Abstract
Frequent overexpression of epidermal growth factor receptor (EGFR) in non‐small‐cell lung cancer (NSCLC) makes EGFR a new therapeutic target. Two specific EGFR tyrosine kinase inhibitors, gefitinib (ZD1839, Iressa) and erlotinib (OSI‐774, Tarceva), have been developed and approved by the US Food and Drug Administration for second‐line and third‐line treatment of advanced NSCLC. Clinical trials have shown considerable variability in the response rate between different patients with NSCLC, which led to the discovery of somatic EGFR‐activating mutations. This brief review summarises the discovery and functional consequences of the mutations, their clinicopathological features and significant implications in the treatment and prognosis of NSCLC.
Lung cancer is one of the most common human cancers and the leading cause of death due to cancer worldwide.1 It claims more lives than colorectal, breast and prostate cancers combined. Fewer than 15% of patients can be cured and have a >5 year survival rate, as the disease is often in an advanced stage at the time of diagnosis and chemotherapy/radiotherapy cannot cure the advanced disease.2 Lung cancer is generally classified into two basic types, small‐cell lung cancer and non‐small‐cell lung cancer (NSCLC). NSCLC accounts for approximately 85% of cases and can be further divided into squamous‐cell carcinoma, adenocarcinoma and large‐cell carcinoma.3 In recent years, adenocarcinoma has replaced squamous‐cell carcinoma as the most common histological subtype of NSCLC in the US and many other parts of the world.4,5
The pathogenesis of lung cancer is yet to be fully understood. Multiple environmental factors are implicated in lung carcinogenesis, such as outdoor and indoor pollution, diet and bacterial and viral infections.6 Cigarette smoke has been widely accepted as the major cause of lung cancer and a linear dose‐response relationship has been established between risk of lung cancer and the amount of cigarettes consumed.7,8,9 The occurrence of lung cancer is also closely associated with the accumulation of multiple genetic and/or epigenetic changes.10,11,12 Better understanding of molecular mechanisms underlying the pathogenesis of lung cancer would provide pivotal guidance for the treatment and prevention of cancer.
EGFRs and lung cancer
Human tumours often express high levels of epidermal growth factor (EGF)‐related receptors, which include EGFR/HER1, c‐erbB2/HER2, c‐erbB3/HER3 and c‐erbB4/HER4 (table 1).16,17,18,19,20 All these receptors share a common extracellular ligand‐binding domain, a hydrophobic transmembrane domain and a multifunctional intracellular domain that has an ATP‐binding site and tyrosine kinase activity (HER3, however, does not have tyrosine kinase activity). Receptor activation is generally initiated by the binding of EGF‐related ligands and receptor homodimerisation or heterodimerisation, which activate the intrinsic tyrosine kinase activity of the receptors and lead to autophosphorylation of the tyrosine residues. These residues further activate downstream signalling cascades, such as the Ras‐Raf‐MAP‐kinase, PI3K‐Akt and STAT pathways, which have strong regulatory effects on cell proliferation, differentiation, survival and migration.21,22,23
Table 1 The family of human epidermal growth factor (EGF)‐related receptors.
| Gene location | Molecular weight | Protein distribution | |
|---|---|---|---|
| c‐erb B‐1/EGFR | 7p12.3–p12.1 | 170 kDa | A large variety of cell type or tissues except haemopoietic cells13 |
| c‐erb B‐2/HER‐2 | 17q21.1 | 185 kDa | A number of human secretory epithelial tissues14 |
| c‐erb B‐3/HER‐3 | 12q13 | 160 kDa | Several human tumour cell lines15 |
| c‐erb B‐4/HER‐4 | 2q33.3–q34 | 180 kDa | Lining epithelia of the gastrointestinal, urinary, reproductive, breast cancer cell lines, and normal skeletal muscle, heart, pituitary, brain and cerebellum16 |
EGFR is generally expressed at a low level in a wide variety of normal tissues. Excessive expression or activation of EGFR is able to induce malignant transformation.24 Overexpression of EGFR has been observed in 40–80% of NSCLCs,20,25 and it is often associated with aggressive clinical behaviours, such as advanced stage, increased metastatic rate, higher tumour proliferation rate and poor prognosis.26,27,28,29 Other than overexpression, a mutant form of EGFR (EGFRvIII) with constitutive tyrosine kinase activity has been identified in NSCLC30,31,32 and is implicated in lung tumorigenesis.33,34
Given that EGFR is often overexpressed in NSCLC and given its potential role in lung carcinogenesis, EGFR has been considered a rational target molecule for the treatment of NSCLC. Specific EGFR tyrosine kinase inhibitors (EGFR‐TKIs), gefitinib and erlotinib, have been approved by the US Food and Drug Administration as monotherapy treatment for advanced or metastatic NSCLC.35,36 Both agents adversely compete with ATP for the critical ATP‐binding site located in the intracellular domain and inhibit receptor phosphorylation.
