Usher syndrome (USH) (OMIM 276901) is an autosomal recessive disorder characterised by hearing impairment associated with retinitis pigmentosa and in some cases vestibular dysfunction. This disease accounts for approximately 50% of individuals with combined deafness and blindness in developed countries. The estimated prevalence of USH ranges from 3.8 to 6.2/100 000.1,2,3
Phenotypically, three clinical types of Usher syndrome have been defined according to the severity of hearing impairment, age of retinitis pigmentosa onset and the presence or absence of vestibular response. Usher syndrome type I (USH1) is the most serious type, characterised by severe to profound congenital sensorineural hearing loss, balance deficiency and prepubertal onset of retinitis pigmentosa leading to blindness. USH2 is characterised by moderate to severe hearing impairment, normal vestibular function and later onset of retinal degeneration than USH1. USH3 displays progressive hearing loss, retinitis pigmentosa and variable vestibular phenotype.
Six loci for USH1 (USH1B–USH1G) have been mapped and, to date, five genes have been identified.4,5 The MYO7A gene was found to be responsible for USH1B6 and is the most common subtype of USH1, accounting for approximately 50% of cases.7,8,9 Defects in MYO7A also cause autosomal dominant non‐syndromic sensorineural hearing impairment (DFNA11) (MIM 601317),10 autosomal recessive deafness (DFNB2) (MIM 600060)11,12 as well as atypical types of Usher syndrome which are clinically similar to USH3.13
The MYO7A gene has 49 exons, of which 48 are coding, and spans approximately 87 kb of genomic sequence on chromosome 11q13.5. The encoded protein is an unconventional myosin, the myosin VIIA,14 predicted to consist of 2215 amino acids and has a molecular mass of 254 kDa. This protein contains three typical domains: the N terminal head or motor; the neck or regulatory domain consisting of five IQ motifs; and the tail which begins with a short coiled coil domain followed by two large repeats, each containing a MyTH4 and a FERM domain, separated by a SH3 domain.15
In humans, myosin VIIA is expressed in a variety of tissues: the inner ear, retina, testis, lung and kidney.16 Different roles have been postulated for myosin VIIA in the inner ear, such as participation in signal transduction in hair cells and differentiation and organisation of hair cell stereocilia.17 In the human retina, myosin VIIA displays an active function in the migration of RPE melanosomes, phagocytosis of photoreceptor cell outer segment tips and opsin transport through the cilium of these photoreceptors.18,19
The MYO7A gene has been studied in USH1 patients from different origins. Results indicate there is a high diversity of mutations4 distributed along the gene with no indication of a hot spot of mutations. In the present study, we report the results of screening for mutations in the MYO7A gene in 40 probands with USH1 from Spain, Italy, Turkey, Morocco and the Czech Republic.
Materials and methods
Subjects
In total, 40 individuals affected by USH1 were collected as part of a large study on the genetics of Usher syndrome. Twenty five Spanish families were recruited from the Federación de Asociaciones de Afectados de Retinosis Pigmentaria del Estado Español (FAARPEE) and also from Ophthalmology and ENT Services of several Spanish Hospitals. Similarly, 10 families were studied from Italy, two from Turkey, one from the Czech Republic and two from Morocco.
Patients were considered as suffering from USH1 on the basis of profound and congenital hearing loss, vestibular areflexia and retinitis pigmentosa. Clinical confirmation of this diagnosis was not available for nine of these patients.
Fifty unrelated healthy Spanish individuals were screened as controls to evaluate the presence and frequency of novel changes found in the patient samples.
Mutation screening
Genomic DNA was extracted from peripheral blood of affected individuals and their family members by standard methods. Polymerase chain reaction (PCR) was performed for all 49 exons, including intron–exon boundaries of the MYO7A gene (primer sequence and PCR conditions are available from the authors on request).
Single strand conformational polymorphism (SSCP) analysis was used to screen all amplified fragments from all affected individuals on polyacrylamide gel electrophoresis. DNA samples with different known polymorphisms were available. Those fragments with a migration pattern different from normal and polymorphic controls were directly sequenced in both forward and reverse modes on an automated sequencer (ABI‐PRISM, model 3130x1) to confirm sequence change location.
After SSCP analysis, in an attempt to detect missed mutations, the whole gene was sequenced directly in patients found to be heterozygous for only one pathogenic variant.
