Abstract
Background
A limited number of studies aimed at investigating the possible association of Y‐chromosome haplogroups with microdeletions of the azoospermia factors (AZFs) or with particular infertile phenotypes, but definitive conclusions have not been attained. The main confounding elements in these association studies are the small sample sizes and the lack of homogeneity in the geographical origin of studied populations, affecting, respectively, the statistical power and the haplogroup distribution.
Materials and methods
To assess whether some Y‐chromosome haplogroups are predisposing to, or protecting against, azoospermia factor c (AZFc; b2/b4) deletions, 31 north Italian patients carrying the AZFc b2/b4 microdeletion were characterised for 8 Y‐chromosome haplogroups, and compared with the haplogroup frequency shown by a north Italian population without the microdeletion (n = 93).
Results and discussion
A significant difference was observed between the two populations, patients with microdeletions showing a higher frequency of the E haplogroup (29.3% vs 9.7%, p<0.01). The geographical homogeneity of the microdeleted samples and of the control population, controlled at microgeographical level, allows the possibility that the geographical structure of the Y genetic variability has affected our results to be excluded.
Conclusion
Thus, it is concluded that in the north Italian population Y‐chromosome background affects the occurrence of AZFc b2/b4 deletions.
Y‐chromosome long‐arm microdeletions are found in 5–10% of men with severe oligospermia and non‐obstructive azoospermia, and encompass one or more azoospermia factor (AZF) loci. Deletions of the azoospermia factor c (AZFc) region are clearly among the most commonly known molecular causes of spermatogenic failure in men.1 These deletions are caused by homologous recombination between the 229‐kb‐long b2 and b4 amplicons2 and span 3.5 Mb. Eight different gene families are removed by AZFc deletions, including all members of the DAZ gene family, which represents the stronger candidate for the AZFc phenotype.1,2,3,4,5,6,7 Although all AZFc deletions are essentially identical in molecular extension, people carrying these microdeletions present variable infertile phenotypes, suggesting the involvement of environmental factors and/or other genetic regions. Furthermore, the function of the AZF genes in human spermatogenesis and the role of the Y‐chromosome background in the predisposition to occurrence of deletions is still largely unknown.
At present, around 250 Y single‐nucleotide polymorphisms have been discovered and their phylogenetic relationships are well known.8 These polymorphic markers of the male‐specific region of the Y chromosome define monophyletic groups of the Y chromosome, which hereafter we will name as “haplogroups”.
A limited number of studies have investigated the possible association of Y‐chromosome haplogroups with microdeletions or with a particular infertile phenotype,9 but the contribution of predisposing factors or genetic background to causing deletions is still debated. In particular, only three studies have investigated the possible association between Y‐chromosome haplogroups and AZF deletions,10,11,12 all of them failing to establish important associations. These works studied such associations in an European population involving 73 microdeleted samples of heterogeneous geographical origin,10 in a northwestern European population involving 50 patients11 and in a Japanese population, more geographically localised but represented by a very low number of people with microdeletions (six patients).12 All the previous studies that found some suggestion of an association with Y‐chromosome haplogroups dealt with infertility. They reported a considerable over‐representation of the haplogroup K(xL,N,O1,O3c,P) in Danish men with reduced sperm count, which did not reach significance probably because of a small sample size,13 and D2b Y lineage in Japanese men with reduced sperm count,14 not confirmed by a later study.12
However, these association studies require particular attention to two principal factors: (1) the geographical structure of the Y‐chromosome variations in the population under investigation, because the Y‐chromosome genetic variability is highly geographically structured and the Y‐haplogroup distribution changes over different geographical areas15; and (2) the number and selection criteria of the patient and control groups.
To assess whether some Y‐chromosome haplogroups predispose to, or protect against, AZFc deletion, we have defined and compared Y‐chromosome haplogroup distribution in a group of unrelated Italian infertile men harbouring the b2/b4 deletion (n = 41, 31 of whom were from north Italy) and in a control group represented by fertile men without microdeletions (fathers of at least one child) from north Italy (n = 93).