Gefitinib and EGFR mutations
The effectiveness of gefitinib has been evaluated by two randomised and double‐blinded Phase II clinical trials in patients with advanced NSCLC.37,38 The results suggest that gefitinib is a well‐tolerated oral EGFR‐TKI. It has meaningful antitumour activity and brings about considerable improvement in cancer‐related symptoms in certain subgroups of patients (approximately 10–19%). Females, non‐smokers, Japanese people and patients with lung adenocarcinoma generally have a higher response rate than males, smokers, people of European origin and patients with other histological types of NSCLC.35,37,38 To determine whether somatic mutations in the EGFR gene play a causal role in response to TKI treatment, two research groups have systematically sequenced all 28 exons of EGFR and identified several important activating mutations that show striking correlation with gefitinib response.39,40 This discovery has been claimed as the most significant molecular event in lung cancer.41 It has greatly stimulated research in this area worldwide, and a number of other novel mutations have been identified (table 2).
Table 2 Mutations identified in exons 18–21 of the EGFR gene (RefSeq NM_005228)*.
| Sequence change | Amino acid change | Reference |
|---|---|---|
| Exon 18 | ||
| 2126A→T | E709V | 42, 43 |
| 2126A→C | E709A | 44, 45 |
| 2126A→G | E709G | 44, 45 |
| 2125G→A | E709K | 45 |
| 2155G→A | I715S | 43 |
| 2155 G→T | G719C | 40, 42–47 |
| 2155 G→A | G719S | 39, 43–45 |
| 2156 G→C | G719A | 42–44, 47, 48 |
| 2159 C→T | S720F | 43, 48 |
| Exon 19 | ||
| 2225T→C | V742A | 49 |
| Del (2236–2244)+2245G→C+2248G→C | E746‐R748 del with E749Q, A750P | 47 |
| Del (2235–2249)/del (2236–2250) | E746‐A750 del | 39, 40, 42–50 |
| Del (2235–2236)+ | E746‐A750 del with I, P ins | 48 |
| Del (2242–2248)+2241A→C | ||
| Del (2237–2251)+2252C→T | E746‐A750 del with V ins | 43 |
| Del (2237–2251) | E746‐T751 del with A ins | 44–46, 48, 50 |
| Del (2235–2236)+ | E746‐T751 del with I ins | 43, 48 |
| Del (2239–2252)/ | ||
| Del (2235–2252)+ | ||
| 2254T→A+2255C→T | ||
| Del (2235–2236)+ | E746‐T751del with I, P ins | 48 |
| Del (2242–2248)+2241A→C | ||
| 2237–2238 AA→TC+ | E746‐T751 del with V ins | 43, 49 |
| Del (2239–2253) | ||
| Del (2484–2501) | E746‐S752 del with D ins | 44 |
| Del (2237–2254)+2255C→T | E746‐S752 del with V ins | 45, 48, 50 |
| Del (2239–2247) | L747‐E749 | 46 |
| Del (2239–2247)+2248G→C | L747‐E749 del with P ins | 39, 42, 43, 45 |
| Del (2240–2248)+2239T→C | L747‐T750 del with P ins | 43, 48 |
| Del (2238–2252)/ | L747‐T751 del | 43, 44, 48, 49 |
| Del (2239–2253)/ | ||
| Del (2240–2254) | ||
| Del (2240–2251) | L747‐T751 del with S ins | 40, 46, 48 |
| Del (2239–2256) | L747‐S752 del | 44, 48 |
| Del (2239–2256)+2258 C→A | L747‐S752 del with Q ins | 43 |
| Del (2238–2255)+2237A→T | L747‐S752 del, E746V | 39 |
| Del (2240–2257) | L747‐P753 del with S ins | 39, 40, 42–46, 48–50 |
| Del (2240–2257) | L747S, R748‐P753 del | 47 |
| Del (2254–2277) | S752‐I759 del | 39, 47 |
| 2273A→G | E758G | 49 |
| Exon 20 | ||
| 2308 ins GCCATA | M766‐A767 with AI ins | 47 |
| 2308 ins CCAGCGTGG+ | A767‐S768 with SVA ins | 47 |
| 2310C→T silent | ||
| 2303G→T | S768I | 43–45, 47 |
| Dup (2549–2557) | S768‐D770 dup | 44 |
| 2308–2316 ins GCCAGCGTG | ASV770‐772 ins | 43 |
| 2320–2322 ins CAC | H774 ins | 43 |
| 2317–2222 ins AACCCC+ | NP773‐774 ins, H775Y | 43 |
| 2223 C>T | ||
| 2320–2325 ins CCCCAC | PH774‐775 ins | 43 |
| 2320–2328 ins AACCCCCAC | NPH774‐776 ins | 43 |
| 2326C→T | R776C | 51 |
| 2308 ins (CCAGCGTGG)+ | ins779 ASV+P782R | 48 |
| 2310C→T+ 2315C→G | ||
| 2311 ins (GCGTGGACA)+ | ins780 SVD+P782R | 48 |
| 2315C→G | ||
| 2369 C→T | T790M | 52–55 |
| Exon 21 | ||
| 2743T→G | L833V | 44 |
| 2750A→T | H835L | 44 |
| 2758C→G | L838V | 44 |
| 2513T→C | L838P | 49 |
| 2520C→T | A840A | 49 |
| 2527G→A | V843I | 49 |
| 2551G→A | V851I | 49 |
| 2572C→A | L858M | 46 |
| 2573T→G | L858R | 39, 40, 42–50 |
| 2582T→A | L861Q | 40, 42–45 |
| 2593G→A | E856L | 49 |
| 2612C→T | A871V | 49 |
*Only mutations reported with both nucleotide and amino acid changes are summarised. The beginning of the coding sequence is defined as +1.
