Abstract
Background
Grey zone or intermediate alleles are one of the three recognised classes of the X‐linked fragile X mental retardation 1 (FMR1) gene showing intergenerational instability. These classes are defined according to the number of CGG repeats in the FMR1 5′‐untranslated region. Although large CGG expansions (>200 repeats) cause a neurodevelopmental anomaly through silencing of the gene, resulting in a deficit of FMR1 specific protein, smaller expansions (approximately 55–200 repeats) are associated with an increased transcription and late‐onset specific phenotypes. Those alleles with a CGG repeat number ranging between approximately 41 and 55 are relatively poorly defined with regard to both transcriptional and translational activity, and also potential phenotypic effects.
Methods and results
Based on a sample of 33 males carrying FMR1 alleles within the grey zone range, defined here as 41–60 CGGs, we show an increased transcriptional activity relative to that seen in common alleles (5–40 CGGS). This is the first study to report a significant relationship between FMR1 mRNA levels and CGG repeat number within the grey zone range (p<0.001). From a piecewise linear regression model, the threshold for onset of the increase in mRNA levels as a function of CGG repeat size has been determined at approximately 39 repeats (standard error (SE) 3.24), and that for the reduction in the rate of this increase at approximately 54 repeats (SE 4.27).
Conclusions
The ambiguities associated with the definition and transcription dynamics of the FMR1 gene within the grey zone range are dealt with. There may be specific phenotypes associated with the toxic “gain‐of‐function” effect of raised mRNA.
There are three recognised allelic classes of expanded CGG trinucleotide repeat in the fragile X mental retardation 1 (FMR1) gene. The full mutation, (>200 repeats), is associated with a severe developmental defect defined as fragile X syndrome. This defect is caused by transcriptional silencing of the FMR1 gene, leading to a gross deficit of a specific protein, fragile X mental retardation protein (FMRP), and subsequent synaptic abnormality.1,2 The premutations, ranging from approximately 55 to 200 repeats3 and unstable when transmitted from mothers to offspring4 may cause slight depletion in FMRP levels, which is proportional to the size of CGG repeat,5,6,7 and elevation of mRNA levels.8 Apart from some behavioural and learning problems associated with lowered FMRP levels,9 phenotypic involvement in premutation carriers comprise premature ovarian failure (POF) in females,10 or late‐onset neurodegeneration manifested as tremor/ataxia, predominantly in males.11,12
The third class of intermediate or grey zone alleles is poorly defined, with the range of 45–54 CGG repeats recommended in the guidelines from the American College of Medical Genetics, which is based on the claim that alleles below this size showed “no meiotic or mitotic instability”.3 However, both boundaries for the grey zone range have varied among studies, extending from 34 or 35 CGG repeats for the lower boundary 13,14 to 58/60 repeats for the upper boundary 15,16,17,18 Data on the molecular phenotypes of these grey zone alleles are scarce. The earliest report of increased transcription was based on two grey zone carriers.6 A later study, based on human cell lines transfected with the FMR1 5′‐untranslated region containing CGG repeat lengths ranging in size from 0 to 99 repeats and a downstream reporter (luciferase) gene, showed an increase in transcription levels for constructs possessing either premutation or grey zone alleles.19 In contrast, the most recent study including both male and female grey zone carriers having 41–60 CGG repeats found neither a significant increase in transcript levels nor the relationship of these levels with the number of CGG repeats.17
The aim of this study was to determine the levels of the FMR1 gene transcript, and the relationship of these levels with the CGG repeat size, in grey zone carriers identified in our 2004 fragile X survey of schoolchildren in Tasmania, Australia.16 In the present study, grey zone alleles are defined as those ranging in size from 41 to 60 CGG repeats, which allows direct comparison with the findings of a similar study.17 We also performed the same analysis on alleles ranging in size from 41 to 54 repeats. We report a significant elevation of mRNA within the grey zone allele range, compared with these levels in common‐size alleles, and a significant association between the levels of FMR1 mRNA and CGG repeat size in grey zone alleles. Moreover, based on a regression spanning CGG repeat sizes ranging from common to premutation, we suggest a more accurate definition of the onset, as well as the dynamics, of this relationship across the smaller expansion allele categories.
