Abstract
Objective
To obtain penetrance data for Huntington's disease when DNA results are in the range of 36–39 CAG repeats and assess the consistency of reporting the upper allele from two reference centres.
Method
Data were collected anonymously on age of onset or age last known to be unaffected from a cohort of individuals with results in this range. DNA samples were re‐analysed in two reference centres. Kaplan‐Meier analysis was used to construct an age of onset curve and penetrance figures.
Results
Clinical data and concordant DNA results from both reference centres were available for 176 samples; penetrance figures (and 95% confidence intervals) for this cohort, at age 65 and 75 years, were 63.9% (55.5% to 73.2%) and 74.2% (64.2% to 84.2%), respectively. Inclusion of 28 additional subjects for whom repeat DNA results were unavailable, obtained from only one reference centre, or discrepant by one repeat within this range, gave penetrance data (including 95% confidence intervals) at ages 65 and 75 years of 62.4% (54.4% to 70.4%) and 72.7.% (63.3% to 82.1%), respectively. 238 duplicate results were available from the reference centres; 10 (4.2%) differed by one CAG repeat in the reporting of the upper allele and in two (0.84%) of these cases the discrepancy was between 39 and 40 repeats.
Conclusion
When DNA results are in this range, a conservative approach is to say that there is at least a 40% chance the person will be asymptomatic at age 65 years and at least a 30% chance the person will be asymptomatic at age 75 years.
Keywords: Huntington's disease, penetrance, 36–39 repeats
Huntington's disease is an autosomal dominant progressive neurodegenerative disorder characterised by a movement disorder (often choreiform in nature), a disturbance of affect and a selective cognitive deficit. In 1993, an unstable expansion of a CAG sequence in the first exon of the gene IT15 was identified as the pathological mutation.1 Soon after the identification of the gene, it was realised that there was a negative correlation between the age of onset and the size of the CAG repeat length, but the wide spread of age of onset for each repeat size effectively made this of limited clinical value; these data have been summarised in reviews.2,3
A number of reports have shown that in the size range of 36–39 repeats the age of onset could either be very late or not occur at all.4,5,6 In 1998, the American College of Medical Genetics and the American Society of Human Genetics (ACMG/ASHG) suggested that service laboratories should use the following in reporting results: under 27 repeats, unequivocally normal; 27–35 repeats, normal but may expand in future generations; 36–39 repeats, abnormal but associated with reduced penetrance; 40 or more repeats, abnormal.7 At that time empiric penetrance risks were unavailable. A study by Brinkman et al using direct observations and a Kaplan‐Meier analysis failed to give an adequate estimate of the reduced penetrance because of small numbers in the reduced penetrance size range of 36–39 repeats.8 The same group analysed a larger data set but extrapolated the information on age of onset curves from data obtained in the range of 41–56 repeats; the authors argued that individuals with results in this size range may not present to medical attention and a direct approach may overestimate the penetrance of these alleles.9 For the most part, service laboratories only investigate samples of DNA from families in which Huntington's disease has occurred or is strongly suspected. This study was undertaken to obtain an age of onset curve for Huntington's disease with allele sizes of 36–39 repeats using a direct observational approach.
Methods
Sample collection
Ethical approval (Trent MREC02/4/062) was granted for obtaining completely anonymised clinical data and aliquots of DNA from genetics departments offering diagnostic and predictive tests for Huntington's disease. Collaborating centres were given data sheets with unique identifying numbers which were used to report gender, age of onset of Huntington's disease or the age at which the individual was last known to be unaffected and brief pedigree details for their results in the 36–39 repeat range. This form was returned together with an aliquot of DNA labelled with the same unique number. No record was kept of the genetic department which sent the data and samples. The DNA samples were then re‐analysed in two UK laboratories at Sheffield and Edinburgh, hereafter called the reference centres. It was not possible to trace any discrepancies back to an individual or a collaborating centre.