Genotyping methods
Two important factors affect the detection of somatic EGFR mutations in clinical cancer samples. The first is the availability of the tumour genome. There is no doubt that frozen surgical tumour samples45 and tumour paraffin blocks47 are the best samples for mutation analysis, as they are directly resected from corresponding tumours and can provide sufficient tumour nucleic acids for genotyping. However, a large proportion of patients with NSCLC are not eligible for surgery on diagnosis. Therefore, non‐surgical specimens, such as diagnostic biopsy and effusion drainage, are probably as important as surgical specimens in these patients with advanced cancer. Pleural effusion56 and needle biopsy/aspiration49 have been successfully managed for mutation screening. Asano et al57 even showed the feasibility of detecting EGFR mutations with the use of soluble DNA extracted from pleural fluid.
The second factor affecting mutation detection is the purity of the tumour genome. Usually clinical cancer samples contain a large proportion of normal cells, which make up a strong background of wild‐type alleles and seriously dilute the signal from biologically important somatic mutations. Therefore, the sensitivity of genotyping methods is of great importance for the detection of mutations.
Among a number of reported methods, PCR‐based direct sequencing is the most commonly used.39,40,43,44,47 With the help of cloning technology, even samples presenting difficulty in direct sequencing can be sequenced using primers located on vectors. Moreover, tumour RNA can be used for genotype determination, as all the reported EGFR‐activating mutations are exonic.42,55 However, RNA is usually more difficult to handle than genomic DNA, because of its rapid degradation and limited quantity.
Single‐strand conformation polymorphism (SSCP) assay is another important method used for EGFR mutation screening. SSCP has been considered to be more sensitive than direct sequencing in mutation analysis.58,59 The two large studies performed by Marchetti et al46 and Sonobe et al48 have reported that SSCP assays not only confirmed all the EGFR mutations detected by direct sequencing but also identified additional mutations that were missed in sequencing analysis. Therefore, SSCP assay could be a reliable method for large‐scale diagnostic screening for EGFR mutations in clinical samples.
For detection of known EGFR mutations, a number of methods have been developed, including restriction fragment length polymorphism and length analysis of fluorophore‐labelled PCR products,60 peptide nucleic acid–locked nucleic acid PCR clamp,61 mutant‐allele‐specific amplification62 and mutant‐enriched PCR.57 The restriction fragment length polymorphism–capillary electrophoresis method can not only detect the mutations but also estimate gene amplification based on the relative height of the mutation peak to the germline peak. The peptide nucleic acid–locked nucleic acid PCR clamp, mutant‐allele‐specific amplification and mutant‐enriched PCR have high sensitivity. They are able to distinguish even one EGFR mutant tumour cell in the presence of up to 1000–2000 normal cells.57,61,62
The pattern and functional consequence of EGFR mutations
Three common types of EGFR mutation—in‐frame deletion, insertion and missense mutation—have been identified. Most of the mutations are located in the tyrosine kinase‐coding domain (exons 18–21). Amino acids 746–753 encoded by exon 19 and amino acid 858 encoded by exon 21 are the two mutation hotspots, comprising >80% of the mutations. All the identified mutations are of somatic origin, and not present in the germline genome.
EGFR mutations have been proposed as an early event in lung carcinogenesis. They are not correlated with the classification of tumour stage.46 Well or moderately differentiated tumours have more EGFR mutations than poorly differentiated tumours.48 Some of the mutations can even be detected in respiratory epithelia with normal histology.63 The oncogenic characteristics of EGFR mutants have recently been proved by anchorage‐independent growth and focus formation in transfected cells and tumour formation in immunocompromised mice.64
EGFR mutants (deletion in exon 19 and L858R) are capable of enhancing EGF‐dependent receptor activation (Tyr1068).40 The downstream signalling pathways Akt and STAT are also selectively activated,65,66 and these have an important anti‐apoptotic function. When mutant EGFR expression is suppressed by specific small interfering RNA or when Akt and STAT pathways are blocked by specific inhibitors, rapid and massive apoptosis occurs. A similar event also happens when EGFR‐TKIs are applied to mutant NSCLC cell lines.51,65,66 All these suggest that excessive EGFR signalling plays a critical role in tumorigenesis in patients harbouring an EGFR mutation, and mutant EGFRs drive the growth of cancer cells and maintain their malignant phenotype by the selective activation of Akt and STAT signalling pathways.