Materials and methods
Sample
The study was approved by the human ethics committees of all the relevant institutions. The carriers of grey zone and premutation aged 5–22 years were Caucasian males of European descent living in Australia. All 33 grey zone carriers (41–60 CGGs) were identified in our screening study of 1253 males attending both government and non‐government schools throughout Tasmania.16 Twenty subjects were identified from among the special educational needs (SEN) students attending normal schools, 10 were identified from among the non‐SEN students, and three were brothers of SEN students previously identified as grey zone carriers. Moreover, 12 carriers of the premutation were identified through cascade testing of the Tasmanian and Victorian fragile X families initially ascertained through clinical admissions of fragile X survey full mutation probands. Five normal controls (with CGGs ranging from 21 to 39) were recruited from among the brothers of grey zone male carriers identified in the Tasmanian survey, and 15 (with the same CGG range) came from a sample anonymously tested at the Tassone Laboratory, University of California at Davis, California, USA. The size of the CGG repeat and the level of FMR1 transcript (mRNA) were assessed for all individuals included in the study in Tassone Laboratory, San Leandro, California, USA.
Molecular assays
DNA analysis
Genomic DNA was isolated from peripheral blood leucocytes (5 ml of whole blood) using standard methods (Puregene Kit; Gentra, Minneapolis, Minnesota, USA). For Southern analysis, 5–10 µg of isolated DNA was digested by EcoRI and NruI endonucleases, digested samples were separated on a 0.8% agarose/TRIS acetate gel. The FMR1 genomic probe StB12.3, labelled with Dig‐11‐dUTP by polymerase chain reaction (PCR; PCR dig synthesis Kit, Roche Diagnostics, Indianapolis, Indiana, USA), was used as described earlier.20
Genomic DNA was also amplified by PCR, using primers c and f, as described in Saluto et al.21 PCR products were separated on 6% denaturing polyacrylamide gels. A dig‐end labelled oligonucleotide, (CGG)10, was used as a probe. Analysis and calculation of repeat size for Southern and PCR analysis were carried out using an Alpha Innotech FluorChem 8800 Image Detection System (San Leandro, California, USA).
FMR1 mRNA levels
Total RNA was isolated from 3 ml of peripheral blood leucocytes using Tempus Blood RNA tubes (Applied Biosystems, Foster City, California, USA). Reverse transcriptase reactions and quantitative (fluorescence) RT‐PCR measurements were carried out as described earlier.8 All quantifications of FMR1 mRNA were performed using a 7900 Sequence detector (Applied Biosystems) using primers and probes as described by Tassone et al.8 mRNA levels were scored relative to the reference glucuronidase gene, and were standardised in relation to the normal population mean of 1.26 (standard error (SE) 0.04).
All quantifications of FMR1 mRNA were repeated for three different RNA concentrations (500, 250, and 125 µg) and incorporated standards for each determination to compensate for any changes in reaction efficiency. For each sample (whole‐cell RNA derived from leucocytes) and each amplicon, quantitative PCR reactions were performed in duplicate for each starting total RNA concentration, for a total of 12 reactions. An additional 12 reactions were run using a standard lymphoblastoid line as a secondary control for a total of 24 reactions per sample analysed as described by Tassone et al.8 SEs for the outcome of repeated (raw) measures in each individual ranged from 0.01 to 0.21, with most values <0.10.
Statistical analysis
The SEs of the mRNA were not significantly related to the number of CGG repeats over the region up to 80 repeats that contains the grey zone and the lower end of premutation, but was significant for the full range. To stabilise the variance, the mRNA levels were transformed by taking the natural logarithm as described in Allen et al.17 Comparison of mRNA levels between the grey zone and normal samples was carried out using the non‐parametric Mann–Whitney U test and t test, for raw and transformed data, respectively.
This relationship between CGG repeat number and mRNA levels was initially explored by a non‐parametric local regression.22 This procedure required no parametric assumptions about the relationship between the number of CGG repeats and the mRNA levels. Instead, it estimates the relationship for each CGG value by computing a weighted moving average of the mRNA levels over neighbouring CGG values. The weights and the proportion of data included as neighbours were determined by cross validation. The moving average over the nearest 70% of the data to each point gave the best fit to the data. The initial analysis detected three positive linear curves, with two distinct breakpoints separating them, one at approximately 35 repeats and the other at approximately 55 repeats.