DNA analysis in the reference centres
PCR amplification of the CAG repeat in the IT15 gene was performed after the method of Warner et al10 but modified to use a fluorescently labelled HD1 primer. Fluorescently labelled products were run on a 48 capillary Applied Biosystems 3730 DNA analyser (Applied Biosystems, Foster City, CA) and CAG repeat sizes determined by direct visual analysis of the fluorescent profile using Applied Biosystems GeneMapper software.
Statistical analysis
All individuals, including both those affected and asymptomatic at‐risk, were included to calculate the cumulative probability of disease onset by a particular age using Kaplan‐Meier survival analysis. Individuals with a CAG repeat length of 36–39 repeats were the cohort at risk from birth to diagnosis of disease or last known contact age (censored). The Kaplan‐Meier survival curves were compared by the log‐rank test statistic. An arbitrary level of 5% statistical significance (two‐tailed) was assumed.
Results
Penetrance data
A total of 263 samples were received: 110 were from males and 151 from females, and in two cases the gender was not reported. Eight samples were excluded because the clinical data were missing, 11 samples sent in error with under 36 repeats were eliminated, and 40 samples were excluded because they were over 40 repeats. Of the remaining 204 samples, there were 16 cases where no DNA was sent or a result could not be obtained in either reference centre, there were five cases where a result was obtained from only one reference centre, and there were seven cases where the results from the reference centres differed by one repeat within the reduced penetrance range (these 28 cases were included in table 1 based on the result from the referring laboratory agreeing with the result from one of the reference centres). The effect of removing theses 28 samples is shown in table 2.
Table 1 Summary of the data including 28 samples for which repeat analysis was absent, partial or discrepant by one CAG repeat.
| CAG repeat size | Number in study | Affected | Mean age of onset (years) | Range in age of onset (years) | Unaffected | Mean age (years) | Range (years) |
|---|---|---|---|---|---|---|---|
| 36 | 21 | 10 | 59.4 | 38–79 | 11 | 42.5 | 23–68 |
| 37 | 46 | 22 | 60.3 | 48–74 | 24 | 47.5 | 21–85 |
| 38 | 52 | 17 | 55.7 | 40–75 | 35 | 48.2 | 25–76 |
| 39 | 85 | 43 | 62.1 | 31–89 | 42 | 48.7 | 20–79 |
| Total | 204 | 92 | 112 |
Table 2 Summary of data based on exclusion of 28 samples for which repeat analysis was absent, partial or discrepant by one CAG repeat.
| CAG repeat size | Number in study | Affected | Mean age of onset (years) | Range in age of onset (years) | Unaffected | Mean age (years) | Range (years) |
|---|---|---|---|---|---|---|---|
| 36 | 18 | 9 | 59.4 | 38–79 | 9 | 42.5 | 23–68 |
| 37 | 39 | 19 | 60.6 | 48–74 | 20 | 48.5 | 21–85 |
| 38 | 46 | 13 | 52.0 | 40–75 | 33 | 48.0 | 25–76 |
| 39 | 73 | 35 | 64.1 | 38–89 | 38 | 48.7 | 20–79 |
| Total | 176 | 76 | 100 |
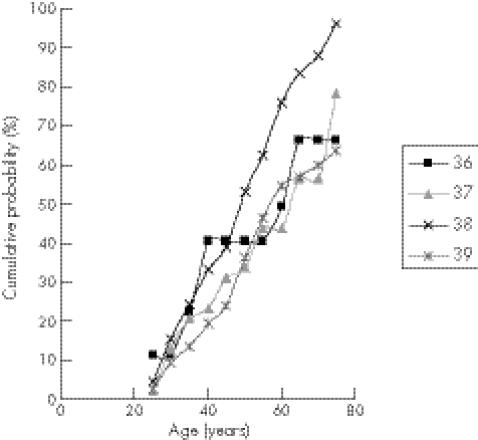
There were no significant differences between the different age of onset curves when the analysis included the 28 samples with repeat results which were absent, obtained from one reference centre or discrepant by one CAG repeat (log‐rank test = 5.7, df = 3, p = 0.13). However, there were significant differences between the curves when these samples were excluded (log‐rank test = 8.5, df = 3, p = 0.036) (fig 1).