To discard the possible known deletion of exons 47–49,20 one PCR was performed in patients who were not heterozygous for any variation in this region, as described by Jaijo et al.8
Prediction of splice scores
Isocoding changes in coding regions and intronic variants were analysed to assess their likelihood of creating or eliminating splice sites using the BDGP splice site prediction program (available at http://www.fruitfly.org/seq_tools/splice.html) and the Splice View program (http://bioinfo.itb.cnr.it/oriel/splice‐view.html). Intronic variant sequences were considered pathological when loss of splicing acceptor or donor activity was predicted and they were not present in control samples. Exonic variants were analysed using the RESCUE‐ESE program (http://genes.mit.edu/burgelab/rescue‐ese) to detect a possible alteration in predicted ESE.
Results
The MYO7A gene was screened for mutations in 40 patients with USH1 from different countries. In this study, 19 likely pathogenic changes were detected: two nonsense mutations, one putative splice site mutation, one insertion and five deletions (one of them in frame) and 10 missense mutations (table 1). Thirteen of these changes have not been described previously. None of the novel missense mutations, the putative splice site mutation or deletion inframe were found in the control sample.
Table 1 Summary of myosin VIIA mutations detected in this study.
| Nucleotide change | Codon change | Exon | Domain | No. of families | Origin |
|---|---|---|---|---|---|
| Missense | |||||
| c.77C>A* | p.A26E | 3 | Motor | 1 | Italy |
| c.395C>T | p.P132L | 5 | Motor | 1 | Spain |
| c.721C>G | p.R241G | 7 | Motor | 3 | Italy |
| c.1097T>C | p.L366P | 11 | Motor | 1 | Italy |
| c.3134T>C/ c.5507T>C | p.I1045T/p.L1836P | 25/40 | Tail | 1 | Italy |
| c.3652G>A | p.G1218R | 29 | Tail | 1 | Italy |
| c.3719G>A† | p.R1240Q | 29 | Tail | 1 | Italy |
| c.4475C>T | p.A1492V | 34 | Tail | 1 | Spain |
| c.6610G>C | p.A2204P | 49 | Tail | 1 | Spain |
| Nonsense | |||||
| c.1884C>A‡ | p.C628X | 16 | Motor | 1 | Spain |
| c.5581C>T§ | p.R1861X | 40 | Tail | 1 | Spain |
| Insertion/deletion | |||||
| c.655_660del | p.I219_H220del | 7 | Motor | 1 | Morocco |
| c.986dupG | p.N330QfsX5 | 9 | Motor | 1 | Italy |
| c.3764delA | p.K1255RfsX8 | 30 | Tail | 1 | Spain |
| c.4297delC | p.Q1433SfsX116 | 32 | Tail | 1 | Spain |
| c.5835_5838delCTTT | p.F1946SfsX23 | 42 | Tail | 1 | Italy |
| c.6025delG* | p.A2009PfsX32 | 44 | Tail | 1 | Italy |
| Putative splice site mutation | |||||
| c.2283‐1G>T¶ | — | 20 | Neck | 1 | Morocco |
On SSCP analysis three patients were found to be carriers of a single missense mutation. The remaining exons were then directly sequenced to detect any possible second mutation missed by the first technique. A second mutation was only found in one patient, who was compound heterozygous for p.R241G and the p.G1218R change, detected by sequencing. Furthermore, the 2 kb deletion reported by Adato et al20 was screened but was not detected in any of the patients.
Mutations in both alleles were identified in 12 families (six homozygous and six compound heterozygous) whereas only one putative pathogenic variant was found in two families. Genotypes of the USH1 patients are shown in table 2.
Table 2 Genotypes of patients with Usher syndrome type 1B.
| Genotype/patient | Mutation 1 | Mutation 2 | Origin |
|---|---|---|---|
| Homozygous | |||
| RP‐1264 | c.5835_5838delCTTT | c.5835_5838delCTTT | Italy |
| RP‐1283* | p.P132L | p.P132L | Spain |
| RP‐1301 | c.2283‐1G>T | c.2283‐1G>T | Morocco |
| RP‐1340* | p.I219_H220del | p.I219_H220del | Spain |
| RP‐1356 | p.I1045T/p.L1836P | p.I1045T/p.L1836P | Italy |
| RP‐1462* | p.R241G | p.R241G | Italy |
| Compound heterozygous | |||
| RP‐1252* | c.3764delA | p.R1861X | Spain |
| RP‐1266* | p.A26E | p.L366P | Italy |
| RP‐1268 | p.N330QfsX5 | p.R1240Q | Italy |
| RP‐1269 | p.R241G | c.6025delG | Italy |
| RP‐1271 | p.R241G | p.G1218R | Italy |
| RP‐1436 | p.C628X | c.4297delC | Spain |
| Heterozygous | |||
| RP‐1293 | p.A1492V | ND | Spain |
| RP‐1426 | p.A2204P | ND | Spain |
ND, not determined.