To locate these breakpoints more precisely, a piecewise linear model23 was applied to the data. This model fitted a constant mean to normal individuals below the first threshold θ1, allowed a slope for grey zone individuals between θ1 and the second threshold θ2 and another slope for premutation individuals above θ2, (table 1). The significance of the breakpoints was assessed, as suggested in Muggeo,23 by testing for differences in the slopes using the Wald statistic. We also applied ordinary linear regression models to analyse both data in the grey zone range only, and the whole data, in order to compare the fit of these models with piecewise linear models. The adequacy of regression models was assessed by examining a plot of residuals versus fitted values, and a Q–Q normality plot of residuals. All analyses were carried out using the publicly available R statistical computing package, V.1.9.1,24 where package “locfit” was used for the local regression, package “segmented” for the piecewise linear regression and package “base” for other analyses.
Table 1 Selected results of fitting piecewise linear regression model to the data from all three allelic categories: normal, grey zone and premutation, with β1 = 0 and thresholds at θ1 and θ2.
| Category | Model for mean | Parameters of interest | Estimate | SE | 95% CI* |
|---|---|---|---|---|---|
| Normal | α | α | 0.194 | 0.047 | (0.106 to 0.289) |
| Grey zone | α−β2θ1+β2xj | β2 | 0.037 | 0.013 | (0.011 to 0.063) |
| Premutation | α−β2θ1−β3θ2+(β2+β3)xj | β2 + β3 | 0.008 | 0.002 | (0.0040 to 0.012) |
*Wald‐based confidence intervals.
Results
The results in tables 1–3 and in figs 1 and 2 are presented for grey zone alleles, defined as having 41–60 CGG repeats.
Table 2 Summary statistics for mRNA levels in normal and grey zone samples for raw data, log‐transformed data.
| Category | CGG range | Mean | SD | Min | Max | |
|---|---|---|---|---|---|---|
| Normal | 17–39 repeats | Raw | 1.233 | 0.221 | 0.850 | 1.590 |
| (n 20) | Transformed | 0.194 | 0.184 | −0.163 | 0.464 | |
| GZ | 41–60 repeats | Raw | 1.765 | 0.418 | 0.960 | 2.620 |
| (n 33) | Transformed | 0.539 | 0.249 | −0.041 | 0.963 |
GZ, grey zone.
p value <0.001 for comparison between grey zone and normals using two‐sample Mann–Whitney U test (for raw data) and t test (for transformed data).
Table 3 Results of fitting simple linear regression model to data on mRNA levels by CGG repeat sizes ranging from 41 to 60.
| Parameter | Estimate | SE | p Value |
|---|---|---|---|
| α | −1.065 | 0.342 | 0.004 |
| β | 0.033 | 0.007 | <0.001 |
n = 33, R2 = 0.417.
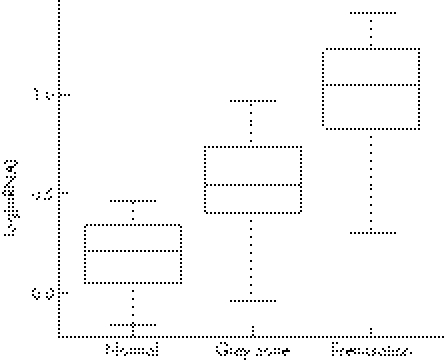
Figure 1 Box plots showing distributions of the log‐transformed values of mRNA levels in three fragile X mental retardation 1 allele categories: normal (17–39 CGG repeats), grey zone (41–60 CGG repeats) and premutation (61–140 CGG repeats).