Patients and methods
Patients
Among infertile men screened in our centre for Y‐chromosome microdeletions, we studied 41 patients with the b2/b4 microdeletion. The patients with microdeletion were all Italian, and for 31 of them it was possible to ascertain their north Italian origin. The control group was collected from among healthy donors represented by 93 fertile men (fathers of at least one child). We paid particular attention to the geographical origin of the patient and control groups, to exclude the geographical factor as a cause of possible different haplogroup frequencies. Thus, as most of the patients carrying the b2/b4 microdeletions (31/41) were from north Italy, we collected the control population from the same geographical area. All the individuals were unrelated and of north Italian origin back to the grandfather. Informed consent was obtained from each participant; the study was approved by the University of Padova Institutional Review Board.
Molecular analyses
The b2/b4 deletion was characterised through a two‐step sequence‐tagging site (STS) screening. The first step included sY254 and sY255 (both in the DAZ gene) screening, generally adopted as the standard procedure for the detection of the AZFc deletions, according to the laboratory guidelines for molecular diagnosis.16 A more detailed characterisation of the deletion breakpoints was performed in a second step, through the screening of the STS flanking and within the most common AZFc deletion (b2/b4): sY1197 (for proximal breakpoint) and sY1125 (for distal breakpoint), and sY1192 and sY1190 (internal).
All the members of the control population were analysed with the STSs sY254 and sY255 for the b2/b4 deletion, and with sY1191 and sY1291 for the partial AZFc gr/gr deletion.17
Each participant was screened for seven Y‐chromosome binary markers (12f2, M89, M09, M45, M96, M145 and TAT) defining eight Y haplogroups (table 1 and fig 1).
Table 1 Haplogroup frequencies in north Italian population with microdeletion and controls.
| Sample | Y haplogroups (%) | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Y (xF,DE) | F (xK,J) | P | K (xP) | N3 | J | D | E | ||
| Controls | 0 (0) | 14 (15) | 51 (54.8) | 5 (5.4) | 0 (0) | 14 (15) | 0 (0) | 9 (9.7) | 93 |
| Microdeletions | 0 (0) | 5 (16.1) | 15 (48.4) | 0 (0) | 0 (0) | 1 (3.2) | 0 (0) | 10 (32.3)* | 31 |
Values are n (%).
*p<0.05 vs controls after Bonferroni correction.
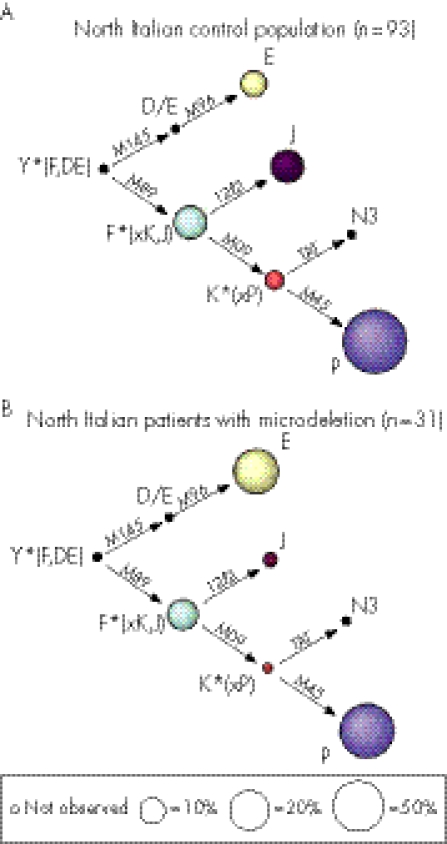
Figure 1 Haplogroup frequencies in the north Italian control population (A) and in the Italian patients with microdeletions (B) and north Italian patients with microdeletions (C). The size of the circle is proportional to the haplogroup frequency (as in table 1).