*Patients in whom segregation analysis was performed.
One putative splice site mutation was observed in this study. This change comprised a variant in the −1 position at intron 19. An in silico analysis was performed to predict whether c.2283‐1G>T could affect the normal splicing of this region. Analysis of the normal allele with the Splice View program gave a score of 0.84 for the acceptor splice site; however, the splice sequence was not recognised for the mutant allele.
One patient was found to carry two missense changes in the homozygous state, p.I1045T in exon 25 and p.L1836P in exon 40. These were novel changes that were not found in the 100 control chromosomes, making it difficult to predict the pathogenic significance of these variations.
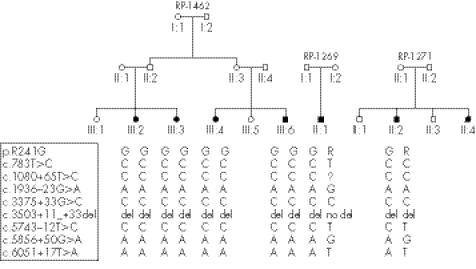
All mutations were private with the exception of the novel variation p.R241G found in three Italian families. Intragenic haplotypes were constructed using the polymorphic variants detected in these patients and the same haplotype linked to the mutation was observed (fig 1).
Figure 1 Pedigree of the Italian families. Haplotypes linked to the common mutation p.R241G are shown.
A non‐described variant was found in exon 33. It consisted of a tandem repetition of a 70 base sequence comprising the last 17 bases of exon 33 and the flanking 53 intronic nucleotides (sequence: AGC CTA CAA ATT CTC AGg tac ccc gca gcc tgc aat gct ccc agt ccc ttg ctc tgt agc tcc agc cca c). The consensus sequence (NM_000260; UCSC Genome Browser) shows that this region is repeated three times but in USH1 patients, three, four and five copies have been found in variable proportions: 42%, 4% and 54%, respectively. Screening for this variant in the control population detected a similar rate for the number of repetitions.
Furthermore, previously unreported non‐pathological intronic or exonic base substitutions were found. Novel variants that do not fall into the likely pathogenic category are summarised in table 3.
Table 3 Novel non‐pathological variants found in this study.
| Exon | Polymorphism | Frequency |
|---|---|---|
| 14 | c.1555‐71G>A | 1/80 |
| 14 | c.1555‐42T>C | 1/80 |
| 31 | c.4023C>T | 1/80 |
| 33 | 3 Tandem repetition | 34/80 |
| 33 | 4 Tandem repetition | 3/80 |
| 33 | 5 Tandem repetition | 54/80 |
| 40 | c.5636+53C>T | 1/80 |
| 40 | c.5598C>A | 1/80 |
| 41 | c.5637‐27C>G | 1/80 |
| 49 | c.6439‐11C>T | 2/80 |
Allele frequency refers to patient sample.
Discussion
Mutation screening of the human MYO7A gene in 40 unrelated patients with USH1 showed a total of 19 pathogenic variants in the coding sequence and flanking intronic sequences. Mutations were detected in 14 of the 40 families studied, accounting for 35%. No mutation was detected in those patients where a diagnosis of USH1 could not be confirmed. If we considered only patients for whom the USH1 diagnosis was unequivocal, the detection rate increased to 45%.
Thirteen different novel mutations were found in addition to the six previously reported ones. Seven variants were found to be located in the motor domain, one in the first IQ motif of the neck and the other 11 in the tail. Previous studies have reported that the majority of mutations in the MYO7A gene are located within the head domain20; however, mutations reported here display regular distribution along the gene. Furthermore, results described in this work confirm the high diversity of mutations responsible for USH1B already reported in previous studies. Most pathological variants found in this screening proved to be private and no mutational hotspots in MYO7A were observed.
