Abstract
Background
Familial hypobetalipoproteinaemia (FHBL) is a codominant disorder characterised by fatty liver and reduced plasma levels of low‐density lipoprotein (LDL) and its protein constituent apolipoprotein B (apoB). FHBL is linked to the APOB gene in some but not all known cases. In a group of 59 patients with FHBL genotyped for APOB gene mutations, we found three novel splice‐site mutations: c.904+4A→G in intron 8, c.3843−2A→G in intron 24 and c.4217−1G→T in intron 25.
Objective
To assess the effects of these mutations on apoB pre‐mRNA splicing.
Methods
ApoB mRNA was analysed in the liver of one proband and in cells expressing APOB minigenes harbouring the mutations found in the other probands.
Results
In the liver of the c.3843−2A→G carrier, an apoB mRNA devoid of exon 25 was identified, predicted to encode a truncated peptide of 1260 amino acids. The analysis of minigene transcripts in COS‐1 cells showed that the c.904+4A→G mutation caused the formation of an mRNA devoid of exon 8, predicted to encode a short apoB of 247 amino acids. The minigene harbouring the c.4217−1G→T mutation in intron 25 generated an mRNA in which exon 25 joined to a partially deleted exon 26, resulting from the activation of an acceptor site in exon 26; this mRNA is predicted to encode a truncated protein of 1380 amino acids. All these truncated apoBs were not secreted as constituents of plasma lipoproteins.
Conclusion
These findings demonstrate the pathogenic effect of rare splice‐site mutations of the APOB gene found in FHBL.
Familial hypobetalipoproteinemia (FHBL, OMIM 107730) is a codominant disorder characterised by low plasma levels of low‐density lipoprotein‐cholesterol (LDL‐C) and total apolipoprotein B (apoB), the main constituent peptide of very low‐density lipoproteins (VLDLs), low‐density lipoprotein (LDL) and chylomicrons.1,2 The frequency of heterozygotic FHBL has been estimated to be around 1:1000–3000.3,4 FHBL heterozygotes may be asymptomatic or have fatty liver disease as the main clinical manifestation.2,5,6 Mild intestinal lipid malabsorption and oral fat intolerance have been reported in some patients.7 FHBL is a genetically heterogeneous disorder that is primarily caused by mutations in the APOB gene; in some families, FHBL seems to be linked to other genes yet to be identified.2 The best characterised patients with FHBL are those who carry mutations in the APOB gene, specifying the production of truncated forms of apoB of various lengths, which may or may not be secreted into the plasma.2,8 The truncated apoBs are designated according to a centile nomenclature with respect to ApoB‐100, the normal protein of 4536 amino acids corresponding to the full‐length translation product of apoB mRNA, which is synthesised by the liver as a constituent of VLDL.1 Only a single amino acid substitution (p.R463W) in apoB has been associated with FHBL so far.9 A few rare mutations affecting splice sites have been reported as putative pathogenic mutations.10,11,12,13,14,15,16 So far, only one splice‐site mutation has been fully characterised with regard to its effect on apoB mRNA splicing.17 In this study, we describe three novel splice‐site mutations in the APOB gene and demonstrate their effect on apoB pre‐mRNA splicing in liver biopsy specimens and in cells transfected with apoB minigenes harbouring these mutations.
Patients and methods
The patients with FHBL described in this report were identified during a survey of 59 consecutive individuals with fatty liver associated with low plasma LDL‐C and apoB and with the clinical diagnosis of definite or probable heterozygotic FHBL. The systematic sequence of APOB gene in these patients led to the identification of putative pathogenic mutations in 33 patients (26 carriers of nonsense/frameshift mutations resulting in truncated apoBs of various sizes, 2 carriers of the R463W substitution and 4 carriers of splice‐site mutations). In this study, we focus on the carriers of splice‐site mutations.
Proband FHBL‐19 was a 38‐year‐old woman who had moderate hepatomegaly, persistently increased serum liver enzymes and low plasma lipids (see Results section). Ultrasound examination of the liver revealed fatty liver. The proband's father was found to have fatty liver and low plasma lipids (total cholesterol 116 mg/dl, LDL‐C 55 mg/dl, apoB 29 mg/dl and triglycerides 49 mg/dl).