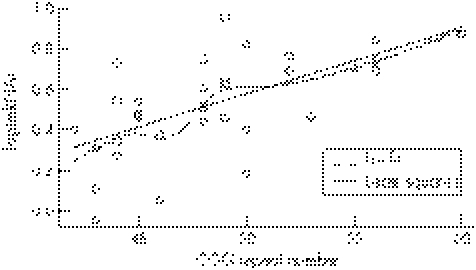
Figure 2 Non‐parametric local fit (Locfit) and least squares fit to the log‐transformed mRNA levels versus CGG repeat numbers for grey zone data. For the least squares, the fitted model is Yj = −1.065 + 0.0329Xj and R2 = 0.4173. p value for the slope <0.001.
mRNA levels
The first question dealt with was whether or not FMR1 mRNA levels are increased in grey zone carriers compared with carriers of normal alleles. Summary statistics for the raw and log‐transformed mRNA levels (table 2) show increased levels in the grey zone compared with the normal sample, the difference being highly significant (p<0.001 for both transformed and non‐transformed values, two‐tailed test). Significant increase (p<0.001) in the levels of mRNA was also found in 28 carriers of grey zone alleles within the 41–54‐repeat range compared with normal alleles (data not shown). Additionally, box plots in fig 1 show that the mRNA levels in the grey zone sample lie clearly between those in the normal and premutation samples. Similar results were obtained for grey zone alleles within the 41–54‐repeat range (data not shown).
Relationship between length of CGG repeat and mRNA levels
Table 2 shows the results of fitting simple linear regression to data on mRNA levels by CGG repeat size in the grey zone range, with the p value for the slope (β) = 0.001. The regression slope (β) was similar for the range 41–54, with the p value <0.006 (data not shown). In fig 2, both the local regression (broken line) and ordinary least square linear regression (solid line) fitted values for both these measures are plotted. The local regression fit confirms the linear model, although there is some deviation from linearity in the local regression that can be attributed to the low number of observations in the range of 45–50 CGG repeats.
Fitted local regression (broken line) of the log‐transformed mRNA levels on the number of CGGs for the total sample, spanning repeat sizes from normal to premutation (fig 3), shows that mRNA levels are unrelated to CGG repeat size within the normal range, but that there is a steep increase within the grey zone range slowing down within the premutation range.
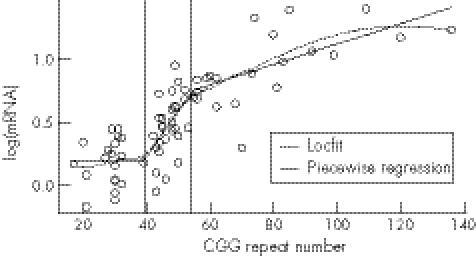
Figure 3 Non‐parametric local fit (Locfit) and piecewise regression fit to the log‐transformed mRNA levels versus CGG repeat numbers for data from all three allelic categories: normal, grey zone and premutation.
Initial analysis of the piecewise linear regression model with two breakpoints confirmed that the regression for the normal sample was not significant (β1 = 0.005; p value = 0.571). The model was therefore re‐fitted using a constant for this line segment. This model gave a better fit to the whole data than either the linear or quadratic ordinary regression models. The fitted piecewise regression of mRNA levels on CGG repeats is plotted in fig 3 (solid line), with vertical lines showing the estimated breakpoints. The first threshold is at approximately 39 CGG repeats (θ1 = 39.16; SE = 3.236; and 95% confidence interval (CI) 32.82 to 45.50), and the second is at approximately 54 repeats (θ2 = 54.31; SE = 4.274; and 95% CI 45.93 to 62.69). Table 3 shows the parameter estimates for all three segments of the regression line defined by these breakpoints. p Values for the difference‐in‐slope parameters were both <0.05 (β2 = 0.037, p = 0.005 and β3 = −0.029, p = 0.003), implying that both breakpoints were significant.