All the biallelic markers, except 12f2 and M89, have been typed through denaturing high‐performance liquid chromatography (DHPLC); the typing of TAT through DHPLC, described here for the first time, was carried out by deriving novel primer sequences—namely, TATbF: 5′‐GGAAAAATACACTACGTC‐3′ and TATbR: 5′‐CCCTAATCCTTTTGAGCCA‐3′ (Invitrogen, Milan, Italy). A PCR was carried out in a 35 μl reaction volume containing 100 ng genomic DNA, 1.4 U AmpliTaq Gold (Applied Biosystems, Brunchburg, New Jersey, USA), 1× PCR Gold buffer (Applied Biosystems), 25 mM MgCl2 (Applied Biosystems), 2 mM of each dNTP (Amersham Biosciences, Piscataway, New Jersey, USA) and a mix of forward and reverse primers (1 μmol/l each). Samples were denatured for 10 min at 94°C using AmpliTaqGold (Applied Biosystems), amplified for 35 cycles of 94°C for 45 s, 54°C for 45 s and 72°C for 1 min, and given a final extension at 72°C for 4 min. The markers M09, M45, M96 and M145 were typed by DHPLC with standard methods18; the 12f2 insertion/deletion polymorphism was typed according to Rosser et al19 by coamplification of a fragment within the deleted region and a PCR control. M89 was sequenced on samples that were not classified by the first screening (ABI PRISM 3730 XL DNA Sequencer, Applied Biosystems).
Statistical analyses
The difference in haplogroup distribution between the control and the microdeleted samples was statistically tested through Fisher's exact probability test on contingency table for single haplogroup20 after Bonferroni correction and through the exact test of population differentiation21 for differentiation between populations. The analysis of molecular variance was performed to calculate the level of genetic structure (ΦST) of the whole dataset, and gene diversities were calculated to compare the level of variability within each group; these calculations and gene diversity were performed with Arlequin V.2.0.22
The analysis was performed for the whole microdeleted sample (hereafter referred to as “Italian microdeleted”, 41 individuals) and the north Italian subgroup (“north Italian microdeleted”, 31 individuals).
Results and discussion
The control population was screened for Y microdeletions through STS analysis, as described in the previous section. The normal presence of STSs sY254, sY255, sY1191 and sY1291 indicated that the control individuals did not carry b2/b4 or gr/gr microdeletions. The comparison of the haplogroup distribution in the control population and in patients with microdeletions showed a similar haplogroup distribution (table 1 and fig 1), and no Y‐chromosome haplogroup was more prevalent in one population than in the other. The gene diversity is comparable, being 0.65 (0.04) in the control, 0.62 (0.05) in the Italian microdeleted and 0.66 (0.05) in the north Italian microdeleted groups.
However, a more detailed analysis raised some interesting observations (fig 1 and table 2). First, the ΦST value is significant at the 5% level, which is high considering that the samples share the same geographical origin (control group and north Italian microdeleted ΦST = 5.44%; p<0.05). Moreover, the two populations are significantly different (p<0.05; table 2) under Fisher's exact test of population differentiation.21
Table 2 Results of the differentiation between patients with microdeletion and control group.
| Statistics | North Italian microdeleted/north Italian control |
|---|---|
| ΦST | 5.44%* |
| Exact test differentiation | 0.01<p<0.05† |
*p<0.05.
†p refers to the significance of the exact test of differentiation.
