located at either end of DAZ, were chosen as the test loci. Standard curves were generated by amplifying a standard DNA without AZFc deletions, and the relative DAZ copy number of the test to the standard were measured.
Figure 2 The +/− STS pattern to identify complete or partial deletions in AZFc.6,8,9 The names (left) and product sizes in bp (right) of the adopted STSs are shown. The black box indicates the patient with azoospermia (Azoo.) in the pedigree. The age information (y.o., years old) has also been given under the box.
Y chromosome haplogrouping
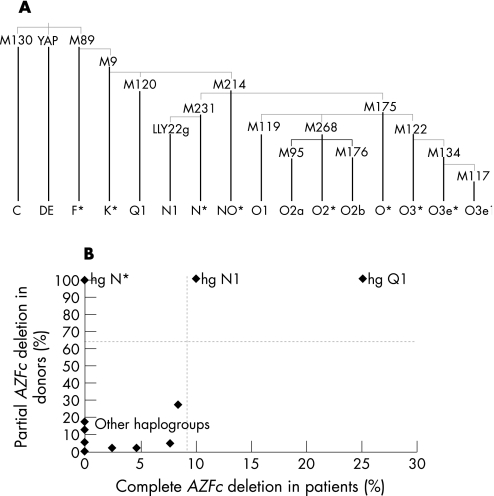
Y chromosome haplogroups were defined using 16 binary markers: M9, M89, M95, M117, M119, M120, M122, M130, M134, M175, M176 (SRY+465), M214, M231, M268, LLY22g and YAP (M1).29,30,31 LLY22g was typed using the protocol kindly provided by Y. Xue and C. Tyler‐Smith (Wellcome Trust Sanger Institute, personal communication). These 16 markers defined 16 Y haplogroups (fig 3A) following the nomenclature recommended by the Y Chromosome Consortium (YCC) and its update.31,32,33 In contrast to the previous report that M231 was phylogenetically equivalent to LLY22g in defining haplogroup N,26 we found that M231 defined a new haplogroup, N*. Therefore, following the YCC naming rules, we renamed the haplogroup defined by LLY22g as N1.33 Haplogroup N1 can be further divided into four sub‐haplogroups by three additional markers: N1*‐LLY22g, N1a‐M128, N1b‐P43 and N1c‐Tat.25,26,29,34
Figure 3 (A) Y chromosome phylogeny. The markers typed in this study are indicated in their defined branches. (B) Scatter plot of deletion frequencies in the Y haplogroups. Frequency of complete AZFc deletion in patients (X axis) and frequency of partial AZFc deletions in controls (Y axis). As there is no haplogroup K* or haplogroup NO* found in healthy controls, these two haplogroups are not shown in this plot.
Statistical analysis
Rousset's exact test of population differentiation between infertile patients and healthy controls was performed by using Arlequin software.35 A Markov chain of 10 000 steps and the statistical significance level of p<0.05 were used. Differences in deletion frequencies between Y chromosome haplogroups or between infertile patients and healthy controls were examined using Fisher's exact test, with statistical significance set at of p<0.05.
RESULTS
Genesis of complete AZFc deletions in pedigrees
Seven pedigrees of complete AZFc deletion were studied, in which the probands lacked AZFc (complete AZFc deletion), whereas their fathers and other paternal relatives do not. In one pedigree, we found that the paternal relatives of the patient were gr/gr‐deleted; neither the gr/gr nor the b2/b3 deletion was found in the other six pedigrees. The patient in the gr/gr‐deleted pedigree is azoospermic. His brother has normal spermatogenesis, and his nephew (9 years old) and father (66 years old, assumed healthy) did not attend semen analysis (fig 2). Y chromosome haplogrouping indicated that this pedigree belongs to haplogroup O1. SFV analysis of DAZ gene indicated the absence of DAZ1/2 in the gr/gr deletions of this pedigree. After the confirmation of paternity using Y chromosome markers, we concluded that the complete AZFc deletion was derived from a gr/gr‐deleted Y chromosome in this pedigree.
Distribution of AZFc deletions in patients and controls
We investigated the distributions of partial AZFc deletions in 296 infertile patients with spermatogenesis failure and in 280 healthy controls (tables 1 and 2). In all, 24 (8.1%) gr/gr and 26 (8.8%) b2/b3 deletions were found in patients, and 20 (7.1%) gr/gr and 18 (6.4%) b2/b3 deletions were observed in controls. No significant difference in deletion frequency between patients and controls was found for either the gr/gr or b2/b3 deletion.