EGFR gene amplification is present in approximately 30–40% of patients with NSCLC,67,68 and it is associated with a susceptibility to gefitinib.68,69,70 The association between EGFR mutations (deletion in exon 19 and L858R in exon 21) and an increased copy number of the EGFR gene has been shown in both preclinical66,71 and clinical investigations,50,70,71 which could partially throw light on gefitinib sensitivity in patients with EGFR gene amplification. Genetic instability has been proposed to be the cause of gene amplification and the facilitator of the incidence of EGFR mutations.71
Factors predisposing to EGFR mutations in NSCLC
The distribution of EGFR mutations in NSCLC has been intensively investigated (table 3). In general, EGFR mutations are more common in patients of oriental origin, in females and in patients with a history of never smoking or with adenocarcinoma.73
Table 3 Overview of 10 large studies on EGFR mutations in non‐small‐cell lung cancer (NSCLC).
| Ethnicity | Subjects (n) | Mutation rate (%) | Deletions in exon 19 (%) | L858R (%) | Mutation rate (%) | |||
|---|---|---|---|---|---|---|---|---|
| Non‐smokers vs smokers | Female vs male | Adenocarcinoma vs other NSCLC | ||||||
| Marchetti46 | White | 860 | 5 | 46 | 46 | 20 vs 2 | 19 vs 2 | 10 vs 0 |
| Eberhard47 | White | 228 | 13 | 62 | 24 | 25 vs 12 | 13 vs 13 | 15 vs 11 |
| Cappuzzuo67 | White | 89 | 17 | 53 | 47 | 46 vs 12 | 26 vs 12 | 20 vs 10 |
| Shigematsu43 | White | 158 | 8 | 46 | 39 | 29 vs 3 | 20 vs 1 | 16 vs 1 |
| East Asian | 361 | 30 | 56 vs 14 | 55 vs 19 | 48 vs 3 | |||
| Kosaka55 | Japanese | 277 | 40 | 47 | 44 | 66 vs 22 | 59 vs 26 | 49 vs 2 |
| Sonobe48 | Japanese | 154 | 39 | 57 | 37 | 79 vs 16 | 70 vs 19 | 55 vs 0 |
| Tokumo72 | Japanese | 120 | 32 | 50 | 47 | 69 vs 15 | 57 vs 20 | 45 vs 3 |
| Huang44 | Taiwanese | 101 | 39 | 33 | 51 | 47 vs 18 | 44 vs 32 | 55 vs 3 |
| Sugio62 | Japanese | 469 | 29 | 45 | 55 | 54 vs 19 | 46 vs 21 | 42 vs 1 |
| Yokoyama45 | Japanese | 349 | 29 | 40 | 39 | 54 vs 14 | 54 vs 15 | 42 vs 1 |
Ethnicity
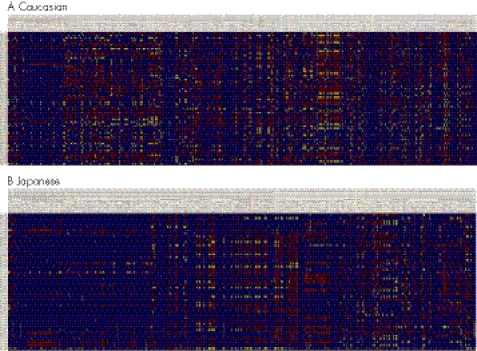
EGFR mutations have significant ethnic variation. The mutation rate is approximately 5–13% among Caucasians,43,46,47 but 30–40% among East Asians (table 3).43,44,55 Although the factors that determine more EGFR mutations in Asians are still an enigma, different genetic backgrounds and living environments could provide informative clues. The international HapMap project (http://www.hapmap.org) has genotyped >300 single‐nucleotide polymorphisms across the EGFR gene in Caucasian, Japanese, Chinese Han and Yoruba populations. Considerable interethnic variations in the prevalence of single‐nucleotide polymorphisms have been observed between Japanese and Caucasian populations (fig 1). It remains unknown if the germline variations are correlated with the occurrence of EGFR somatic mutations. However, the association between germline and somatic variations in upper gastrointestinal familial adenomatous polyposis has been reported.74
Figure 1 Genotype distribution of germline EGFR polymorphisms in Caucasian and Japanese populations. The genotype datasets are downloaded from the HapMap project and displayed in Visual Genotype (VG) format (http://pga.gs.washington.edu). The columns are the polymorphic sites. The rows are the arrays of samples. Blue represents a homozygote for the common allele, red represents a heterozygote (both common and rare allele) and yellow represents a homozygote for the rare allele.