As we can exclude the possibility that different geographical origins have affected the haplogroup distribution, especially for the north Italian microdeleted, these results suggest a probable role of the Y‐chromosome background on the Y‐microdeletion event. To assess whether the selected control groups were representative of the north Italian population, we compared the observed frequencies with data from Italian populations, showing perfect data matching23 (C Capelli, personal communication). The most frequently observed haplogroup, P, is the same in the control group (55%) and in the two groups with microdeletions (48% in north Italian). This haplogroup (or more precisely its sub‐branch M173‐derived, named R) is the most common haplogroup in Europe.15 In our survey, we did not observe D and N3 haplogroups. F*(xK,J) was observed at frequencies between 15% and 17% in all the samples, even if it represents the second most frequent haplogroup, with J, in the control population but not in patients with microdeletions. Important differences between the control groups and the groups with deletions were seen for the distribution of the E haplogroup.
The J haplogroup varied between the control group (15%) and the group with microdeletions (3%). However, the difference is not significant (p<0.05). However, the difference of the E lineage frequency is significant, reaching 10% in the reference population and increasing dramatically in the microdeleted samples (32%, p<0.01; table 1).
The E lineage mean frequency in north Italy is about 11%, a frequency similar to that found in our control population24,25 (C Capelli, personal communication 2006). These observations reinforce the importance of the findings of this study, as the high frequency of the E haplogroup observed in the sample with microdeletions (32% in north Italian microdeleted) has not yet been reported in any Italian population. Such a high frequency would not be expected, even if the patients are predominantly from south Italy, where the maximum E lineage is <25% (C Capelli, personal communication 2006).
Our results on the E haplogroup suggest that this particular background is more prone to rearrangements, and in particular to b2/b4 microdeletions, than other haplogroups. One would have expected that selection would have acted to lower its frequency, but this does not seem to be the case. This E haplogroup is distributed over a wide area (eastern and northern Africa, the Near East and Europe)25 and the most recent ancestor with which it is associated is a long time ago (95% CI 21.1 to 25.4 × 1000 years).25 For AZFc and other Y‐chromosome deletions causing spermatogenesis failure, the prevalence of the deleted Y chromosome is <0.03%, about the rate at which the deletions arise,2,6 not high enough for selection to dominate over genetic drift. Moreover, evaluation of the effect of selection and in particular of reduced frequency is difficult as variable infertile phenotypes are associated with the patients with AZFc deletions presented here (from oligozoospermic to azoospermic). As not all of them lead to complete infertility, the effect of selection is probably weakened, as also supported by the evidence of cases of father‐to‐son transmission of microdeletions.1
The counterpart of the E haplogroup‐increased frequency in the microdeleted chromosomes is the reduction of the J haplogroup frequency in the microdeleted samples (one individual, table 1), which is much lower than the 15% observed in the control population, even if the difference is not statistically significant. The Y chromosomes belonging to this haplogroup have already shown that they protect against the occurrence of Y‐chromosome microdeletions because of the molecular characteristics of the J haplogroup. In fact, it does not contain the LIPA4 element that is supposed to facilitate the homologous intrachromosomal recombination leading to AZFa deletions.26
This association study between b2/b4 microdeletions and Y‐chromosome haplogroups is the first with a large number of patients with microdeletions and in which the geographical origin of both the patient and the control populations are highly comparable at a microgeographical level. This factor is important for all the studies involving Y‐chromosome distribution as it is usually distributed over clines even in regional studies,27 and this obviously greatly affects the comparison of haplogroup frequencies from two populations if they do not originate from the same area. This bias could have hindered possible associations in previous studies performed on a small number of patients or on subjects from different geographical areas. We thus present here a reliable significant signal of association between the b2/b4 microdeletion occurrence and haplogroup E, occurring at higher frequency in the north Italian microdeleted than in the non‐microdeleted north Italian population.
We thus conclude that in the north Italian population, Y‐chromosome background affects differently the occurrence of AZFc b2/b4 deletions, with haplogroup E probably predisposing to the occurrence. Studies on other well‐defined populations and further molecular analyses for greater identification of E sub‐lineages, and possibly microsatellite variability, will help to clarify the implication of our results also for infertility status.