Proband FHBL‐20 was a 40‐year‐old man with hepatomegaly, slightly increased serum liver enzymes and low plasma lipids (see Results section). Ultrasound examination of the liver revealed the presence of fatty liver, which was confirmed by liver biopsy (steatosis in 70% of hepatocytes with no signs of inflammation, necrosis and iron deposits). The proband's sister also had fatty liver (documented by liver biopsy), associated with low plasma lipids (total cholesterol 81 mg/dl, LDL‐C 29 mg/dl, apoB 14 mg/dl and triglycerides 29 mg/dl). The proband's 7‐year‐old son was found to have fatty liver and low plasma lipids (total cholesterol 96 mg/dl, LDL‐C 38 mg/dl, apoB 21 mg/dl and triglycerides 33 mg/dl).
Proband FHBL‐22 was an 8‐year‐old girl admitted to hospital for obesity, moderate hepatomegaly and premature puberty. She had fatty liver and low plasma lipids (see Results section). No family member was available for study.
Proband FHBL‐28 was a 17‐year‐old boy who was found to have hepatomegaly, persistently increased serum liver enzymes and low plasma lipids (see Results section). Liver biopsy showed a marked macrovescicular steatosis and mild focal perisinusoidal fibrosis. No iron deposits were observed. The proband's father was found to have fatty liver, slightly increased serum liver enzymes and low plasma lipids (total cholesterol 85 mg/dl, LDL‐C 35 mg/dl, apoB 22 mg/dl and triglycerides 81 mg/dl).
In all these patients with FHBL, the other routine laboratory tests, including hepatitis B virus and hepatitis C virus infection, were either within the normal range or negative.
The group of controls included 100 healthy individuals of both sexes randomly selected among blood donors with normolipidaemia.
This study was approved by and performed in accordance with the ethical standards of the Universities of Trieste, Genova, and Modena and Reggio Emilia, Italy. Informed and written consent was obtained from the probands (or in the case of children from their parents) and their family members as well as from controls.
Analysis of plasma lipoproteins and apolipoproteins
Plasma total cholesterol, triglycerides, high‐density lipoprotein‐cholesterol, LDL‐C, apolipoprotein A‐I and apoB were measured as reported previously.7 Plasma lipoproteins were separated by continuous density gradient ultracentrifugation and apolipoproteins by sodium dodecyl sulphate/polyacrylamide gel electrophoresis.7
Analysis of APOB gene
Genomic DNA was isolated from peripheral blood. The entire APOB gene‐coding region, including the 5′‐flanking region and at least 50 base pairs of intronic sequence at each intron–exon boundary (AY324608.1, GI:32187678), were amplified by PCR, as described previously.7,18 Sequences were detected on an Applied Biosystems 3100 DNA sequencer and results were analysed with ABI PRISM SeqScape software (Applied Biosystems, Warrington, UK). One hundred controls were screened for the splice‐site mutations found in patients with FHBL, by sequencing the appropriate APOB gene regions.
Analysis of apoB mRNA in liver biopsy
Total RNA was extracted (RNeasy Minikit, Qiagen, Hilden, Germany) from liver biopsy specimens from proband FHBL‐20 (heterozygotic for c.3843–2A→G in intron 24) and from a surgical biopsy specimen from a patient who had been operated on for cholecystectomy without liver damage (control liver). RNA was retrotranscribed and a region of apoB cDNA (GenBank accession number NM_000384) spanning from the 3′‐end of exon 24 to the 5′‐end of exon 26 was amplified by PCR. Primers and amplification conditions are given in supplementary methods (available at http:// jmg.bmj.com/supplemental).
Construction of APOB minigenes and their expression in transfected cells
To investigate the effects of splice‐site mutations in intron 8 (proband FHBL‐22) and intron 25 (proband FHBL‐19; see Results section), we adopted an in vitro strategy, as suitable liver samples from these patients were not available. This procedure included, as a first step, the construction of wild‐type and mutant APOB minigenes by the amplification of the appropriate APOB gene regions (encompassing the splice sites harbouring the mutations) from the probands' genomic DNA. The primers and the conditions used for genomic DNA amplification are specified in supplementary table 1s (available at http://jmg.bmj.com/supplemental). The minigene encompassing intron 8 contained the 3′‐end of exon 7 (98 bp), intron 7, exon 8, intron 8, exon 9, intron 9 and the 5′‐end of exon 10 (165 bp). The minigene encompassing intron 25 contained the 3′‐end of exon 25 (278 bp), intron 25 and 5′‐end of exon 26 (1363 bp). The minigenes were cloned in the pTargeT (Promega, Madison, Wisconsin, USA) expression vector and then transfected into COS‐1 cells as described in supplementary methods (available at http://jmg.bmj.com/supplemental).