Figure 1 Cumulative age of onset curves for Huntington's disease alleles in the range of 36–39 repeats based on 176 samples for which clinical data and concordant DNA results were available from two reference laboratories.
Table 3 shows the cumulative probability of the age of onset of Huntington's disease by age and CAG repeat size for ages 65–75 years.
Table 3 Cumulative probability (%) of age of onset of Huntington's disease by CAG repeat size based on the 176 results with concordant DNA results from both reference centres.
| Age (years) | 36 (repeats) | 37 (repeats) | 38 (repeats) | 39 (repeats) | 36–38 (repeats) | 36–39 (repeats) |
|---|---|---|---|---|---|---|
| 65 | 66.3 (34.4 to 98.2) | 56.4 (36.4 to 76.4) | 83.5 (71.2 to 95.8) | 56.9 (44.2 to 69.6) | 70.1 (58.7 to 81.5) | 63.9 (55.5 to 73.2) |
| 70 | 66.3 (34.4 to 98.2) | 56.5 (36.4 to 76.4) | 88.2 (75.8 to 100) | 59.8 (46.7 to 72.9) | 74.1 (61.8 to 86.4) | 67.1 (58.2 to 75.9) |
| 75 | 66.3 (64.2 to 84.2) | 78.2 (46.4 to 100) | 96.1 (86.3 to 100) | 63.8 (49.9 to 77.7) | 87.0 (72.9 to 100) | 74.2 (64.2 to 84.2) |
Data in parentheses are 95% confidence intervals.
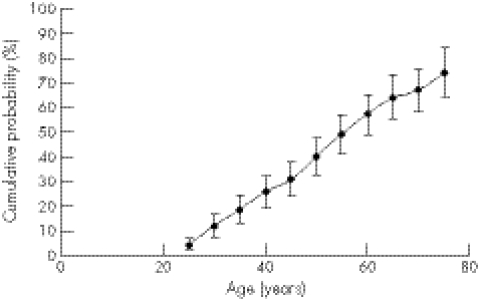
A trend of increasing penetrance with increase in repeat size was expected. Table 3 shows this was not the case: the penetrance figures for 38 repeats were significantly higher (fig 1). The effect was still present when the data for 36–38 repeats were aggregated as penetrance was higher than for 39 repeats alone. This may be the effect of studying small numbers: there were 13 affected cases, with a mean age of onset of 52.0 years, and 33 unaffected cases in the first analysis (table 2). When the additional 28 samples were included, the number of affected cases increased to 17, with a mean age of onset of 55.7 years, and the number of unaffected cases increased to 35; this resulted in the penetrance for 38 repeats still being high but the statistical significance between the curves was lost. Therefore, only one age of onset curve was drawn (fig 2).
Figure 2 Combined age of onset curve for Huntington's disease alleles in the range of 36–39 repeats based on 176 samples for which clinical data and concordant DNA results were available from two reference laboratories. Error bars represent the 95% confidence interval. The curve for the combined data including an additional 28 cases (see text) plotted almost exactly on top of this curve and is not shown.
When the analysis was based strictly on the original protocol of concordant data between the two reference centres, penetrance figures (and 95% confidence intervals) for ages 65 and 75 years for individuals with results in the range of 36–39 repeats were 63.9% (55.5% to 73.2%) and 74.2% (64.2% to 84.2%), respectively. When the additional 28 samples were included, penetrance figures (and 95% confidence intervals) at ages 65 and 75 years were 62.4% (54.4% to 70.4%) and 72.7% (63.3% to 82.1%), respectively.
As sib‐sib pairs tend to have a similar age at onset for given expanded repeat sizes,11 we assessed the possible bias introduced by the inclusion of first degree relative pairs (sib‐sib and parent‐child) by randomly selecting one of the pair for exclusion from the survival analysis. There was no significant difference in the results obtained, suggesting that their inclusion did not introduce major bias.
Issues at the 36 repeat and 40 repeat boundaries
There was no issue at the 36 repeat boundary; the reference centres found no case sent as ⩾36 repeats to have a lower repeat number.