Acknowledgements
We thank Dr Chris Tyler‐Smith and Dr Yali Xue for providing control samples for the DHPLC setting and for fruitful discussion on the typing method. We thank all the DNA donors and patients without whom this work would not have been possible. The financial support of the Italian Ministry of University (grant 20050625729) is gratefully acknowledged. BA, AF and DZ are supported by the University of Padova.
Abbreviations
AZF - azoospermia factor
AZFc - azoospermia factor c
DHPLC - denaturing high performance liquid chromatography
STS - sequence‐tagging site
Footnotes
Competing interests: None declared.
References
- 1.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev 200122226–239. [DOI] [PubMed] [Google Scholar]
- 2.Kuroda‐Kawaguchi T, Skaletsky H, Brown L G, Minx P J, Cordum H S, Waterston R H, Wilson R K, Silber S, Oates R, Rozen S, Page D C. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 200129279–286. [DOI] [PubMed] [Google Scholar]
- 3.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page D. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA‐binding protein gene. Nat Genet 199510383–393. [DOI] [PubMed] [Google Scholar]
- 4.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, Kohn F M, Schill W B, Farah S, Ramos C, Hartmann M, Hartschuh W, Meschede D, Behre H M, Castel A, Nieschlag E, Weidner W, Grone H J, Jung A, Engel W, Haidl G. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet 19965933–943. [DOI] [PubMed] [Google Scholar]
- 5.Oates R D, Silber S, Brown L G, Page D C. Clinical characterization of 42 oligospermic or azoospermic men with microdeletion of the AZFc region of the Y chromosome, and of 18 children conceived via ICSI. Hum Reprod 2002172813–2824. [DOI] [PubMed] [Google Scholar]
- 6.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates R D, Page D C, Rozen S. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 200271906–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferlin A, Tessari A, Ganz F, Marchina E, Barlati S, Garolla A, Engl B, Foresta C. Association of partial AZFc region deletions with spermatogenic impairment and male infertility. J Med Genet 200542209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Y Chromosome Consortium A nomenclature system for the tree of human Y‐chromosomal binary haplogroups. Genome Res 200212339–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krausz C, Quintana‐Murci L, Forti G. Y chromosome polymorphisms in medicine. Ann Med 200436573–583. [DOI] [PubMed] [Google Scholar]
- 10.Paracchini S, Stuppia L, Gatta V, Palka G, Moro E, Foresta C, Mengua L, Oliva R, Ballesca J L, Kremer J A, van Golde R J, Tuerlings J H, Hargreave T, Ross A, Cooke H, Huellen K, Vogt P H, Tyler‐Smith C. Y‐chromosomal DNA haplotypes in infertile European males carrying Y‐microdeletions. J Endocrinol Invest 200023671–676. [DOI] [PubMed] [Google Scholar]
- 11.Quintana‐Murci L, Krausz C, Heyer E, Gromoll J, Seifer I, Barton D E, Barrett T, Skakkebaek N E, Rajpert‐De Meyts E, Mitchell M, Lee A C, Jobling M A, McElreavey K. The relationship between Y chromosome DNA haplotypes and Y chromosome deletions leading to male infertility. Hum Genet 200110855–58. [DOI] [PubMed] [Google Scholar]
- 12.Carvalho C M, Fujisawa M, Shirakawa T, Gotoh A, Kamidono S, Freitas Paulo T, Santos S E, Rocha J, Pena S D, Santos F R. Lack of association between Y chromosome haplogroups and male infertility in Japanese men. Am J Med Genet A 2003116152–158. [DOI] [PubMed] [Google Scholar]
- 13.Krausz C, Quintana‐Murci L, Rajpert‐De Meyts E, Jorgensen N, Jobling M A, Rosser Z H, Skakkebaek N E, McElreavey K. Identification of a Y chromosome haplogroup associated with reduced sperm counts. Hum Mol Genet 2001101873–1877. [DOI] [PubMed] [Google Scholar]