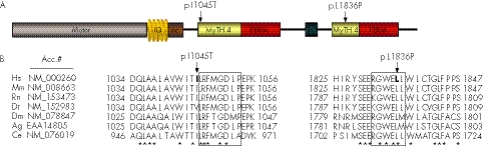
The missense variants p.I1045T and p.L1836P were found in a homozygous state in the same patient. Both changes are located in MyTH4 domains which contain four highly conserved regions (MGD, LRDE, RGW and ELEA) present in different myosins. The Ile1045 precedes the MGD region of the first MyTH4 domain and is a quite commonly conserved residue among other species. On the other hand, the Leu1836 is an amino acid found in the RGW region of the second MyTH4 domain (fig 2). The implication of both amino acids in important regions of the MyTH4 domains together with the fact that neither of these missense variants were found in control samples implies that we cannot discard the possibility that these changes may exert a pathological effect.
Figure 2 (A) Myosin VIIA protein structure, where the different domains and mutations p.I1045T and p.L1836P are indicated. (B) Amino acid sequence alignment of myosin VIIA orthologues derived from seven different species: Homo sapiens (Hs), Mus musculus (Mm), Rattus norvegicus (Rn), Danio rerio (Dr), Drosophila melanogaster (Dm), Anopheles gambiae (Ag) and Caenorhabditis elegans (Ce). Blocks show location of the MGD and RGW regions. Bold indicates the substituted residue in the USH1 patient. * Indicates the conserved amino acid in the seven species. Consensus MGD region: (K/R)(F/Y)MGDhP; consensus RGW region: RGWxLH; where x is any residue and h is an hydrophobic amino acid.
One novel mutation, the p.R241G, was found in three USH1 Italian families who came from the same region in southern Italy. One was homozygous for this mutation and two families were compound heterozygous with another mutation, the c.6025delG, described in the Spanish population,24 and the previously unreported p.G1218R variant. Haplotypes were constructed using the intragenic variants observed in these patients. The same haplotype linked to the mutation was observed in these families, suggesting a common origin.
The variation c.2283‐1T>G was found in the homozygous state in one USH1 patient. This change has recently been found in another study in the heterozygous state with a missense mutation.9 In silico analysis of the efficiency of the splice site indicated a pathogenic effect, given the lack of acceptor splice site recognition. Nevertheless, the effect of this mutation should be confirmed by in vitro analysis. Roux et al9 found this sequence variant in a family from Algeria and in our study it was found in a patient from Morocco. As both families come from Magreb, it is very tempting to speculate a common origin for this mutation; however, only haplotype analysis can elucidate this question.
The change p.Y1719C has been described previously in several studies.22,25 This variant was considered pathological because the region where it is located is highly conserved and because it was not found in control samples. This variant was found by Nájera et al24 in a control population and thus its pathological significance was called into doubt. Jaijo et al8 reported this variant in three USH1 probands. These patients were directly sequenced for mutations in the MYO7A gene but a second mutation was not found. Finally, Roux et al9 also found this change in control samples and considered it as a non‐pathogenic variant. In this report, the variant p.Y1719C has been found in two patients and designated as a polymorphic variant without pathological significance.
USH1 patients were found to carry different numbers of copies for a tandem repetition of 70 bp in exon/intron 33. A high proportion of three and five copies were observed while four repeats were only found in three patients. This region comprises the donor splice site of exon 33 and therefore the control population was screened in order to discard the possible pathological implication of the number of repeats in splicing. The frequency observed in control samples for three, four and five copies was the same as in the USH1 population, and thus this variant has been considered as non‐pathogenic.
Two patients were found to carry only one mutation: p.A1492V and p.A2204P. Although we directly sequenced the remaining exons and the 2 kb deletion was not found, a second mutation could not be identified. The probability that these patients are carriers of a MYO7A mutation and that Usher syndrome is caused by mutations in another USH gene is low (estimated at approximately 1.5%).26 Possible explanations are that the second mutation is located deep in an intronic region or in the promoter, which we did not screen. One should also bear in mind that with the screening method used in the present study, large deletions or inversions would also escape detection.
Screening for USH1 genes in patients with USH1 has recently been performed in different populations and the prevalence observed for MYO7A ranges from 39% to 55%.7,9 In our series (Jaijo et al8 and the present study), the MYO7A gene is implicated in approximately 45% of USH1 patients. It is worth noting that we were able to find 14 of the 20 expected mutations in the Italian patients, yielding a 70% detection rate in MYO7A. This would represent the highest percentage ever detected but because of the small sample size and the fact that three of them share a common mutation, a greater number of families must be studied to determine the real rate of MYO7A mutations in the Italian population.