Discussion
This study is the first to report significantly raised levels of mRNA in a sample of males carrying intermediate size, or grey zone, FMR1 alleles, compared with the carriers of normal alleles. This finding deals with the controversy concerning the functioning of the slightly expanded FMR1 alleles by showing that transcription is indeed disturbed in a significant proportion of these alleles. Earlier studies have shown that the raised mRNA levels reflect increased transcriptional activity rather than an increased mRNA stability.8,19 Using the transcription inhibitor actinomycin D, it was shown that the elevation of the mRNA transcript cannot be explained by its increased stability.8 Moreover, direct evidence for an increased transcription rate has recently been obtained using a nuclear run‐on experiment.25
We also show that there is a relationship of these levels with the size of CGG repeat in the grey zone class, and that the sharp increase in mRNA levels corresponding to the number of CGG repeats seen in the grey zone range is significantly reduced in the premutation range. Our data show a statistically significant trend for mRNA level to be a linear function of the repeat size in the grey zone sample, but there is also a considerable variation in this sample, and an overlap with the normal range. The onset of the transcription increase indicated by our data is at approximately 39 repeats; however, more observations, especially at the upper end of the “normal” alleles, are required to estimate this threshold more precisely, and we do not recommend, at this stage, its use as the sole criterion of classification of small expansion alleles.
Our results confirm the findings from two earlier studies,6,19 but are discrepant with those from the study of Allen et al,17 in which no significant relationship was detected between mRNA and CGG repeat number in grey zone alleles of identical range. The possible reasons for this discrepancy include a smaller grey zone sample of 21 males of unspecified composition (compared with 33 individuals of European descent in this study) and/or different procedures used to assess mRNA levels. The procedures involved in running each individual test in the present study ensured high reliability and minimised the inherent variation. It is less likely, however, that the discrepancy reflects difference in an ascertainment between the two studies. Although more than half of the individuals in our grey zone sample were identified in a sample pre‐selected for SEN, mRNA level, and not the behavioural phenotype, was the primary trait in this study. Furthermore, we found no correlation between any cognitive/behavioural scores and either mRNA or CGG repeat size in this sample (own unpublished data).
Based on the discovery of late‐onset tremor/ataxia in some males carrying a premutation gene and showing elevation of mRNA, a toxic “gain‐of‐function” effect of excessive FMR1 mRNA on the brain was considered.12,26,27. It seems that the POF occurring in a significant proportion of female premutation carriers may also be attributed to a similar effect on ovarian cells.10 If indeed raised levels of FMR1 mRNA cause late‐onset abnormal conditions in premutation carriers, we can speculate that the raised levels observed in carriers of grey zone alleles, even if more modest, could have a similar effect. Such an assumption is supported by a significant association recently reported, in population‐based studies, between POF and grey zone alleles.14,15 Taken together with the present results, it seems that screening studies of both POF and late‐onset neurodegenerative conditions with tremor/ataxia should consider the intermediate, as well as premutation, FMR1 allele sizes. The effect of grey zone on neurodevelopment is, however, still controversial, with some studies showing a noticeable increase of grey zone carriers among SEN children,18,28 but others failing to do so.13,16,29,30
Our results also deal with the ambiguity currently associated with the definition of grey zone alleles. The threshold for the onset of a progressive increase in FMR1 transcription was determined to be approximately 39 CGGs. Although more data are required to narrow the error margins, it seems that the lower bound for the grey zone currently recommended at 453 may have to be revisited. As for the upper bound, our data suggest a continuous scale of involvement, rather than the clear‐cut separation of grey zone from premutation that has been recommended.3
The present data also showed that the rate of increase in mRNA levels relative to the number of CGGs tends to slow in the premutation range, an observation that appears at variance with the hypothesis that slight FMRP deficit in premutation5,6 leads to the transcriptional enhancement.7,8 However, the results from a more recent study indicate that this enhancement may result from a direct (cis) effect of the CGG element.19
In conclusion, our results, in combination with some phenotypic effects of grey zone alleles reported earlier, on the one hand, and the transmission instability of these alleles,31 on the other, suggest that premutation–grey zone differences are quantitative rather than qualitative.
It is recommended, however, that our findings be verified on a larger, population‐based, sample of grey zone carriers, and that future studies cover the whole range of both grey zone and premutation alleles, to allow clarification of the demarcation lines within a broad category of small expansions.
Acknowledgements
We thank the study participants and their families for their contribution. We also thank Ms Katherine Heading for maintaining contact with the families of participants, and for organising blood collection from participants and their relatives. We also thank Ms Alison Jacob of the Department of Education Tasmania for support and help in the identification of grey zone carriers in the original survey of Tasmanian schoolchildren.