Mutation nomenclature
Mutations are described according to mutation nomenclature.19,20 Nucleotide numbers are derived from apoB cDNA sequence (GenBank accession number NM_000384). Amino acid sequence changes in apoB are described according to the National Center for Biotechnology Information reference sequence (NP_000375.2, GI:105990532).
Computational analysis of APOB gene mutations
An information theory‐based software tool (http://splice.cmh.edu) was used to interpret non‐coding sequence variations within functional sequence elements such as splice sites.21
Results
Plasma lipids and lipoproteins
Table 1 shows the plasma lipids and lipoproteins in patients with FHBL. All had low plasma levels of total cholesterol, LDL‐C and apoB (below the fifth centile of the control population of similar age and sex). A similar plasma lipid profile was observed in some first‐degree relatives of the probands (when available for study; see Methods section), thus confirming the vertical transmission of the plasma lipid abnormality and the clinical diagnosis of FHBL. The analysis of apoB in plasma lipoproteins separated by density‐gradient ultracentrifugation5,7,18 failed to reveal the presence of truncated apoBs secreted into the plasma (data not shown)
Table 1 Plasma lipids and lipoproteins of FHBL probands.
| Proband | Age (years)/sex | BMI (kg/m2) | TC (mg/dl) | TG (mg/dl) | LDL‐C (mg/dl) | HDL‐C (mg/dl) | Apo A‐I (mg/dl) | ApoB (mg/dl) |
|---|---|---|---|---|---|---|---|---|
| FHBL‐19 | 38/F | 29.3 | 88 | 46 | 35 | 44 | 124 | 22 |
| FHBL‐20 | 43/M | 23.8 | 123 | 100 | 59 | 44 | 169 | 27 |
| FHBL‐22 | 8/F | 27.3 | 54 | 24 | 14 | 35 | 104 | 12 |
| FHBL‐28 | 17/M | 25.0 | 90 | 61 | 36 | 42 | 150 | 28 |
Apo, apolipoprotein; BMI, body mass index; F, female; FHBL, familial hypobetalipoproteinaemia; HDL‐C, high‐density lipoprotein‐cholesterol; LDL‐C, low‐density lipoprotein‐cholesterol; M, male; TC, total cholesterol; TG, triglyceride.
Sequence of APOB gene
The sequence of the APOB gene revealed the presence of three potentially pathogenic mutations affecting splice sites: (1) c.904+4A→G in intron 8 (proband FHBL‐22); (2) c.3843−2A→G in intron 24 (probands FHBL‐20 and FHBL‐28); and (3) c.4217−1G→T in intron 25 (proband FHBL‐19). When the family members were screened to define the cosegregation of the splice‐site mutations with the FHBL phenotype, only patients with hypobetalipoproteinaemia were found to carry the mutations. No carriers of these mutations were found among 100 healthy controls.
Other sequence variants in the coding region and introns found in our probands are listed in supplementary tables 2s–5s (available at http://jmg.bmj.com/supplemental). Besides silent mutations or common amino acid polymorphisms, we detected two rare non‐conservative amino acid substitutions: c.5905A→C, p.Ser1942Arg found in FHBL‐22 and c.11809G→A, p.Glu3910Lys found in probands FHBL‐20 and FHBL‐28. Cosegregation analysis in families of probands FHBL‐20 and FHBL‐28 showed that the Glu3910Lys substitution cosegregated with the splice‐site mutation (c.3843−2A→G). Cosegregation analysis was not possible in the case of proband FHBL‐22 as no family member was available for study.
Computational analysis of the APOB gene mutations
To evaluate the effect of the splice‐site mutations, we applied the automated splice‐site analysis to 2 kb genomic DNA fragments encompassing the region harbouring each specific mutation.21 This analysis indicated that (1) c.904+4A→G in intron 8 reduced the information content of the corresponding site from 11.4 to 8.8 bits, rendering it “functionally leaky ”; and (2) c.3843−2A→G in intron 24 and c.4217−1G→T in intron 25 decreased the information content of the sites (from 9.1 to 0.9 bits and from 8.7 to 0.0 bits, respectively), rendering them “functionally inactive”.