A total of 40 samples were excluded from the analysis because one or both reference centres gave a result of ⩾40 repeats. In three cases a repeat result could not be obtained from both reference centres and in one case the collaborating centre sent in a sample with 43 repeats. The style of reporting varied between collaborating centres: six gave a range which included 40 or more repeats; however, there were 24 cases in which the collaborating centres unequivocally reported a result in the reduced penetrance range and both reference centres gave a result of 40 repeats. In addition, there were two cases where the collaborating centres reported 39 repeats, but there was discordance of one allele between the reference centres: one gave a result which was 39 repeats and the other gave a result of 40 repeats.
There were four samples with two abnormal alleles; essentially, one was in the low penetrance range and the other was over 40 repeats. In one of these, three alleles were observed; the collaborating centre gave a result of 17, 36 and 41. This was confirmed in the reference centres although there was a discrepancy of one repeat in the reporting of the lower allele. In the other three samples there was concordance with the collaborating and reference centres giving results of 37 and 43 repeats, 38 and 42 repeats, and 38 and 45 repeats.
Discrepancies in reporting the upper alleles
It was not possible to obtain a repeat DNA result from both reference centres for 24 of the 263 cases sent from the collaborating centres. One sample without adequate clinical data was excluded from the penetrance study but the reporting centre gave the result as 36 and 37 repeats whereas the reference centres gave the result as 38 and 38 repeats, and 38 and 38 or 38 and 39 repeats. Excluding this sample, there were 238 cases where duplicate results were obtained from both reference centres and of these 10 (4.2%) differed by one CAG repeat in the reporting of the upper allele. This would only have made a difference to the interpretation of the result in the two cases (0.84%) where the discrepancy was between 39 and 40 repeats.
Discussion
A large body of data concerning the 36–39 repeat range was analysed in this study. The excess of samples from females may represent the fact that more females come forward for predictive tests.12 These data were generated from individuals with a family history of Huntington's disease and should not be applied in the case of an allele of this size being detected in an otherwise healthy individual; for example, testing the partner of an at‐risk individual as part of an exclusion test or for pre‐implantation diagnosis.
Identification of a reduced penetrance allele relies on either a symptomatic individual coming to medical attention or an asymptomatic individual deciding to have a predictive test and the result being in the range of 36–39 repeats. This may result in an overestimate of the penetrance as more affected individuals may come to attention compared with elderly asymptomatic individuals requesting a predictive test. In an earlier study, Langbhen et al extrapolated information on penetrance based on age of onset curves in a group of individuals with 41–56 repeats.9 They estimated penetrance data at age 75 years of 14%, 26%, 45% and 60% at 36, 37, 38 and 39 repeats, respectively. This method makes allowance for missing alleles in this range, but assumptions have to be made. If, in addition to missing alleles, there are other factors to account for the data in the range of 36–40 repeats not fitting their mathematical model, then it is possible that the number of missing alleles was overestimated and the penetrance underestimated.
The bias towards an overestimate of penetrance in this report is shown by the effect of having relatively few affected individuals with 38 repeats compared with asymptomatic individuals. It may be that this occurred by chance; however, as expected, there was a trend towards increasing the number of samples with each CAG repeat size. It is difficult to identify a plausible alternative explanation but the possible existence of one must remain open for the present.
A discrepancy of one CAG repeat within the four main ACMG/ASHG groups is not clinically significant. There were 24 cases where the collaborating centre unequivocally reported a low penetrance allele and both reference centres gave a result of 40 repeats. Taken together with the 176 results in table 2, this represents 12.0% of cases unequivocally intended to be reported as being in the reduced penetrance range and having concordant results from the reference centres. Although high, it is to be remembered that these samples were reported from as early as 1994 and multiple laboratory methods were used. It is interesting to note that in this large cohort there was a discrepancy of one CAG repeat in reporting the upper allele between the reference centres in 4.2% of cases; in two (0.84%) of these cases the discrepancy was at the 39 and 40 repeat boundary which would represent a misclassification. If the 24 samples intended to be reported in the reduced penetrance range were included in the analysis, penetrance figures (and 95% confidence intervals) would have been 49.4% (38.0% to 60.8%) at age 65 years and 55.8% (42.7% to 68.9%) at age 75 years.