- 14.Kuroki Y, Iwamoto T, Lee J, Yoshiike M, Nozawa S, Nishida T, Ewis A A, Nakamura H, Toda T, Tokunaga K, Kotliarova S E, Kondoh N, Koh E, Namiki M, Shinka T, Nakahori Y. Spermatogenic ability is different among males in different Y chromosome lineage. J Hum Genet 199944289–292. [DOI] [PubMed] [Google Scholar]
- 15.Jobling M A, Tyler‐Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 20034598–612. [DOI] [PubMed] [Google Scholar]
- 16.Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of Y‐chromosomal microdeletions. State of the art 2004. Int J Androl 200427240–249. [DOI] [PubMed] [Google Scholar]
- 17.Repping S, Skaletsky H, Brown L, van Daalen S K, Korver C M, Pyntikova T, Kuroda‐Kawaguchi T, de Vries J W, Oates R D, Silber S, van der Veen F, Page D C, Rozen S. Polymorphism for a 1.6‐Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 200335247–251. [DOI] [PubMed] [Google Scholar]
- 18.Underhill P A, Shen P, Lin A A, Jin L, Passarino G, Yang W H, Kauffman E, Bonne‐Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd J R, Mehdi S Q, Seielstad M T, Wells R S, Piazza A, Davis R W, Feldman M W, Cavalli‐Sforza L L, Oefner P J. Y chromosome sequence variation and the history of human populations. Nat Genet 200026358–361. [DOI] [PubMed] [Google Scholar]
- 19.Rosser Z H, Zerjal T, Hurles M E, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley D G, Brede G, Cooper G, Corte‐Real H B, de Knijff P, Decorte R, Dubrova Y E, Evgrafov O, Gilissen A, Glisic S, Golge M, Hill E W, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko S A, Krumina A, Kucinskas V, Lavinha J, Livshits L A, Malaspina P, Maria S, McElreavey K, Meitinger T A, Mikelsaar A V, Mitchell R J, Nafa K, Nicholson J, Norby S, Pandya A, Parik J, Patsalis P C, Pereira, Peterlin B, Pielberg G, Prata M J, Previdere C, Roewer L, Rootsi S, Rubinsztein D C, Saillard J, Santos F R, Stefanescu G, Sykes B C, Tolun A, Villems R, Tyler‐Smith C, Jobling M A. Y‐chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 2000671526–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slatkin M. Linkage disequilibrium in growing and stable populations. Genetics 1994137331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond M, Rousset F. An exact test for population differentiation. Evolution 1995491280–1283. [DOI] [PubMed] [Google Scholar]
- 22.Schneider S, Roessli D, Excoffier L. Arelquin: a software for population genetics data analysis release 2.0. 2000
- 23.Previdere C, Stuppia L, Gatta V, Fattorini P, Palka G, Tyler‐Smith C. Y‐chromosomal DNA haplotype differences in control and infertile Italian subpopulations. Eur J Hum Genet 19997733–736. [DOI] [PubMed] [Google Scholar]
- 24.Semino O, Magri C, Benuzzi G, Lin A A, Al‐Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner P J, Zhivotovsky L A, King R, Torroni A, Cavalli‐Sforza L L, Underhill P A, Santachiara‐Benerecetti A S. Origin, diffusion, and differentiation of Y‐chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 2004741023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruciani F, La Fratta R, Santolamazza P, Sellitto D, Pascone R, Moral P, Watson E, Guida V, Colomb E B, Zaharova B, Lavinha J, Vona G, Aman R, Cali F, Akar N, Richards M, Torroni A, Novelletto A, Scozzari R. Phylogeographic analysis of haplogroup E3b (E‐M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 2004741014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt P H. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet 200092563–2572. [DOI] [PubMed] [Google Scholar]
- 27.Cavalli‐Sforza L L, Menozzi P, Piazza A.The history and geography of human genes. Princeton, NJ: Princeton University Press, 1994