Analysis of apoB mRNA splicing in human liver
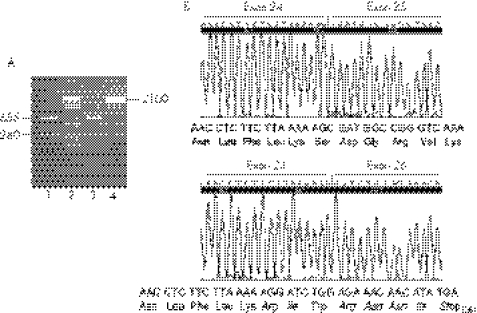
To verify the prediction of automated splice‐site analysis, we analysed the apoB mRNA in the liver biopsy specimens of proband FHBL‐20 (c.3843−2A→G in intron 24) and a control. Reverse transcription PCR amplification of a cDNA region spanning from the 3′‐end of exon 24 to the 5′‐end of exon 26 generated a single fragment of 655 bp in the control liver and two fragments of 655 and 280 bp in the proband's liver (fig 1A). The nucleotide sequence showed that in the 655 bp fragments, exon 24 was followed by exon 25 (fig 1B, upper panel) and the latter by exon 26 (data not shown), indicating that these fragments were generated by the normal apoB allele. In the 280 bp fragment, exon 24 was followed by exon 26 with the complete skipping of exon 25 (fig 1B, lower panel; r.3843_4216del). The joining of exon 24 to exon 26 causes a shift in the reading frame, leading to a premature termination codon. The predicted translation product of this mRNA is a protein of 1260 amino acids containing a stretch of seven novel amino acids at its carboxyl‐terminus (fig 1B, lower panel; p.Ser1254ArgfsX7). This short truncated apoB was designated apoB‐27.77, according to the centile nomenclature. We assumed that the same abnormal apoB mRNA was also present in proband FHBL‐28 heterozygotic for the same splice‐site mutation and whose liver biopsy specimen was not available for study.
Figure 1 Apolipoprotein (apo)B mRNA in the liver of proband familial hypobetalipoproteinaemia (FHBL)‐20, heterozygotic for the c.3843−2A→G mutation in intron 24. (A) Agarose gel electrophoresis of the reverse transcription and polymerase chain reaction (RT‐PCR) amplification of apoB cDNA region spanning from the 3′‐end of exon 24 to the 5′‐end of exon 26. Lane 1: proband; lane 2: DNA size marker; lane 3: control; lane 4: PCR product of the same APOB gene region amplified from genomic DNA. (B) Electropherograms of a partial sequence of the two apoB cDNAs present in the proband's liver. The upper panel shows the sequence of the normal 655 bp RT‐PCR fragment, encompassing the junction between exons 24 and 25; the lower panel shows the sequence of the abnormal 280 bp RT‐PCR fragment, encompassing the junction between exons 24 and 26. This junction produces a frameshift, leading to the insertion of seven novel amino acids (in italics) and a premature stop codon.
Analysis of transcription products of APOB minigenes in COS‐1 cells
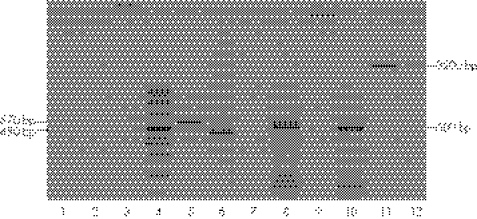
As in the other FHBL probands no liver samples were available for apoB mRNA analysis, we investigated the in vitro transcription products of APOB minigenes harbouring the splice‐site mutations found in these probands. The transcription product generated by APOB minigene harbouring the c.904+4A→G in intron 8 consisted of a single band of approximately 480 bp as opposed to a 570 bp band generated by the corresponding wild‐type minigene (fig 2, lanes 5 and 6). The sequence showed that, in the abnormal 480 bp band, exon 7 was followed by exon 9 with the complete skipping of exon 8 (r.819_904del, supplementary figure 1 available at http://jmg.bmj.com/supplemental). The exon 7–exon 9 junction causes a frameshift, leading to the insertion of two novel amino acids followed by a premature stop codon at position 248 (p.Asn246LysfsX2). The predicted mutant protein is a short peptide of 247 amino acids designated apoB‐5.44.