The discrepancy in data between the reference centres in this study can be compared with results from a European pilot quality assessment scheme in which a variation of ±1 CAG in the range up to 40 repeats and ±3 CAG repeats for results over 40 repeats was allowed.13 Thirteen laboratories returned results on six DNA samples from five families. Despite the allowed limits, 6.2% of alleles were reported outside these limits and in one case there would have been a significant misdiagnosis between the normal and pathological range.
Another potential bias in this study is that there may be some samples reported in the collaborating centre as 40 repeats which could have been reclassified as 39 repeats. This information is unknown and could not be obtained without dramatically increasing the size of the study.
Although the mean age of onset and CAG repeat size are negatively correlated, for any particular CAG repeat size there is wide spread in age of onset. This is reflected in this data set as the spread of ages of onset for both 39 repeats and the whole cohort was 31–89 years (table 1). Apart from the CAG repeat length, other factors, both genetic and non‐genetic, influence the age of onset.14
A patient with an abnormal predictive test result cannot be given accurate information regarding age of onset based on the repeat size. There is variation between laboratories and clinicians as to how results are reported; some laboratories do not report the actual repeat size but only the main category as set out by the ACMG/ASHG; others report the actual repeat size to the clinician who may or may not disclose it to the family. The reason for caution in disclosing the actual result is that it does not add useful information for the individual and has the potential to be misinterpreted.
The onset of Huntington's disease is insidious and difficult to determine accurately. While every effort was made to check the DNA result, age of onset data were based on estimates from multiple collaborating centres; it is likely that the rigour of that estimation varied both within and between centres.
Given these various constraints and the bias towards overestimating penetrance, we would argue that broad‐based conservative statements are made regarding age of onset. We suggest that if a result is found in the range of 36–39 repeats in the context of a predictive test, there is at least a 40% chance the person will be asymptomatic at age 65 years and at least a 30% chance the person will be asymptomatic at age 75 years.
Key points
It is well established that reduced penetrance may occur in Huntington's disease when the CAG repeat result in the IT15 gene is in the range of 36–39 repeats.
Based on a direct observational study, we suggest that if an individual has a predictive test result in this range, the chance of them being asymptomatic at 65 years of age is at least 40% and the chance of them being asymptomatic at 75 years of age is at least 30%.
Using two reference laboratories, a discrepancy of one CAG repeat occurred in the reporting of the upper allele in a small number of cases.
Acknowledgements
We thank Mrs Val McCormack and Mrs Margaret Steele for help with data entry. A large number of individuals helped in obtaining clinical data and retrieving samples. We would also like to thank Leslie Snell for help with obtaining samples from Australia and Ben Wymer for technical support.
Footnotes
This work was funded from a grant by Action Medical Research.
Competing interests: None declared.
References
- 1.Huntington's Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 199372971–983. [DOI] [PubMed] [Google Scholar]
- 2.Harper P S, Jones L. Huntington's disease: genetic and molecular studies. In: Bates G, Harper P, Jones L, eds. Huntingtons disease, 3rd edn. Oxford: Oxford University Press, 2002123–124.