Smoking status and sex
Cigarette smoke is the most significant risk factor for lung cancer. N‐nitrosamines and polycyclic aromatic hydrocarbons, the two major classes of tobacco‐related carcinogens, can produce as many as 100 mutations per cell genome by means of formation of DNA adducts.75 Cigarette smoke, however, is not the mutagen of EGFR, as EGFR mutations are usually more frequent in patients who never smoke or have a low exposure to cigarette smoke (table 3). Risk factors associated with non‐smoking lung cancer, such as pre‐existing lung diseases, a family history of cancer, passive smoking, indoor cooking fumes and occupational exposures76 could affect the occurrence of EGFR mutations. More studies are required to identify the causal mutagens and elucidate the potential mutagenic capability.
EGFR mutations are more frequent in female than in male patients, especially among Asians (table 3). This difference could be correlated with their distinct life styles and smoking habits. Generally, women tend to be non‐smokers or light smokers, and they play a heavier role in housework, such as cooking and cleaning, whereas those men who smoke tend to be heavy smokers and are more often involved in social activities. If the occurrence of EGFR mutations is associated with potential indoor mutagens, there is no doubt that women would have a higher mutation rate than men. On the other hand, female endocrine factors could also play a role in EGFR mutations.
Histology of NSCLC
EGFR mutations are more frequent in lung adenocarcinoma than in other histological types of NSCLC (table 3). As the predominant subtype of NSCLC, adenocarcinoma usually originates from a peripheral airway compartment, either surface epithelium or bronchial mucosal glands.77 It is likely that the specific cellular milieu in these cell types is more susceptible to the influence of mutagens that induce EGFR mutations compared with other cell types.
Clinical significance of EGFR mutations
EGFR mutations have been considered the best predictive marker for response to treatment with EGFR‐TKIs in NSCLC. A number of retrospective studies have indicated that patients with the mutations have higher objective response rates to gefitinib than patients with the wild type (table 4). The first prospective clinical trial has recently been conducted among 75 patients with chemotherapy‐naive advanced NSCLC in Japan. Among 25 patients with EGFR mutations, 16 received gefitinib monotherapy and nine received standard chemotherapy. In agreement with the retrospective studies, the group receiving gefitinib had a 75% response rate to gefitinib.78 Another recent prospective Phase II trial of gefitinib has shown similar results.79
Table 4 EGFR mutations, clinical tyrosine kinase inhibitor response and survival.
| Mutation/EGFR‐TKI responders (%) | ORR* (mutations vs wild type) | TTP* (months; mutations vs wild type) | Median survival (months; mutation vs wild type) | |
|---|---|---|---|---|
| Lynch40 | 8/9 (89) | — | — | — |
| Paez39 | 5/5 (100) | — | — | — |
| Huang44 | 7/9 (78) | — | — | — |
| Cappuzzo67 | 8/12 (67) | 53% vs 5% | 9.9 vs 2.6 (p = 0.02) | 20.8 vs 8.4 (p = 0.09) |
| Eberhard47 | 8/26 (31) | 53% vs 18% | 8 vs 5 (p<0.001) | Not reached vs 10 (p<0.001) |
| Mitsudomi42 | 24/26 (92) | 83% vs 10% | — | Significantly longer (p = 0.005) |
| Taron50 | 16/22 (73) | 94% vs 13% | — | Not reached vs 9.9 (p<0.001) |
| Shih49 | 20/23 (87) | 69% vs 9% | 9 vs. 2.2 (p = 0.001) | 13.9 vs 4.8 (p = 0.013) |
| Takano70 | 32/35 (91) | 82% vs 11% | 12.6 vs 1.7 (p<0.001) | 20.4 vs 6.9 (p<0.001) |
*ORR, objective response rate (complete response or partial response); TTP, time to progression.
—, data are not available.
Sensitivity to gefitinib or erlotinib is also associated with the type of mutation and the existence of additional mutations. Patients with deletion mutation in exon 19 have higher response rates than those with L858R in exon 21.42,80 An additional mutation in exon 22 (K884E) is able to make tumour cells sensitive to gefitinib but resistant to erlotinib.81 Further laboratory and clinical studies are required to elucidate the underlying mechanisms.
Furthermore, EGFR mutations have been considered a favourable prognostic indicator in NSCLC. Patients with the mutations have improved time‐to‐progression and longer overall survival than those with the wild type (table 4).42,49,50,67,70 However, two Japanese studies have found conflicting results, namely that there is no significant difference in survival between the groups with and without the mutations.43,62 This discrepancy could reflect the fact that other prognostic factors besides EGFR mutations also play an important role in survival among patients with NSCLC.