Table 1 Distribution of the subjects and complete/partial AZFc deletions in Y chromosome haplogroups.
| Group | n | Y chromosome haplogroup | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | DE | F* | K* | Q1 | N1 | N* | NO* | O1 | O2a | O2* | O2b | O* | O3* | O3e* | O3e1 | ||
| All | |||||||||||||||||
| Patient | 296 | 25 | 3 | 2 | 1 | 8 | 20 | 3 | 1 | 52 | 5 | 12 | 3 | 1 | 82 | 35 | 43 |
| Control | 280 | 24 | 1 | 2 | 0 | 10 | 13 | 1 | 0 | 49 | 8 | 11 | 3 | 1 | 70 | 39 | 48 |
| gr/gr deletion only | |||||||||||||||||
| Patient | 24 | 1 | 1 | 6 | 2 | 3 | 4 | 5 | 2 | ||||||||
| Control | 20 | 1 | 10 | 2 | 1 | 3 | 2 | 1 | |||||||||
| b2/b3 deletion only | |||||||||||||||||
| Patient | 26 | 3 | 18 | 3 | 2 | ||||||||||||
| Control | 18 | 3 | 13 | 1 | 1 | ||||||||||||
| Complete AZFc deletion only | |||||||||||||||||
| Patient | 14 | 2 | 2 | 1 | 4 | 1 | 2 | 2 | |||||||||
| Control | 0 | ||||||||||||||||
Table 2 SFV absence of DAZ genes in partial AZFc deletions.
| Group | n | gr/gr deletion | b2/b3 deletion | ||
|---|---|---|---|---|---|
| DAZ1/2 absence | DAZ3/4 absence | DAZ1/2 absence | DAZ3/4 absence | ||
| Patient | 50 | 17 | 7 | 1 | 25 |
| Control | 38 | 10 | 10 | 0 | 18 |
According to the published data of SFV or SNV analysis of the DAZ gene, it was proposed that only partial AZFc deletions with DAZ1/2 SFV absence seem to be associated with impairment of spermatogenesis, and absence of the DAZ3/4 SFV may have a limited effect on spermatogenesis.9,10,14,15 To further characterise the partial deletion subtypes, we typed the SFV at sY587, which can distinguish DAZ1/2 from DAZ3/4 (table 2). In contrast to the gr/gr deletion, most b2/b3 deletions showed DAZ3/4 SFV absence, which was consistent with previous reports and the arrangement mechanism of the b2/b3 deletion (fig 1C, D).10 Although there were more DAZ1/2 SFV absences in patients than in controls, no significant difference in the frequency of partial deletion subtypes was found between these two groups.
We also observed 14 (4.7%) complete AZFc deletions in patients, whereas none was found in healthy controls. This is consistent with previous reports that complete AZFc deletions lead to azoospermia or oligozoospermia with few exceptions.6,27
Haplogroups with high frequency of partial AZFc deletions
To explore the relationship of deletions and Y haplogroups, we first typed 16 Y chromosomal markers to define 16 haplogroups in patients and controls. Deletion frequencies varied greatly among the Y haplogroups (table 1). High frequencies of partial AZFc deletions were found in haplogroups Q1, N1 and N*.
Haplogroup Q1 was found to be gr/gr‐deleted. All 10 controls carry gr/gr deletions. Six of eight (75%) patients are gr/gr‐deleted, and the other two patients (25%) carry complete AZFc deletions. Therefore, we speculated that the ancestor of haplogroup Q1 was a gr/gr deletion carrier and that these two complete deletions were derived from gr/gr‐deleted Y chromosomes of Q1.
In haplogroup N1, all the tested men carry AZFc deletions, complete or partial. All 13 controls and 18 of 20 (90%) patients carry b2/b3 deletions, and the other two patients (10%) carry a complete AZFc deletion, which is consistent with the reported fixation of b2/b3 deletion in this haplogroup.9,10 Again, we speculated that these complete deletions were derived from b2/b3‐deleted ancestors of haplogroup N1. In this study, we also typed three further markers (M128, P43 and Tat) to subdivide haplogroup N1. One N1a‐M128 control and five N1c‐Tat subjects (four patients and one control) were found. Although no N1b‐P43 was detected in this study, it was found that both of two tested N1b‐P43 men of unknown spermatogenic phenotype had b2/b3 deletions (P. A. Underhill, Stanford University, personal communication). The b2/b3 deletions in haplogroup N1 were shown to be widely distributed throughout all the N1 sub‐haplogroups, which suggests that they have a common origin and are derived from the b2/b3‐deleted ancestor of N1.
Similar to N1, all three patients and one control in haplogroup N* have b2/b3 deletions. As N1 was phylogenetically derived from N*, the b2/b3 deletions in these two haplogroups could share the same deletion ancestor.