Abbreviations
FMR1 - fragile X mental retardation 1
FMRP - fragile X mental retardation protein
PCR - polymerase chain reaction
POF - premature ovarian failure
SEN - special educational needs
Footnotes
Funding: This study was supported by the National Institute of Child Health and Human Development grant HD 36071 to RJH and DZL, and the National Health and Medical Research Council of Australia Project Grant No 330400 to DZL, RMH, and FT.
Competing interests: None declared.
References
- 1.Pieretti M, Zhang F, Fu Y H, Warren S T, Oostra B A, Caskey C T, Nelson D L. Absence of expression of the FMR1 gene in the fragile X syndrome. Cell 199166817–822. [DOI] [PubMed] [Google Scholar]
- 2.Irwin S A, Galvez R, Greenough W T. Dendritic spine structural anomalies in fragile X mental retardation syndrome. Cereb Cortex 2000101038–1044. [DOI] [PubMed] [Google Scholar]
- 3.Maddalena A, Richards C S, McGinnis M J, Brothman A, Desnick R J, Grier R E, Hirsch B, Jacky P, McDowell G, Popovich B, Watson M, Wolff D J. Technical standards and guidelines for fragile X: the first series of disease‐specific supplements to the Standards and Guidelines for Clinical Genetic Laboratories of the White American College of Medical Genetics. Quality Assurance Subcommittee of the Laboratory Practice Committee. Genet Med 20013200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y H, Kuhl D P A, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J, Holden J J, Fenwick R G, Warren S T. Variation of the CGG repeat X site results in genetic instability. Resolution of Sherman paradox. Cell 1991671047–1058. [DOI] [PubMed] [Google Scholar]
- 5.Tassone F, Hagerman R J, Taylor A K, Mills J B, Wood S, Gane L, Hagerman P J. Clinical involvement and protein expression in individuals with the FMR1 premutation. Am J Med Genet 200091144–152. [DOI] [PubMed] [Google Scholar]
- 6.Kenneson A, Zhang F, Hagedorn C H, Warren S T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate‐length and premutation carriers. Hum Mol Genet 2001101449–1454. [DOI] [PubMed] [Google Scholar]
- 7.Primerano B, Tassone F, Hagerman R J, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in Fragile X patients with premutations. RNA 200281482–1488. [PMC free article] [PubMed] [Google Scholar]
- 8.Tassone F, Hagerman R J, Taylor A K, Gane L W, Godfrey T E, Hagerman P J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile X syndrome. Am J Hum Genet 2000666–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loesch D Z, Huggins R M, Hagerman R J. (2004) Phenotypic variation and FMRP levels in fragile X. Ment Retard Dev Disabil Res Rev 20041031–41. [DOI] [PubMed] [Google Scholar]
- 10.Allingham‐Hawkins D J, Babul‐Hirji R, Chityat D, Holden J J A, Yang K T, Lee C, Hudson R, Gorwill H, Nolin S L, Glicksman A, Jenkins E C, Brown W T, Howard‐Peebles P N, Becchi C, Cummings E, Fallon L, Seitz S, Black S H, Vianna‐Morgante A M, Costa S S, Otto P A, Mingroni‐Netto R C, Murray A, Webb J, Vieri F. Fragile X premutation is a significant risk factor for premature ovarian failure: the international collaborative POF in fragile X study‐preliminary data. Am J Med Genet 199983322–325. [PMC free article] [PubMed] [Google Scholar]
- 11.Hagerman R J, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman P J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology 200157127–130. [DOI] [PubMed] [Google Scholar]
- 12.Loesch D Z, Churchyard A, Brotchie P, Marot M, Tassone F. Evidence for, and a spectrum of, neurological involvement in fragile X premutation: FXTAS and beyond. Clin Genet 200567412–417. [DOI] [PubMed] [Google Scholar]
- 13.Patsalis P C, Sismani C, Hettinger J A, Holden J J, Lawson J S, Chalifoux M, Wing M, Walker M, Leggo J. Frequencies of ‘grey zone' and premutation‐size FMR1 CGG‐repeat alleles in patients with developmental disability in Cyprus and Canada. Am J Med Genet 199984195–197. [PubMed] [Google Scholar]