Secondary EGFR mutations and acquired resistance to gefitinib
Gefitinib has brought significant treatment response among patients with NSCLC with EGFR mutations. However, the mean duration of response is generally 6–8 months.37,38 Most of these patients eventually relapse and are resistant to further treatment with EGFR‐TKIs. The acquired resistance is closely associated with the development of a secondary mutation in exon 20, that is, a substitution of methionine for threonine at amino acid position 790 (T790M).52,53 Pao et al52 have reported that three of six patients with acquired resistance to TKIs harbour the T790M mutation in the progressing tumours. T790M is believed to abrogate the binding of TKIs to the ATP‐kinase‐binding pocket and lead to the continued activation of ErbB‐3/PI3K/Akt signalling.53,82 The scenario is analogous to the secondary mutations in BCR‐ABL and KIT that confer acquired resistance to imatinib (Gleevec) in chronic myeloid leukaemia and gastrointestinal stromal tumour.83,84 T790M has been considered to be present only in relapsed tumours and its appearance to be secondary to treatment of TKIs. Nevertheless, rare T790M mutation has been detected in untreated tumour55 and even in germline DNA, and it is associated with family aggregation of NSCLC.54 Therefore, T790M could play a role in lung tumorigenesis.
Association of EGFR mutations with K‐ras and p53 gene mutations
K‐ras is a key molecule in the signalling pathways, which regulate cellular proliferation and transformation. The K‐ras mutation is one of the major genetic changes detected in lung cancer. The mutations are more common in adenocarcinomas, are closely correlated with cigarette smoke and are predominantly the guanine‐to‐thymine transversion at codon 12.85,86 Cigarette smoke has been proposed as the causal mutagen. P53 is another well‐characterised gene in lung cancer. Most of the mutations are located in evolutionally conserved regions, and the mutation spectra are different between smokers and non‐smokers. The relationship between the type of p53 mutations and the histology of lung cancer is still unclear.8788
The correlation between EGFR mutations and K‐ras mutations has been intensively investigated. Generally, EGFR mutations are present only in lung adenocarcinomas which do not harbour K‐ras mutations.43,46,47,48,55,62 This observation might imply that there are different subsets of lung adenocarcinomas, which possess different mutation spectra and causal mutagens. By contrast, mutations in the EGFR gene and the p53 gene happen independently,48,55 but the status of gene–gene interaction and its influence on lung carcinogenesis and treatment response remains unknown.
Summary and future research directions
The discovery of somatic EGFR mutations has been claimed as a big victory of molecular medicine.41 It has had a considerable effect on the treatment of NSCLC. The strong associations between the mutations and gefitinib responsiveness and favourable prognosis provide forceful support for an individual genotype‐based therapeutic strategy. By now, a number of genotyping methods have been developed. However, most of them are technically complicated and need substantial research resources and expertise, which make routine clinical screening difficult and impractical. Developing a set of simple and accurate genotyping methods would be of great importance for translating this bench discovery into clinical application. On the other hand, further basic studies are required to investigate whether other functional consequences are present downstream of EGFR signalling pathways and whether additional predictive biomarkers are available. For patients with acquired resistance to EGFR‐TKIs, monitoring the EGFR mutation status in recurrent tumours is crucial for revealing the molecular mechanisms of drug resistance and developing new generations of TKIs. Finally, more large prospective studies to evaluate the therapeutic and prognostic value of EGFR mutations are required in patients with EGFR‐mutation‐enriched NSCLC. Prospective study is usually superior to retrospective study in bias and confounder control and therefore can provide more accurate assessment of the significance of EGFR mutations in the clinical treatment of NSCLC and of the possibility that EGFR‐TKIs might replace conventional chemotherapy as the preferred antitumour drugs for the subset of patients with NSCLC.
Gealy R, Zhang L, Siegfried JM, et al. Comparison of mutations in the p53 and K‐ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomarkers Prev 1999;8(Pt 1):297–302.
Abbreviations
EGF(R) - epidermal growth factor (receptor)
NSCLC - non‐small cell lung cancer
PCR - polymerase chain reaction
SSCP - single‐strand conformation polymorphism
TKI - tyrosine kinase inhibitor
Footnotes
Competing interests: None.