In contrast to the 100% fixation of AZFc deletions in haplogroups N*, N1 and Q1, the frequency of partial AZFc deletions was low (averaging 8.7% in patients and 5.5% in controls) in the other haplogroups.
Copy number variation of the DAZ gene in partial AZFc deletions
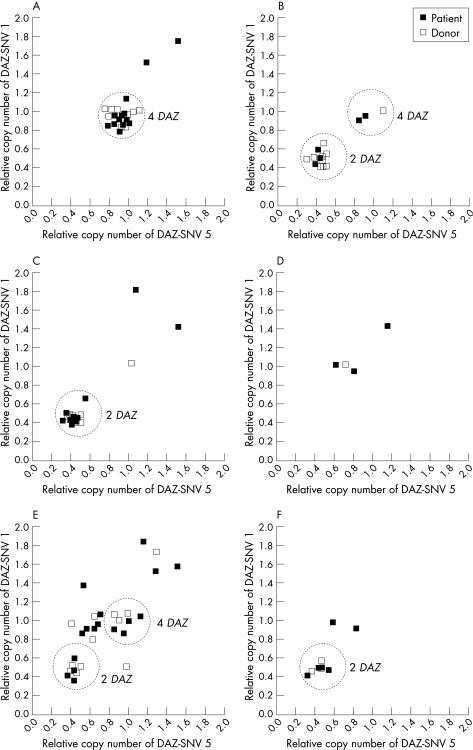
A previous report showed that there are four DAZ gene copies in AZFc on most human Y chromosomes.36 The frequent rearrangements in AZFc can change DAZ copy numbers. The gr/gr deletion and b2/b3 deletion commonly reduce DAZ copy from four to two,8,9,10 whereas partial duplication in AZFc was able to raise DAZ copy to >4 in non‐deletions or compensate for DAZ copy reduction caused by partial deletion.36,37DAZ copy numbers were examined using quantitative real‐time PCR analysis of two loci in the DAZ gene (fig 4).
Figure 4 Scatter plot of the copy number of DAZ‐SNVs in the test DNAs relative to the standard DNA. (A) Non‐deleted samples; (B) gr/gr deletions in haplogroup Q1; (C) gr/gr deletions in non‐Q1 haplogroups; (D) b2/b3 deletion in haplogroup N*; (E) b2/b3 deletion in haplogroup N1; (F) b2/b3 deletion in non‐N haplogroups. Filled box indicates patient and open box indicates healthy control.
In total, 26 (13 patients and 13 controls) of 28 randomly selected non‐deletions were shown to have the same DAZ copy number as the standard (fig 4A). Considering the previous observation that most Y chromosomes bear four DAZ copies,36 these 26 non‐deletions and the standard were assumed to have four DAZ copies. In addition, an increase in DAZ copies was found in two patients without deletions, but no increase was found in controls. This suggests that partial duplication of AZFc occurred in these two patients. No reduction in DAZ copy number was found in patients without deletions.
All 44 gr/gr deletions were examined. In haplogroup Q1, 13 (4 patients and 9 controls) of 16 men with gr/gr deletions bore two DAZ copies, which is consistent with the fact that commonly half of the four DAZ copies are removed by the gr/gr deletion (fig 4B).8 The other two patients and one control carrying the gr/gr deletion were shown to have four DAZ copies, suggesting that partial duplications followed gr/gr deletion and compensated for the reduction in DAZ copy number caused by the gr/gr deletion.37 There were 28 gr/gr deletions (18 patients and 10 controls) in non‐Q1 haplogroups, 25 of which bore two DAZ copies (fig 4C). DAZ duplications were also found in three men, including one control with four DAZ copies, one patient with six DAZ copies and one patient with a complex DAZ substructure (four DAZ‐SNV 1 copies but >4 DAZ‐SNV 5 copies). As DAZ‐SNV 1 and 5 are at opposite ends of DAZ genes, the unequal copy numbers of these two loci may indicate duplication of incomplete DAZ.
In contrast to the fact that DAZ copy numbers are reduced to two in most gr/gr deletions, partial duplication of DAZ copy may have occurred in many b2/b3 deletions. In haplogroup N*, all the four men (three patients and one control) with b2/b3 deletions bore at least four DAZ copies, according to the DAZ‐SNV 1 locus (fig 4D). In haplogroup N1, 10 (five patients and five controls) of 31 patients with deletions had two DAZ copies, and the other 21 subjects (13 patients and 8 controls) had >2 copies according to the DAZ‐SNV 1 or 5 locus (fig 4E). Of the nine subjects with b2/b3 deletions in the non‐N haplogroups, three patients and four controls had two DAZ copies and the other two patients had >2 copies of the DAZ‐SNV 1 locus (fig 4F).