Figure 2 Analysis of the transcription product of the APOB minigene harbouring the c.904+4A→G mutation in intron 8. RNA isolated from COS‐1 cells, transfected with wild‐type and mutant APOB minigenes,was reverse transcribed and the exon 7–exon 10 region of apoB cDNA was amplified by polymerase chain reaction (PCR). Untransfected cells (lanes 1–3); DNA size marker (lane 4); apolipoprotein (apo)B cDNA in cells transfected with wild‐type (lane 5) and mutant (lane 6) minigenes; “mock” reverse transcription (RT)‐PCR in cells transfected with mutant (lane 7) and wild‐type (lane 9) minigenes; cDNA of neomycin resistance gene in cells transfected with mutant (lane 8) and wild‐type (lane 10) minigenes; PCR amplification of mutant and wild‐type minigenes (lanes 11 and 12). “Mock” RT‐PCR, RT‐PCR performed without the addition of reverse transcriptase to the reaction mixture.
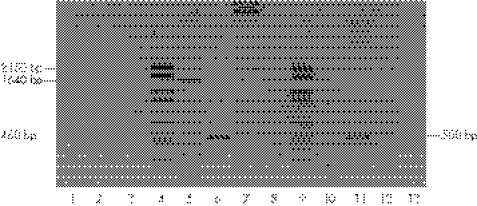
The transcription product generated by the APOB minigene harbouring the c.4217−1G→T mutation in intron 25 consisted of a single band of approximately 460 bp as opposed to a 1640 bp band generated by the wild‐type counterpart (fig 3, lanes 5 and 6). The sequence analysis of the 460 bp fragment showed that exon 25 joined directly to the thymidine at position c.5397 (located 1181 bp from the beginning of exon 26) (r.4217_5395del; fig 4). This abnormal splicing eliminates 1180 bp of exon 26 expected to produce a deletion of 392 amino acids in the apoB. The partial deletion of exon 26 creates the codons for two new amino acids at the junction with exon 25 (valine at position 1379 and threonine at position 1380) followed by a premature stop codon at position 1381 (p.Gly1379_Ser1772→ValfsX3). The predicted mutant protein is a peptide of 1380 amino acids, designated apoB‐30.42.
Figure 3 Analysis of transcription product of mutant APOB minigene harbouring the c.4217−1G→T mutation in intron 25. RNA isolated from COS‐1 cells, transfected with wild‐type and mutant APOB minigenes, was reverse transcribed and the cDNA region spanning from the 3′‐end of exon 25 to the 5′‐end of exon 26 was amplified by polymerase chain reaction (PCR). Untransfected cells (lanes 1–3); DNA size marker (lane 4); apolipoprotein B (apoB) cDNA in cells transfected with wild‐type (lane 5) and mutant (lane 6) minigenes; PCR amplification of mutant (lane 7) and wild‐type (lane 8) minigenes; cDNA of neomycin resistance gene in cells transfected with mutant (lane 10) and wild‐type (lane 11) minigenes; “mock” reverse transcription‐PCR in cells transfected with mutant (lane 12) and wild‐type (lane 13) minigenes. “Mock” RT‐PCR, RT‐PCR performed without the addition of reverse transcriptase to the reaction mixture.
Figure 4 Partial nucleotide sequence of the cDNA generated by the mutant APOB minigene harbouring the c.4217–1G→T mutation in intron 25. The upper panel shows the junction between exons 25 and 26 in the cDNA generated by the wild‐type minigene. The lower panel shows that in mutant cDNA, exon 25 joins the partially deleted exon 26 (exon 25 joins the c.5397T corresponding to position 1181 from the beginning of exon 26). This junction causes a frameshift, leading to a premature stop codon.
Discussion
Functional analysis of APOB gene splice‐site mutations in liver biopsy specimens
In this study, we demonstrated the pathogenic effect of three novel splice‐site mutations of the APOB gene in FHBL. Until now, only six splice‐site mutations of the APOB gene had been reported as a putative cause of FHBL.10,11,12,13,14,15 They were located in highly conserved splice‐site regions of introns 1, 5, 7, 13 and 24, and were assumed to affect the apoB pre‐mRNA splicing process; however, with the exception of the mutation in the donor splice site of intron 24 (IVS24+2T→C),17 their biological effect on apoB pre‐mRNA splicing has not been investigated. The ideal situation to investigate such an effect would be the ex vivo analysis of apoB mRNA in liver or intestinal biopsy specimens from patients with FHBL harbouring these mutations. However, this approach is often precluded as suitable tissue samples are not easily available.