References
- 1.Parkin D M. Global cancer statistics in the year 2000. Lancet Oncol 20012533–543. [DOI] [PubMed] [Google Scholar]
- 2.Greenlee R T, Hill‐Harmon M B, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin 20015115–36. [DOI] [PubMed] [Google Scholar]
- 3.Brambilla E, Travis W D, Colby T V, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J 2001181059–1068. [DOI] [PubMed] [Google Scholar]
- 4.Travis W D, Travis L B, Devesa S S. Lung cancer. Cancer 199575(Suppl)191–202S. [DOI] [PubMed] [Google Scholar]
- 5.Franceschi S, Bidoli E. The epidemiology of lung cancer. Ann Oncol 199910(Suppl 5)3–6. [DOI] [PubMed] [Google Scholar]
- 6.Lam W K. Lung cancer in Asian women—the environment and genes. Respirology 200510408–417. [DOI] [PubMed] [Google Scholar]
- 7.Doll R, Peto R. Cigarette smoking and bronchial carcinoma: dose and time relationships among regular smokers and lifelong non‐smokers. J Epidemiol Community Health 197832303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akiba S, Hirayama T. Cigarette smoking and cancer mortality risk in Japanese men and women—results from reanalysis of the six‐prefecture cohort study data. Environ Health Perspect 19908719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freund K M, Belanger A J, D'Agostino R B, Kannel W B. The health risks of smoking. The Framingham Study: 34 years of follow‐up. Ann Epidemiol 19933417–424. [DOI] [PubMed] [Google Scholar]
- 10.Sekido Y, Fong K M, Minna J D. Progress in understanding the molecular pathogenesis of human lung cancer. Biochim Biophys Acta 19981378F21–F59. [DOI] [PubMed] [Google Scholar]
- 11.Fong K M, Sekido Y, Minna J D. Molecular pathogenesis of lung cancer. J Thorac Cardiovasc Surg 19991181136–1152. [DOI] [PubMed] [Google Scholar]
- 12.Breuer R H, Postmus P E, Smit E F. Molecular pathology of non‐small‐cell lung cancer. Respiration 200572313–330. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu N, Behzadian M A, Shimizu Y. Genetics of cell surface receptors for bioactive polypeptides: binding of epidermal growth factor is associated with the presence of human chromosome 7 in human‐mouse cell hybrids. Proc Natl Acad Sci USA 1980773600–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coussens L, Yang‐Feng T L, Liao Y C, Chen E, Gray A, McGrath J, Seeburg P H, Libermann T A, Schlessinger J, Francke U, Levinson A, Ullrich A. Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 19852301132–1139. [DOI] [PubMed] [Google Scholar]
- 15.Kraus M H, Issing W, Miki T, Popescu N C, Aaronson S A. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci USA 1989869193–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plowman G D, Culouscou J M, Whitney G S, Green J M, Carlton G W, Foy L, Neubauer M G, Shoyab M. Ligand‐specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci USA 1993901746–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl M I, Carpenter G. Role of growth factors and their receptors in the control of normal cell proliferation and cancer. Clin Physiol Biochem 19875130–139. [PubMed] [Google Scholar]
- 18.Carpenter G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu Rev Biochem 198756881–914. [DOI] [PubMed] [Google Scholar]
- 19.Lemoine N R, Lobresco M, Leung H, Barton C, Hughes C M, Prigent S A, Gullick W J, Kloppel G. The erbB‐3 gene in human pancreatic cancer. J Pathol 1992168269–273. [DOI] [PubMed] [Google Scholar]
- 20.Salomon D S, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor‐related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 199519183–232. [DOI] [PubMed] [Google Scholar]
- 21.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell 2000103211–225. [DOI] [PubMed] [Google Scholar]
- 22.Yarden Y, Sliwkowski M X. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 20012127–137. [DOI] [PubMed] [Google Scholar]
- 23.Scagliotti G V, Selvaggi G, Novello S, Hirsch F R. The biology of epidermal growth factor receptor in lung cancer. Clin Cancer Res 200410(12 Pt 2)4227–32S. [DOI] [PubMed] [Google Scholar]
- 24.Mendelsohn J, Baselga J. The EGF receptor family as targets for cancer therapy. Oncogene 2000196550–6565. [DOI] [PubMed] [Google Scholar]
- 25.Ennis B W, Lippman M E, Dickson R B. The EGF receptor system as a target for antitumor therapy. Cancer Invest 19919553–562. [DOI] [PubMed] [Google Scholar]
- 26.Pavelic K, Banjac Z, Pavelic J, Spaventi S. Evidence for a role of EGF receptor in the progression of human lung carcinoma. Anticancer Res 1993131133–1137. [PubMed] [Google Scholar]
- 27.Ohsaki Y, Tanno S, Fujita Y, Toyoshima E, Fujiuchi S, Nishigaki Y, Ishida S, Nagase A, Miyokawa N, Hirata S, Kikuchi K. Epidermal growth factor receptor expression correlates with poor prognosis in non‐small cell lung cancer patients with p53 overexpression. Oncol Rep 20007603–607. [DOI] [PubMed] [Google Scholar]
- 28.Volm M, Rittgen W, Drings P. Prognostic value of ERBB‐1, VEGF, cyclin A, FOS, JUN and MYC in patients with squamous cell lung carcinomas. Br J Cancer 199877663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicholson R I, Gee J M, Harper M E. EGFR and cancer prognosis. Eur J Cancer 200137(Suppl 4)9–15S. [DOI] [PubMed] [Google Scholar]
- 30.Garcia de Palazzo I E, Adams G P, Sundareshan P, Wong A J, Testa J R, Bigner D D, Weiner L M. Expression of mutated epidermal growth factor receptor by non‐small cell lung carcinomas. Cancer Res 1993533217–3220. [PubMed] [Google Scholar]