In spite of the major implication of MYO7A in USH1, recent studies have reported the importance of other USH1 genes, such as CDH23, PCDH15 or USH1C.9 Screening for mutations in these genes in patients without MYO7A mutations must be carried out to determine the extent of the involvement of other genes in our series.
Key points
Usher syndrome type I (USH1) is the most severe form of Usher syndrome and is characterised by profound sensorineural hearing loss, prepubertal onset of retinitis pigmentosa and vestibular dysfunction. Different genes are causative for USH1; however, the MYO7A gene bears the main responsibility.
Our aim was to identify MYO7A gene mutations in Usher syndrome type I patients from diverse origins. Therefore, mutation screening was performed in 40 unrelated patients in all 49 exons of the gene.
19 different mutations were identified, 13 of which were novel. These variants include two nonsense mutations, one putative splice site mutation, one insertion and five deletions in coding sequence and 10 missense mutations.
All mutations were private, with the exception of p.R241G. It is likely that this variant shares a common origin in the southern Italian population.
A wide spectrum of MYO7A mutations have been observed, with this gene being responsible for USH1 in approximately 45% of the families studied.
Acknowledgements
The authors grateful to the patients participating in the study and their family members, and also to the FAARPEE for their help and cooperation. This work was supported by a grant from the Fondo de Investigaciones Sanitarias (PI04/0918), Redes Tematicas de Investigacion Cooperativa (FIS PI05/0612 and FIS PI05/1195), the Integrated Project EVI‐Genoret (Contract No LSHG‐CT‐2005‐512036) and the ONCE. Elena Aller is the recipient of a fellowship from Agencia Valenciana de Ciencia i Tecnologia (CTBPRB/2003/122).
Abbreviations
PCR - polymerase chain reaction
SSCP - single strand conformational polymorphism
USH - Usher syndrome
Footnotes
Competing interests: None.
References
- 1.Rosenberg T, Haim M, Hauch A M, Parving A. The prevalence of Usher syndrome and other retinal dystrophy‐hearing impairment associations. Clin Genet 199751314–321. [DOI] [PubMed] [Google Scholar]
- 2.Hope C I, Bundey S, Proops D, Fielder A R. Usher syndrome in the city of Birmingham—prevalence and clinical classification. Br J Ophthalmol 19978146–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espinos C, Najera C, Millan J M, Ayuso C, Baiget M, Perez‐Garrigues H, Rodrigo O, Vilela C, Beneyto M. Linkage analysis in Usher syndrome type I (USH1) families from Spain. J Med Genet 199835391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed Z M, Riazuddin S, Riazuddin S, Wilcox E R. The molecular genetics of Usher syndrome. Clin Genet 200363431–444. [DOI] [PubMed] [Google Scholar]
- 5.Gerber S, Bonneau D, Gilbert B, Munnich A, Dufier J L, Rozet J M, Kaplan J. USH1A: chronicle of a slow death. Am J Hum Genet 200678357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston M D.et al Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 199537460–61. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang X M, Yan D, Du L L, Hejtmancik J F, Jacobson S G, Nance W E, Li A R, Angeli S, Kaiser M, Newton V, Brown S D, Balkany T, Liu X Z. Characterization of Usher syndrome type I gene mutations in an Usher syndrome patient population. Hum Genet 2005116292–299. [DOI] [PubMed] [Google Scholar]
- 8.Jaijo T, Aller E, Oltra S, Beneyto M, Najera C, Ayuso C, Baiget M, Carballo M, Antinolo G, Valverde D, Moreno F, Vilela C, Perez‐Garrigues H, Navea A, Millan J M. Mutation profile of the MYO7A gene in Spanish patients with Usher syndrome type I. Hum Mutat 200627290–291. [DOI] [PubMed] [Google Scholar]