In this study, we have been able to analyse, for the first time, apoB mRNA in the liver of one proband (FHBL‐20 heterozygotic for c.3843−2A→G mutation affecting the acceptor splice site of intron 24). On the prediction that the function of the acceptor site in intron 24 was abolished with the possible elimination of exon 24 in mature mRNA or activation of cryptic acceptor sites in intron 24 or exon 25, we performed reverse transcription‐PCR amplification of an mRNA region spanning from exon 24 to the 5′‐end of exon 26. Our prediction was confirmed, as the patient's liver contained, besides the normally spliced mRNA encoded by the wild‐type allele, an abnormal mRNA devoid of exon 25, generated by alternative splicing.22 The content of this abnormal mRNA was lower than the mRNA generated by the wild‐type allele (fig 1A). This is not surprising, as mRNAs harbouring premature termination codons are usually degraded more rapidly than their wild‐type counterparts (nonsense‐mediated mRNA decay).23,24 This abnormal mRNA is predicted to encode a truncated apoB of 1260 amino acids (apoB‐27.77).
It is of interest that the unique splice‐site mutation of the APOB gene, functionally characterised so far, was also located in intron 24 where it involved the donor splice site.17 In this case, the in vitro expression of the mutant APOB minigene harbouring the mutation showed a splicing defect with the activation of a cryptic splice site in intron 24, resulting in the insertion of 40 bp of intron 24 into the mature mRNA with an alteration of the translation frame. The predicted translation product of this mRNA was a truncated protein designated apoB‐27.6.17
Functional analysis of APOB gene splice‐site mutations in vitro
To define the effect of the other splice‐site mutations, we adopted an in vitro strategy, based on analysis of the transcription products of mutant APOB minigenes transfected into mammalian cells. In the case of the donor splice‐site mutation in intron 8 (c.904+4A→G), analysis of the transcription products revealed that this mutation abolished the function of the normal site, which was replaced by the canonical donor splice site in intron 7, a condition leading to alternative splicing and the skipping of exon 8 in mature mRNA. The translation product of this mRNA is predicted to be a short protein (apoB‐5.44).
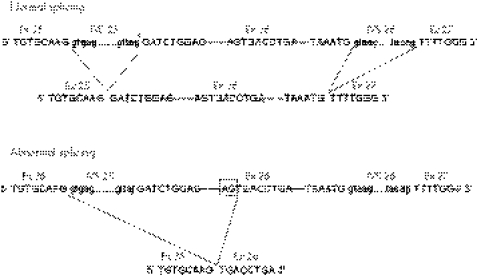
Analysis of the transcription product of the minigene harbouring the mutation in the acceptor splice site of intron 25 (c.4217−1G→T) revealed a more complex situation related to the structure of the APOB gene. One of the peculiar features of this gene is the presence of an exceedingly long exon 26 (7571 bp). The size of this exon prevented us from constructing a minigene encompassing the whole critical gene region (ie, approximately 9000 bp region spanning from 3′‐end of exon 25 to the 5′‐end of exon 27), which could have allowed us to detect transcripts generated by the activation of cryptic acceptor sites along the whole exon 26 or the involvement of the canonical acceptor splice site in intron 26. We constructed a minigene of 2150 bp, whose 3′‐end was located approximately 1360 bp from the start of exon 26, on the assumption that the function of the mutant acceptor splice site in intron 25 could be replaced by some cryptic acceptor splice sites present along exon 26. Indeed, the mRNA generated by the mutant APOB minigene was much shorter than its wild‐type counterpart (fig 3), suggesting the activation of an exonic cryptic splice site. This abnormal mRNA, which contained exon 25 but lacked the first 1180 nucleotides of exon 26, was the result of an abnormal splicing involving a cryptic acceptor splice site located within exon 26 as fig 5 shows. The predicted translation product of this mRNA is a truncated protein designated apoB‐30.42.