It has been recently reported in Han Chinese in Taiwan that partial duplication occurred in one‐third of b2/b3 deletions in haplogroup N‐LLY22g (corresponding to N1 in this study), a frequency that is less than our finding of two‐thirds.37 However, the frequencies of partial duplication are relatively high for in haplogroup N1 in both studies. There may be two reasons for the difference in frequency difference between the two studies. One is the different substructure of haplogroup N1 between Han Chinese in Taiwan and Han Chinese in East China. The other is that imbalanced duplication between DAZ‐SNV 1 and 5 loci may not be detectable by the Southern blot analysis used by Lin et al.37 This possibility is also supported by our observation that partial duplications indicated by both DAZ‐SNV 1 and 5 together were found in 11 of 31 (about one‐third) b2/b3 deletions in haplogroup N1, which is consistent with the recent report.37
Distribution of complete AZFc deletions in Y haplogroups
In total, 14 complete AZFc deletions were found in seven haplogroups of patients, with frequencies of 2.4–100%, of which the three highest were haplogroups NO* (100%), Q1 (25.0%) and N1 (10.0%). Considering the fixation of partial AZFc deletions in haplogroups N*, N1 and Q1, it was suggested that the haplogroups abundant in partial deletions showed a higher frequency of complete deletion than the other haplogroups (fig 3B). As haplogroup NO* contained only one sample (complete AZFc deletion) and the frequency of partial deletion was undetermined for this haplogroup, haplogroup NO* was not included in the following association study.
A significant difference (p<0.04, one‐sided; odds ratio (OR = 4.20), 95% CI 1.21 to 14.5) in the frequency of complete AZFc deletion was found between N*N1Q1 (fixation of partial AZFc deletions) and non‐N*N1Q1 haplogroups (non‐fixation of partial AZFc deletions) (table 3). In contrast to haplogroups N1 and Q1, all the b2/b3 deletions in haplogroup N* were followed by partial duplication. When partial AZFc deletions without duplication were accounted for, a more significant difference (p<0.03, one‐sided; OR = 4.78, 95% CI 1.37 to 16.7) in complete deletion frequency was found between haplogroups N1 and Q1 (high frequency of partial AZFc deletions without duplication) and the non‐N1Q1 haplogroups (relatively low frequency of partial AZFc deletions without duplication) (table 3). The aforementioned observations suggest that complete AZFc deletions are more likely to occur in partially deleted than in non‐deleted AZFc genes.
Table 3 Distribution of the patients of complete AZFc deletion in Y chromosome haplogroups with different frequencies of partial AZFc deletions.
| Haplogroup | Complete AZFc deletion | No complete AZFc deletion | OR (95% CI) | p Value* | |
|---|---|---|---|---|---|
| N1 and Q1 (high frequency of partial AZFc deletions†) | 4 (14.3%) | 24 (85.7%) | 4.78 (1.37 to 16.7) | 0.026 | |
| Non‐N1Q1 (low frequency of partial AZFc deletions) | 9 (3.4%) | 258 (96.6%) | |||
| N*, N1, Q1 (fixation of partial AZFc deletions‡) | 4 (12.9%) | 27 (87.1%) | 4.20 (1.21 to 14.5) | 0.036 | |
| Non‐N*N1Q1 (non‐fixation of partial AZFc deletions) | 9 (3.4%) | 255 (96.6%) |
*Fisher's exact test, one‐sided.
†Excluding the partial AZFc deletions followed by partial duplication.
‡Including the partial AZFc deletions followed by partial duplication.
DISCUSSION
A new gr/gr‐deleted haplogroup: Q1
The previous report of fixation of the gr/gr deletion in haplogroup D2b challenged its effect on male infertility.8 It was suggested that the ancestor of haplogroup D2b was gr/gr‐deleted and this deletion can be successfully transmitted from generation to generation, contradicting a direct association of gr/gr deletion and spermatogenic failure.
In this study, we found that a new haplogroup, Q1, is gr/gr‐deleted. In this group, all the tested healthy controls carried the gr/gr deletion, and all the infertile patient carried either the gr/gr deletion or complete AZFc deletions that were speculated to derive from gr/gr‐deleted ancestors.