- 3.Gusella J F, MacDonald M E. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci 20001109–115. [DOI] [PubMed] [Google Scholar]
- 4.Legius E, Cuppens H, Dierick H, Van Zandt K, Dom R, Fryns J ‐ P, Evers‐Kiebooms G, Decruyenaere M, Demyttenaere K, Marynen P, Cassiman J J. Limited expansion of the (CAG)n repeat of the Huntington gene: a premutation (?). Eur J Hum Genet 1994244–50. [DOI] [PubMed] [Google Scholar]
- 5.Rubinsztein D C, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman J ‐ J, Chotai K, Connarty M, Craufurd D, Curtis A, Curtis D, Davidson M J, Differ A ‐ M, Dode C, Dodge A, Frontali M, Ranen N G, Stine O C, Sherr M, Abbott M H, Franz M L, Graham C A, Harper P S, Hedreen J C, Jackson A, Kaplan J ‐ C, Losekoot M, MacMillan J C, Morrison P, Trottier Y, Novelletto A, Simpson S A, Thielmann J, Whittaker J L, Folstein S E, Ross C A, Hayden M R. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am J Hum Genet 19965916–22. [PMC free article] [PubMed] [Google Scholar]
- 6.McNeil S M, Novoletto A, Srinidhi J, Barnes G, Kornbluth I, Altherr M R, Wasmuth J J, Gusella J F, MacDonald M, Myers R H. Reduced penetrance of the Huntington's disease mutation. Hum Mol Genet 19976775–779. [DOI] [PubMed] [Google Scholar]
- 7.ACMG/ASHG ACMG/ASHG Statement. Laboratory guidelines for Huntington's disease genetic testing. Am J Hum Genet 1998621243–1247. [PMC free article] [PubMed] [Google Scholar]
- 8.Brinkman R R, Mezei M M, Theilmann J, Almqvist E, Hayden M R. The likelihood of being affected with Huntington disease by a particular age, for a specific CAG size. Am J Hum Genet 1997601202–1210. [PMC free article] [PubMed] [Google Scholar]
- 9.Langbhen D R, Brinkman R R, Faulsh D, Paulsen J S, Hayden M R, on behalf of the International Huntington's Disease Collaborative Group A new model for prediction of the age of onset and penetrance for Huntington's disease based on CAG length. Clin Genet 200465267–277. [DOI] [PubMed] [Google Scholar]
- 10.Warner J P, Baron L H, Brock D H. A new polymerase chain reaction (PCR) assay for the trinucleotide repeat that is unstable and expanded on Huntington's disease chromosomes. Mol Cell Probes 19937235–238. [DOI] [PubMed] [Google Scholar]
- 11.Squitieri F, Sabbadini G, Mandich P, Gellera C, Di Maria E, Bellone E, Castellotti B, Nargi E, de Grazia U, Frontali M, Novolletto A. Family and molecular data for a fine analysis of age of onset in Huntington disease. Am J Med Genet 200095366–373. [DOI] [PubMed] [Google Scholar]
- 12.Harper P S, Lim C, Craufurd D, on behalf of the UK Huntington's Disease Prediction Consortium Ten years of presymptomatic testing for Huntington's disease: the experience of the UK Huntington's Disease Prediction Consortium. J Med Genet 200037567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Losekoot M, Bakker B, Laccone F, Stenhouse S, Elles R. A European pilot quality assessment scheme for molecular diagnosis of Huntington's disease. Eur J Hum Genet 19997217–222. [DOI] [PubMed] [Google Scholar]
- 14.The U.S.‐Venezuela Collaborative Research Project Wexler NS, Lorimer J, Porter J, Gomez F, Moskowitz C, Shackell E, Marder K, Penchaszadeh G, Roberts SA, Gayán J, Brocklebank D, Cherny SS, Cardon LR, Gray J, Dlouhy SR, Wiktorski S, Hodes ME, Conneally PM, Penney JB, Gusella J, Cha J‐H, Irizarry M, Rosas D, Hersch S, Hollingsworth Z, MacDonald M, Young AB, Andresen JM, Housman DE, de Young MM, Bonilla E, Stillings T, Negrette A, Snodgrass SR, Martinez‐Jaurrieta MD, Ramos‐Arroyo MA, Bickham J, Ramos JS, Marshall F, Shoulson I, Rey GJ, Feigin A, Arnheim N, Acevedo‐Cruz A, Acosta L, Alvir J, Fischbeck K, Thompson LM, Young A, Dure L, O'Brien CJ, Paulsen J, Brickman A, Krch D, Peery S, Hogarth P, Higgins DS Jr, Landwehrmeyer B. Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci U S A 20051013498–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]