Figure 5 Effect of c.4217–1G→T mutation in intron 25 on the apolipoprotein (apo)B pre‐mRNA splicing. The upper panel shows the abridged APOB gene region spanning from the 3′‐end of exon 25 to the 5′‐end of exon 27 and the normal splicing. The mutation in the acceptor splice site of intron 25 (indicated by an asterisk) abolishes the function of this site. The lower panel shows the abnormal splicing due to the activation of a cryptic acceptor splice site located in exon 26 (AG dinucleotide of this site at position c.5395–5396 is boxed). In the mutant mRNA, exon 25 joins a partially deleted exon 26 (devoid of the first 1180 nucleotides; fig 4). Ex, Exon; IVS, intron.
Key points
We report three novel splice‐site mutations in the apolipoprotein B gene in patients with familial hypobetalipoproteinaemia.
The biological effect of these mutations was investigated in COS‐1 cells transfected with apolipoprotein B minigenes harbouring the mutations as well as in the liver of one patient.
We identified abnormally spliced mRNAs predicted to encode truncated apolipoprotein Bs that are unable to bind lipids and form lipoprotein particles.
Truncated apoBs not detectable in plasma and fatty liver
The truncated apoBs predicted to result from the three splice‐site mutations reported in this study were not detectable in plasma, suggesting that they were not secreted as plasma lipoprotein constituents or lipid‐free proteins. These truncated proteins have a reduced capacity to form lipoprotein particles, being devoid of key lipid‐binding domains present in normal apoB. In the lipid‐poor form, they are probably degraded intracellularly. The resulting defect in lipoprotein secretion is the cause of severe fatty liver observed in our patients, which might evolve towards a more severe liver disease.25
Supplementary methods and tables are available at http://jmg.bmj.com/supplemental
Acknowledgments
This study was supported by a grant from Telethon Foundation (number GGP05042) to PT and by a grant from Fondazione Cassa di Risparmio di Trieste (FCT 01/03) to CT.
Abbreviations
apoB - apolipoprotein B
FHBL - familial hypobetalipoproteinaemia
LDL - low‐density lipoprotein
LDL‐C - LDL‐cholesterol
PCR - polymerase chain reaction
VLDL - very‐low‐density lipoproteins
Footnotes
Competing interests: None.
Informed consent was obtained for publication of patients' details in this report.
Supplementary methods and tables are available at http://jmg.bmj.com/supplemental
References
- 1.Kane J P, Havel R J. Disorders of the biogenesis and secretion of lipoproteins containing the B apolipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, eds. The metabolic and molecular bases of inherited disease. Vol II. 8th edn. New York: McGraw‐Hill, 20012717–2752.
- 2.Schonfeld G, Lin X, Yue P. Familial hypobetalipoproteinemia: genetics and metabolism. Cell Mol Life Sci 2005621372–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linton M F, Farese R V, Young S G. Familial hypobetalipoproteinemia. J Lipid Res 199334521–541. [PubMed] [Google Scholar]
- 4.Welty F K, Lahoz C, Tucker K L, Ordovas J M, Wilson P W, Shaefer E J. Frequency of apo B and apo E gene mutations as causes of hypobetalipoproteinemia in the Framingham offspring population. Arterioscler Thromb Vasc Biol 1998181745–1751. [DOI] [PubMed] [Google Scholar]
- 5.Tarugi P, Lonardo A, Ballarini G, Grisendi A, Pulvirenti M, Bagni A, Calandra S. Fatty liver in heterozygous hypobetalipoproteinemia caused by a novel truncated form of apolipoprotein B. Gastroenterology 19961111125–1133. [DOI] [PubMed] [Google Scholar]
- 6.Whitfield A J, Barrett P H, Robertson K, Havlat M F, van Bockxmeer F M, Burnett J R. Liver dysfunction and steatosis in familial hypobetalipoproteinemia. Clin Chem 200551266–269. [DOI] [PubMed] [Google Scholar]