As the STS absence of sY1291, which is characteristic of a gr/gr deletion, can also be caused by polymorphic loss of o1084/o1085 (sY1291) in haplogroup J,17 we performed the quantitative DAZ copy assay to confirm the gr/gr deletions.23 Most gr/gr deletions of haplogroup Q1 were found to bear two DAZ copies, which is consistent with the fact that two of the four DAZ gene copies are commonly removed by the gr/gr deletion.8 Partial duplication occurred in the other three haplogroup Q1 gr/gr deletions and compensated for the DAZ copy reduction. Therefore, the polymorphic loss of sY1291 in haplogroup Q1 can be excluded, and haplogroup Q1 is a new haplogroup that is gr/gr‐deleted.
It was recently reported that gr/gr deletion and later duplication has been found in seven Han Chinese of haplogroup Q (defined as P36) in Taiwan. Considering two previous observations that non‐deletions and b2/b3 deletions have been found in Q sub‐haplogroups other than Q1 and that haplogroup Q1 is the dominant Q sub‐haplogroup in Han Chinese,8,9,38 it is possible that the gr/gr deletions of Q haplogroup detected by Lin et al in Taiwanese Han Chinese belong to haplogroup Q1, not to other Q sub‐haplogroups.
Haplogroup Q1 is widely distributed in Sino‐Tibetan populations, with a frequency of 1.8% to 7.1%.38 No predisposition to male infertility has been reported in haplogroup Q1. Further examination of the spermatogenic ability of men in haplogroup Q1 is therefore imperative to uncover the role of the gr/gr deletion in spermatogenesis.
A two‐step process of complete AZFc deletion
Because of the rearrangement structure of AZFc, complete AZFc deletion (between amplicon b2 and b4) is assumed to be derived from two types of Y chromosome: a normal one or one with a partial AZFc deletion. In the pedigrees of this study, one of seven (14.3%) complete AZFc deletions was found to be derived from the the gr/gr deletion, which suggested that some complete deletions of AZFc can be caused by a two‐step process (partial AZFc deletion followed by deletion of the rest of the gene). However, it is unknown whether it is easier to generate compete AZFc deletion in a partially deleted AZFc than in a non‐deleted one. If it is easier, there will be a higher frequency of complete AZFc deletions in haplogroups that have a high frequency of partial AZFc deletions than in haplogroups lacking these deletions.
Association of partial AZFc deletions with increased incidence of complete AZFc deletion
In total, 14 (4.7%) complete AZFc deletions were found in our 296 patients with azoospermia or oligozoospermia, which is consistent with the frequency estimate (5–6%) in azoospermia or severe oligozoospermia.12 The distribution of this deletion was found vary between the different haplogroups in this study. Significantly more complete AZFc deletions were found in the haplogroups (N1 and Q1) that had a high frequency of partial AZFc deletion (excluding partial duplication) than in the other haplogroups with fewer partial deletions. This observation suggests an association of partial AZFc deletions with an increased incidence of complete AZFc deletion.
Although the mechanism of this susceptibility of partial AZFc deletions to complete AZFc deletion requires further scrutiny, comparison of the structures of partially deleted AZFc with that of normal AZFc suggested a reason for this predisposition. Complete AZFc deletion has known to be caused by intrachromosomal homologous recombination between b amplicons at opposite ends of the AZFc region.6 The distance between these two b amplicons (that is, the deletion length) is about 3.5 Mb in reference AZFc, 1.9 Mb in the gr/gr deletion and 1.7 Mb in the b2/b3 deletion (fig 1E). The recent genomic survey of deletion polymorphisms in humans reported that the deletion length followed a L‐shape distribution for deletion frequency, suggesting that small deletions have a higher prevalence than larger ones.39 Therefore, the reduction of nearly half in the distance between recombination targets may consequently increase the incidence of complete AZFc deletion and raise the risk of male infertility.
Recently, the susceptibility of haplogroup E to complete AZFc deletion was reported in northern Italian men,40 although its mechanism was unknown. As higher frequencies of the gr/gr deletion (40% in haplogroup E3b2 and 20% in haplogroup E3b3) were reported in the sub‐haplogroups of haplogroup E3b (the dominant E haplogroup in Europeans) than in other haplogroups of Europeans (<16.7%),8,41 the effects of partial AZFc deletions can be considered a candidate cause for this susceptibility.
The possible susceptibility of overdosage of DAZ gene to spermatogenic impairment
It was recently reported that partial AZFc duplications with six DAZ copies was a risk factor of male infertility in Taiwanese Han Chinese.37 In this study, most of the tested men without deletions normally bore four DAZ copies, whereas the non‐deleted men with >4 copies were found only in patients. In contrast to the non‐deleted men, there were commonly two DAZ copies left in partial AZFc deletions. DAZ copy duplication was also found in some partial deletions. Four gr/gr deletions (two patients and two controls) and 21 b2/b3 deletions (13 patients and 8 controls) were shown to have four copies of DAZ‐SNV 1 or 5. No obvious difference was found between patients and controls, which suggested that the DAZ restoration to four copies caused by partial AZFc duplication after deletion had a limited effect on spermatogenesis. However, two gr/gr deletions (both patients) and six b2/b3 deletions (five patients and only one control) were shown to bear >4 copies of DAZ‐SNV 1 or 5. The observation that high DAZ copy number (>4) was over‐represented in patients suggested a possible susceptibility of DAZ overdosage to spermatogenic impairment.