- 14.Bretherick K L, Fluker M R, Robinson W P. FMR1 repeat sizes in the gray zone and high end of the normal range are associated with premature ovarian failure. Hum Genet 2005117376–382. [DOI] [PubMed] [Google Scholar]
- 15.Bodega B, Bione S, Dalpra L, Toniolo D, Ornaghi F, Vegetti W, Ginelli E, Marozzi A. Influence of intermediate and uninterrupted FMR1 CGG expansions in premature ovarian failure manifestation. Hum Reprod 200621952–957. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell R J, Holden J J A, Zhang C, Curlis Y, Slater H R, Burgess T, Kirkby K C, Carmichael A, Heading K D, Loesch D Z. FMR1 (fragile X) alleles in Tasmania: a screening study of the special educational needs (SEN) population. Clin Genet 20056738–46. [DOI] [PubMed] [Google Scholar]
- 17.Allen E G, He W, Yadav‐Shah M, Sherman S L. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum Genet 2004114439–447. [DOI] [PubMed] [Google Scholar]
- 18.Youings S A, Murray A, Dennis N, Ennis S, Lewis C, McKechnie N, Pound M, Macpherson J N, Dennis N R, Morton N E, Jacobs P A. FRAXA and FRAXE: the results of a five year study. J Med Genet 200037415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L ‐ S, Tassone F, Sahota P, Hagerman P J. The (CGG)n repeat element within the 5' untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum Mol Genet 2003123067–3074. [DOI] [PubMed] [Google Scholar]
- 20.Tassone F, Hagerman R J, Garcia‐Arocena D, Khandjian D, Greco C M, Hagerman P J. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J Med Genet 200441e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saluto A, Brussino A, Tassone F, Arduino C, Cagnoli C, Pappi P, Hagerman P, Migone N, Brusco A. An enhanced polymerase chain reaction assay to detect pre‐and full mutation alleles of the fragile X mental retardation 1 gene. J Mol Diagn 20055605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loader C.Local Regression and Likelihood. New York: Springer‐Verlag, 1999
- 23.Muggeo V M R. Estimating regression models with unknown break‐points. Stat Med 2003223055–3071. [DOI] [PubMed] [Google Scholar]
- 24.R Development Core Team R. A language and environment for statistical computing. Vienna, Austria: R Foundation for statistical computing, 2004, http://www.R‐project.org (accessed 27 Oct 2006)
- 25.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman P J. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA 2007 (in press) [DOI] [PMC free article] [PubMed]
- 26.Tassone F, Iwahashi C, Hagerman P J. FMR1 RNA within the intranuclear inclusions of fragile X‐associated tremor/ataxia syndrome (FXTAS). RNA Biol 20041103–105. [DOI] [PubMed] [Google Scholar]
- 27.Jin P, Zarnescu D C, Zhang F, Pearson C E, Lucchesi J C, Moses K, Warren S T. RNA‐mediated neurodegeration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 200339739–747. [DOI] [PubMed] [Google Scholar]
- 28.Mazzocco M M, Sonna N L, Teisl J T, Pinit A, Shapiro B K, Shah N, Reiss A L. The FMR1 and FMR2 mutations are not common etiologies of academic difficulty among school‐age children. J Dev Behav Pediatr 199718392–398. [DOI] [PubMed] [Google Scholar]
- 29.Haddad L A, Aguiar M J B, Costa S S, Mingroni‐Netto R C, Vianna‐Morgante A M, Pena S D. Fully mutated and grey zone FRAXA alleles in Brazilian mentally retarded boys. Am J Med Genet 199984198–201. [DOI] [PubMed] [Google Scholar]
- 30.Crawford D C, Meadows K L, Newman J L, Taft L F, Pettay D L, Gold L B, Hersey S J, Hinkle E F, Stanfield M L, Holmgreen P, Yeargin‐Allsop M, Boyle C, Sherman S L. Prevalence and phenotype consequence of FRAXA and FRAXE alleles in a large ethnically, diverse, special education‐needs population. Am J Hum Genet 199964495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan A K, Crawford D C, Scott E H, Leslie M L, Sherman S L. Paternally transmitted FMR1 alleles are less stable than maternally transmitted alleles in the common and intermediate size range. Am J Hum Genet 2002701532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]