- 7.Tarugi P, Lonardo A, Gabelli C, Sala F, Ballarini G, Cortella I, Previato L, Bertolini S, Cordera R, Calandra S. Phenotypic expression of familial hypobetalipoproteinemia in three kindreds with mutations of apolipoprotein B gene. J Lipid Res 2001421552–1561. [PubMed] [Google Scholar]
- 8.Hooper A J, van Bockxmeer F M, Burnett J R. Monogenic hypocholesterolaemic disorders and apolipoprotein B metabolism. Crit Rev Clin Lab Sci 200542515–545. [DOI] [PubMed] [Google Scholar]
- 9.Burnett J R, Shan J, Miskie B A, Whitfield A J, Yuan J, Tran K, McKnight C J, Hegele R A, Yao Z. A novel nontruncating apoB gene mutation, R463W, causes familial hypobetalipoproteinemia. J Biol Chem 200327813442–13452. [DOI] [PubMed] [Google Scholar]
- 10.Huang L ‐ S, Kayden H, Sokol R J, Breslow J L. ApoB gene nonsense and splicing mutations in a compound heterozygote for familial hypobetalipoproteinemia. J Lipid Res 1991321341–1348. [PubMed] [Google Scholar]
- 11.Talmud P J, Krul E S, Pessah M, Gay G, Schonfeld G, Humphries S E, Infante R. Donor splice mutation generates a lipid‐associated apolipoprotein B‐27.6 in a patient with homozygous hypobetalipoproteinemia. J Lipid Res 199435468–477. [PubMed] [Google Scholar]
- 12.Welty F K, Guida K A, Andersen J J. Donor splice site mutation (210+1 G_C) in the apo B gene causes a very low level of apo B‐100 and LDL cholesterol. Arterioscler Thromb Vasc Biol 2001211864–1865. [PubMed] [Google Scholar]
- 13.Hegele R A, Miskie B A. Acanthocytosis in a patient with homozygous familial hypobetalipoproteinemia due to a novel APOB splice site mutation. Clin Genet 200261101–103. [DOI] [PubMed] [Google Scholar]
- 14.Yue P, Yuan B, Gerhard D S, Neuman R G, Isley W L, Harris W S, Schonfeld G. Novel mutations of apo B cause apo B truncations undetectable in plasma and familial hypobetalipoproteinemia. Hum Mutat 200220110–116. [DOI] [PubMed] [Google Scholar]
- 15.Whitfield A J, Marais A D, Robertson K, Barrett P H R, van Bockxmeer F M, Burnett J R. Four novel mutations in APOB causing heterozygous and homozygous familial hypobetalipoproteinemia. Hum Mutat 200322178. [DOI] [PubMed] [Google Scholar]
- 16.Pulai J, Zakeri H, Kwok P ‐ Y, Kim J H, Wu J, Schonfeld G. Donor splice mutation (665+1GT) in familial hypobetalipoproteinemia with no detectable apo B truncation. Am J Med Genet 199880218–220. [PubMed] [Google Scholar]
- 17.Nemeth‐Slany A, Talmud P, Grundy S M, Patel S B. Activation of a cryptic splice‐site in intron 24 leads to the formation of apolipoprotein B‐27.6. Atherosclerosis 1997133163–170. [DOI] [PubMed] [Google Scholar]
- 18.Tarugi P, Lonardo A, Ballarini G, Erspamer L, Tondelli E, Bertolini S, Calandra S. A study of fatty liver disease and plasma lipoproteins in a kindred with familial hypobetalipoproteinemia due to a novel truncated form of apolipoprotein B (apoB‐54.5). J Hepatol 200033361–370. [DOI] [PubMed] [Google Scholar]
- 19.den Dunnen J T, Antonarakis S E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 2000157–12. [DOI] [PubMed] [Google Scholar]
- 20.den Dunnen J T, Paalman M H. Standardizing mutation nomenclature: why bother? Hum Mutat 200322181–182. [DOI] [PubMed] [Google Scholar]
- 21.Nalla V K, Rogan P K. Automated splicing mutation analysis by information theory. Hum Mutat 200525334–342. [DOI] [PubMed] [Google Scholar]
- 22.Black D L. Mechanisms of alternative pre‐messenger RNA splicing. Ann Rev Biochem 200372291–336. [DOI] [PubMed] [Google Scholar]
- 23.Cartegni L, Chew S L, Krainer A R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 20023285–298. [DOI] [PubMed] [Google Scholar]
- 24.Holbrook J A, Neu‐Yilik G, Hentze M W, Kulozik A E. Nonsense‐mediated decay approaches the clinic. Nat Genet 200436801–808. [DOI] [PubMed] [Google Scholar]
- 25.Bjorkegren J, Beigneux A, Bergo M O, Maher J J, Young S G. Blocking the secretion of hepatic very low density lipoproteins renders the liver more susceptible to toxin‐induce injury. J Biol Chem 20022775476–5483. [DOI] [PubMed] [Google Scholar]