Variable spermatogenic effects of partial AZFc deletions
As described in the introduction, the spermatogenic effects of partial AZFc deletions vary among populations. In our previous study, the gr/gr deletion was not shown to render an increased risk of spermatogenic impairment in East Asians.23 In this study, the sample size was enlarged and the b2/b3 deletion was also typed. We also performed Rousset's exact test of population differentiation,35 and no significant difference in Y haplogroup distribution was found between patients and controls (table 1). Therefore, the genetic stratification of the Y chromosome was excluded. In this study, we showed that neither the gr/gr nor the b2/b3 deletion was found to be associated with spermatogenic impairment.
CONCLUSION
In summary, by examining 296 patients with spermatogenic impairment and 280 healthy controls, we found that a new common haplogroup Q1 is gr/gr‐deleted. In addition, no association of the gr/gr or b2/b3 deletion with spermatogenic impairment was found in the tested population. Both of these results suggest phenotypic variation of partial AZFc deletions across populations. As Y haplogroups are always region‐characteristic,32 the different genetic backgrounds between Y haplogroups may account for this variation among populations. Therefore, Y haplogrouping is strongly recommended for comprehensive analysis of the controversial spermatogenic effects of partial AZFc deletions. Furthermore, we identified a patient in whom further deletion in the gr/gr‐deleted AZFc led to complete loss of AZFc, which suggested a two‐step process of complete AZFc deletion other than de novo complete deletion. Although no significant frequency difference in duplication following partial deletion in AZFc was found between patients and controls, overdosage of DAZ loci was over‐represented in patients, and its susceptibility to spermatogenic impairment can not be excluded. The association of partial AZFc deletions with increased incidence of complete AZFc deletion also provided a new insight into the role of partial AZFc deletions in male infertility. Further examination of this correlation in more populations will shed new light on the mechanism underlying complete deletion of AZFc on the human Y chromosome.
ELECTRONIC DATABASE INFORMATION
Arlequin's Home on the Web, http://lgb.unige.ch/arlequin/
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for sY142, sY160, sY254, sY639, sY1054, sY1125, sY1161, sY1191, sY1197, sY1201, sY1206 and sY1291)
ACKNOWLEDGEMENTS
We thank all the sample donors who made this work possible. We also thank Dr Yali Xue and Dr Chris Tyler‐Smith for kindly providing the unpublished information of the LLY22g marker. We thank Dr Peter Underhill for sharing with us the deletion data in haplogroup N‐P43. This study was supported in part by National Key Project for Basic Research (973) (2002CB512900) and National Natural Science Foundation of China (No.30571582).
Abbreviations
AZFc - azoospermia factor c
STS - sequence tagged site
SFV - sequence family variant
SNV - single nucleotide variant
YCC - Y Chromosome Consortium
Footnotes
Competing interests: None declared.
The first two authors have contributed equally to this study and they should be regarded as joint first authors.