- 9.Roux A F, Faugere V, Le Guedard S, Pallares‐Ruiz N, Vielle A, Chambert S, Marlin S, Hamel C, Gilbert B, Malcolm S, Claustres M. Survey of the frequency of USH1 gene mutations in a cohort of Usher patients shows the importance of cadherin 23 and protocadherin 15 genes and establishes a detection rate of above 90%. J Med Genet 200643763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X Z, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel K P, Brown S D. Autosomal dominant non‐syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet 199717268–269. [DOI] [PubMed] [Google Scholar]
- 11.Liu X Z, Walsh J, Mburu P, Kendrick‐Jones J, Cope M J, Steel K P, Brown S D. Mutations in the myosin VIIA gene cause non‐syndromic recessive deafness. Nat Genet 199716188–190. [DOI] [PubMed] [Google Scholar]
- 12.Weil D, Kussel P, Blanchard S, Levy G, Levi‐Acobas F, Drira M, Ayadi H, Petit C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin‐VIIA gene. Nat Genet 199716191–193. [DOI] [PubMed] [Google Scholar]
- 13.Liu X Z, Hope C, Walsh J, Newton V, Ke X M, Liang C Y, Xu L R, Zhou J M, Trump D, Steel K P, Bundey S, Brown S D. Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet 199863909–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley P M, Weston M D, Chen Z Y, Orten D J, Hasson T, Overbeck L D, Pinnt J, Talmadge C B, Ing P, Mooseker M S, Corey D, Sumegi J, Kimberling W J. The genomic structure of the gene defective in Usher syndrome type Ib (MYO7A). Genomics 19974073–79. [DOI] [PubMed] [Google Scholar]
- 15.Levy G, Levi‐Acobas F, Blanchard S, Gerber S, Larget‐Piet D, Chenal V, Liu X Z, Newton V, Steel K P, Brown S D, Munnich A, Kaplan J, Petit C, Weil D. Myosin VIIA gene: heterogeneity of the mutations responsible for Usher syndrome type IB. Hum Mol Genet 19976111–116. [DOI] [PubMed] [Google Scholar]
- 16.Hasson T, Heintzelman M B, Santos‐Sacchi J, Corey D P, Mooseker M S. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc Natl Acad Sci U S A 1995929815–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam K N, Weil D, Yonekawa H, Wolfrum U, El‐Amraoui A, Petit C. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet 200514347–356. [DOI] [PubMed] [Google Scholar]
- 18.Udovichenko I P, Gibbs D, Williams D S. Actin‐based motor properties of native myosin VIIa. J Cell Sci 2002115445–450. [DOI] [PubMed] [Google Scholar]
- 19.El‐Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci 20051184593–4603. [DOI] [PubMed] [Google Scholar]
- 20.Adato A, Weil D, Kalinski H, Pel‐Or Y, Ayadi H, Petit C, Korostishevsky M, Bonne‐Tamir B. Mutation profile of all 49 exons of the human myosin VIIA gene, and haplotype analysis, in Usher 1B families from diverse origins. Am J Hum Genet 199761813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bharadwaj A K, Kasztejna J P, Huq S, Berson E L, Dryja T P. Evaluation of the myosin VIIA gene and visual function in patients with Usher syndrome type I. Exp Eye Res 200071173–181. [DOI] [PubMed] [Google Scholar]
- 22.Janecke A R, Meins M, Sadeghi M, Grundmann K, Apfelstedt‐Sylla E, Zrenner E, Rosenberg T, Gal A. Twelve novel myosin VIIA mutations in 34 patients with Usher syndrome type I: confirmation of genetic heterogeneity. Hum Mutat 199913133–140. [DOI] [PubMed] [Google Scholar]
- 23.Cuevas J M, Espinos C, Millan J M, Sanchez F, Trujillo M J, Garcia‐Sandoval B, Ayuso C, Najera C, Beneyto M. Detection of a novel Cys628STOP mutation of the myosin VIIA gene in Usher syndrome type Ib. Mol Cell Probes 199812417–420. [DOI] [PubMed] [Google Scholar]
- 24.Nájera C, Beneyto M, Blanca J, Aller E, Fontcuberta A, Millan J M, Ayuso C. Mutations in myosin VIIA (MYO7A) and usherin (USH2A) in Spanish patients with Usher syndrome types I and II, respectively. Hum Mutat 20022076–77. [DOI] [PubMed] [Google Scholar]
- 25.Cuevas J M, Espinos C, Millan J M, Sanchez F, Trujillo M J, Ayuso C, Beneyto M, Najera C. Identification of three novel mutations in the MYO7A gene. Hum Mutat 199914181. [DOI] [PubMed] [Google Scholar]
- 26.Kimberling W J. Estimation of the frequency of occult mutations for an autosomal recessive disease in the presence of genetic heterogeneity: application to genetic hearing loss disorders. Hum Mutat 200526462–470. [DOI] [PubMed] [Google Scholar]