References
- 1.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet 197634119–124. [DOI] [PubMed] [Google Scholar]
- 2.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page D C. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA‐binding protein gene. Nat Genet 199510383–393. [DOI] [PubMed] [Google Scholar]
- 3.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Köhn F M, Schill W B, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, H Behre M, Castel A, Nieschlag E, Weidner W, Gröne H J, Jung A, Engel W, Haidl G. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 19965933–943. [DOI] [PubMed] [Google Scholar]
- 4.Foresta C, Moro E, Ferlin A. Y Chromosome microdeletions and alterations of spermatogenesis. Endocr Rev 200122226–239. [DOI] [PubMed] [Google Scholar]
- 5.Krausz C, Forti G, McElreavey K. The Y chromosome and male fertility and infertility. Int J Androl 20032670–75. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda‐Kawaguchi T, Skaletsky H, Brown L G, Minx P J, Cordum H S, Waterston R H, Wilson R K, Silber S, Oates R, Rozen S, Page D C. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 200129279–286. [DOI] [PubMed] [Google Scholar]
- 7.Skaletsky H, Kuroda‐Kawaguchi T, Minx P J, Cordum H S, Hillier L, Brown L G, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin‐Wollam A, Yang S P, Waterston R H, Wilson R K, Rozen S, Page D C. The male‐specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003423825–837. [DOI] [PubMed] [Google Scholar]
- 8.Repping S, Skaletsky H, Brown L, van Daalen S K, Korver C M, Pyntikova T, Kuroda‐Kawaguchi T, de Vries J W, Oates R D, Silber S, van der Veen F, Page D C, Rozen S. Polymorphism for a 1.6‐Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 200335247–251. [DOI] [PubMed] [Google Scholar]
- 9.Repping S, van Daalen S K, Korver C M, Brown L G, Marszalek J D, Gianotten J, Oates R D, Silber S, van der Veen F, Page D C, Rozen S. A family of human Y chromosomes has dispersed throughout northern Eurasia despite a 1.8‐Mb deletion in the azoospermia factor c region. Genomics 2004831046–1052. [DOI] [PubMed] [Google Scholar]
- 10.Fernandes S, Paracchini S, Meyer L H, Floridia G, Tyler‐Smith C, Vogt P H. A large AZFc deletion removes DAZ3/DAZ4 and nearby genes from men in Y haplogroup N. Am J Hum Genet 200474180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen P. The fragility of fertility. Nat Genet 200129243–244. [DOI] [PubMed] [Google Scholar]
- 12.Noordam M J, Repping S. The human Y chromosome: a masculine chromosome. Curr Opin Genet Dev 200616225–232. [DOI] [PubMed] [Google Scholar]
- 13.de Llanos M, Ballesca J L, Gazquez C, Margarit E, Oliva F. High frequency of gr/gr chromosome Y deletions in consecutive oligospermic ICSI candidates. Hum Reprod 200520216–220. [DOI] [PubMed] [Google Scholar]
- 14.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, Engl B, Foresta C. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 200542209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giachini C, Guarducci E, Longepied G, Degl'Innocenti S, Becherini L, Forti G, Mitchell M J, Krausz C. The gr/gr deletion(s): a new genetic test in male infertility? J Med Genet 200542497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch M, Cram D S, Reilly A, O'Bryan M K, Baker H W, de Kretser D M, McLachlan R I. The Y chromosome gr/gr subdeletion is associated with male infertility. Mol Hum Reprod 200511507–512. [DOI] [PubMed] [Google Scholar]
- 17.Machev N, Saut N, Longepied G, Terriou P, Navarro A, Levy N, Guichaoua M, Metzler‐Guillemain C, Collignon P, Frances A M, Belougne J, Clemente E, Chiaroni J, Chevillard C, Durand C, Ducourneau A, Pech N, McElreavey K, Mattei M G, Mitchell M J. Sequence family variant loss from the AZFc interval of the human Y chromosome, but not gene copy loss, is strongly associated with male infertility. J Med Genet 200441814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hucklenbroich K, Gromoll J, Heinrich M, Hohoff C, Nieschlag E, Simoni M. Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis. Hum Reprod 200520191–197. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho C M, Zuccherato L W, Bastos‐Rodrigues L, Santos F R, Pena S D. No association found between gr/gr deletions and infertility in Brazilian males. Mol Hum Reprod 200612269–273. [DOI] [PubMed] [Google Scholar]
- 20.de Carvalho C M, Zuccherato L W, Fujisawa M, Shirakawa T, Ribeiro‐dos‐Santos A K, Santos S E, Pena S D, Santos F R. Study of AZFc partial deletion gr/gr in fertile and infertile Japanese males. J Hum Genet 200651794–799. [DOI] [PubMed] [Google Scholar]
- 21.Fernando L, Gromoll J, Weerasooriya T R, Nieschlag E, Simoni M. Y‐chromosomal microdeletions and partial deletions of the azoospermia factor c (AZFc) region in normozoospermic, severe oligozoospermic and azoospermic men in Sri Lanka. Asian J Androl 2006839–44. [DOI] [PubMed] [Google Scholar]
- 22.Ravel C, Chantot‐Bastaraud S, El Houate B, Mandelbaum J, Siffroi J P, McElreavey K. GR/GR deletions within the azoospermia factor c region on the Y chromosome might not be associated with spermatogenic failure. Fertil Steril 200685229–231. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Li Z, Wen B, Jiang J, Shao M, Zhao Y, He Y, Song X, Qian J, Lu D, Jin L. A frequent partial AZFc deletion does not render an increased risk of spermatogenic impairment in East Asians. Ann Hum Genet 200670304–313. [DOI] [PubMed] [Google Scholar]
- 24.Wu B, Lu N X, Xia Y K, Gu A H, Lu C C, Wang W, Song L, Wang S L, Shen H B, Wang X R. A frequent Y chromosome b2/b3 subdeletion shows strong association with male infertility in Han‐Chinese population. Hum Reprod 2007221107–1113. [DOI] [PubMed] [Google Scholar]
- 25.Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos F R, Schiefenhovel W, Fretwell N, Jobling M A, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford M H, Ginter E K, Evgrafov O V, Tyler‐Smith C. Genetic relationships of Asians and Northern Europeans, revealed by Y‐chromosomal DNA analysis. Am J Hum Genet 1997601174–1183. [PMC free article] [PubMed] [Google Scholar]
- 26.Rootsi S, Zhivotovsky L A, Baldovic M, Kayser M, Kutuev I A, Khusainova R, Bermisheva M A, Gubina M, Fedorova S A, Ilumae A M, Khusnutdinova E K, Voevoda M I, Osipova L P, Stoneking M, Lin A A, Ferak V, Parik J, Kivisild T, Underhill P A, Villems R. A counter‐clockwise northern route of the Y‐chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet 200715204–211. [DOI] [PubMed] [Google Scholar]
- 27.Tyler‐Smith C, McVean G. The comings and goings of a Y polymorphism. Nat Genet 200335201–202. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization WHO laboratory manual for the examination of human semen and sperm‐cervical mucus interaction. Cambridge, United Kingdom: Cambridge University Press, 1992
- 29.Underhill P A, Shen P, Lin A A, Jin L, Passarino G, Yang W H, Kauffman E, Bonne‐Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd J R, Mehdi S Q, Seielstad M T, Wells R S, Piazza A, Davis R W, Feldman M W, Cavalli‐Sforza L L, Oefner P J. Y chromosome sequence variation and the history of human populations. Nat Genet 200026358–361. [DOI] [PubMed] [Google Scholar]
- 30.Shinka T, Tomita K, Toda T, Kotliarova S E, Lee J, Kuroki Y, Jin D K, Tokunaga K, Nakamura H, Nakahori Y. Genetic variations on the Y chromosome in the Japanese population and implications for modern human Y chromosome lineage. J Hum Genet 199944240–245. [DOI] [PubMed] [Google Scholar]
- 31.Sengupta S, Zhivotovsky L A, King R, Mehdi S Q, Edmonds C A, Chow C E, Lin A A, Mitra M, Sil S K, Ramesh A, Usha Rani M V, Thakur C M, Cavalli‐Sforza L L, Majumder P P, Underhill P A. Polarity and temporality of high‐resolution Y‐chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet 200678202–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jobling M A, Tyler‐Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 20034598–612. [DOI] [PubMed] [Google Scholar]
- 33.Y Chromosome Consortium A nomenclature system for the tree of human Y‐chromosomal binary haplogroups. Genome Res 200012339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karafet T M, Osipova L P, Gubina M A, Posukh O L, Zegura S L, Hammer M F. High levels of Y‐chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter‐gatherer way of life. Hum Biol 200274761–789. [DOI] [PubMed] [Google Scholar]
- 35.Raymond M, Rousset F. An exact test of population differentiation. Evolution 1995491280–1283. [DOI] [PubMed] [Google Scholar]
- 36.Repping S, van Daalen S K, Brown L G, Korver C M, Lange J, Marszalek J D, Pyntikova T, van der Veen F, Skaletsky H, Page D C, Rozen S. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet 200638463–467. [DOI] [PubMed] [Google Scholar]
- 37.Lin Y W, Hsu L C, Kuo P L, Huang W J, Chiang H S, Yeh S D, Hsu T Y, Yu Y H, Hsiao K N, Cantor R M, Yen P H. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat 200728486–494. [DOI] [PubMed] [Google Scholar]
- 38.Su B, Xiao C, Deka R, Seielstad M T, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli‐Sforza L, Chakraborty R, Jin L. Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 2000107582–590. [DOI] [PubMed] [Google Scholar]
- 39.Conrad D F, Andrews T D, Carter N P, Hurles M E, Pritchard J K. A high‐resolution survey of deletion polymorphism in the human genome. Nat Genet 20063875–81. [DOI] [PubMed] [Google Scholar]
- 40.Arredi B, Ferlin A, Speltra E, Zuccarello D, Ganz F, Marchina E, Stuppia L, Krausz C, Foresta C. Y chromosome haplogroups and susceptibility to AZFc microdeletion in an Italian population. J Med Genet 200744205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semino O, Magri C, Benuzzi G, Lin A A, Al‐Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner P J, Zhivotovsky L A, King R, Torroni A, Cavalli‐Sforza L L, Underhill P A, Santachiara‐Benerecetti A S. Origin, diffusion and differentiation of Y‐chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004741023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]