- 31.Pedersen M W, Meltorn M, Damstrup L, Poulsen H S. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti‐cancer therapy. Ann Oncol 200112745–760. [DOI] [PubMed] [Google Scholar]
- 32.Chu C T, Everiss K D, Wikstrand C J, Batra S K, Kung H J, Bigner D D. Receptor dimerization is not a factor in the signalling activity of a transforming variant epidermal growth factor receptor (EGFRvIII). Biochem J 1997324(Pt 3)855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamazaki H, Ohba Y, Tamaoki N, Shibuya M. A deletion mutation within the ligand binding domain is responsible for activation of epidermal growth factor receptor gene in human brain tumors. Jpn J Cancer Res 199081773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, Jung B L, McNamara K, Xia H, Glatt K A, Thomas R K, Sasaki H, Horner J W, Eck M, Mitchell A, Sun Y, Al‐Hashem R, Bronson R T, Rabindran S K, Discafani C M, Maher E, Shapiro G I, Meyerson M, Wong K K. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci USA 20061037817–7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen M H, Williams G A, Sridhara R, Chen G, Pazdur R. FDA drug approval summary: gefitinib (ZD1839) (Iressa) tablets. Oncologist 20038303–306. [DOI] [PubMed] [Google Scholar]
- 36.Siegel‐Lakhai W S, Beijnen J H, Schellens J H. Current knowledge and future directions of the selective epidermal growth factor receptor inhibitors erlotinib (Tarceva) and gefitinib (Iressa). Oncologist 200510579–589. [DOI] [PubMed] [Google Scholar]
- 37.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard J Y, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong R P, Baselga J. Multi‐institutional randomized phase II trial of gefitinib for previously treated patients with advanced non‐small‐cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin Oncol 2003212237–2246. [DOI] [PubMed] [Google Scholar]
- 38.Kris M G, Natale R B, Herbst R S, Lynch T J, Jr, Prager D, Belani C P, Schiller J H, Kelly K, Spiridonidis H, Sandler A, Albain K S, Cella D, Wolf M K, Averbuch S D, Ochs J J, Kay A C. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non‐small cell lung cancer: a randomized trial. JAMA 20032902149–2158. [DOI] [PubMed] [Google Scholar]
- 39.Paez J G, Janne P A, Lee J C, Tracy S, Greulich H, Gabriel S, Herman P, Kaye F J, Lindeman N, Boggon T J, Naoki K, Sasaki H, Fujii Y, Eck M J, Sellers W R, Johnson B E, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 20043041497–1500. [DOI] [PubMed] [Google Scholar]
- 40.Lynch T J, Bell D W, Sordella R, Gurubhagavatula S, Okimoto R A, Brannigan B W, Harris P L, Haserlat S M, Supko J G, Haluska F G, Louis D N, Christiani D C, Settleman J, Haber D A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 20043502129–2139. [DOI] [PubMed] [Google Scholar]
- 41.Marx J. Medicine. Why a new cancer drug works well, in some patients. Science 2004304658–659. [DOI] [PubMed] [Google Scholar]
- 42.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, Hatooka S, Shinoda M, Takahashi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non‐small‐cell lung cancer with postoperative recurrence. J Clin Oncol 2005232513–2520. [DOI] [PubMed] [Google Scholar]
- 43.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba, Fong K M, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth J A, Herz J, Minna J D, Gazdar A F. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 200597339–346. [DOI] [PubMed] [Google Scholar]
- 44.Huang S F, Liu H P, Li L H, Ku Y C, Fu Y N, Tsai H Y, Chen Y T, Lin Y F, Chang W C, Kuo H P, Wu Y C, Chen Y R, Tsai S F. High frequency of epidermal growth factor receptor mutations with complex patterns in non‐small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 2004108195–8203. [DOI] [PubMed] [Google Scholar]
- 45.Yokoyama T, Kondo M, Goto Y, Fukui T, Yoshioka H, Yokoi K, Osada H, Imaizumi K, Hasegawa Y, Shimokata K, Sekido Y. EGFR point mutation in non‐small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci 200697753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese P P, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F. EGFR mutations in non‐small‐cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 200523857–865. [DOI] [PubMed] [Google Scholar]
- 47.Eberhard D A, Johnson B E, Amler L C, Goddard A D, Heldens S L, Herbst R S, Ince W L, Janne P A, Januario T, Johnson D H, Klein P, Miller V A, Ostland M A, Ramies D A, Sebisanovic D, Stinson J A, Zhang Y R, Seshagiri S, Hillan K J. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non‐small‐cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 2005235900–5909. [DOI] [PubMed] [Google Scholar]
- 48.Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking‐independent, lung adenocarcinoma. Br J Cancer 200593355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shih J Y, Gow C H, Yu C J, Yang C H, Chang Y L, Tsai M F, Hsu Y C, Chen K Y, Su W P, Yang P C. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer 2006118963–969. [DOI] [PubMed] [Google Scholar]
- 50.Taron M, Ichinose Y, Rosell R, Mok T, Massuti B, Zamora L, Mate J L, Manegold C, Ono M, Queralt C, Jahan T, Sanchez J J, Sanchez‐Ronco M, Hsue V, Jablons D, Sanchez J M, Moran T. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib‐treated chemorefractory lung adenocarcinomas. Clin Cancer Res 2005115878–5885. [DOI] [PubMed] [Google Scholar]
- 51.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA 200410113306–13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pao W, Miller V A, Politi K A, Riely G J, Somwar R, Zakowski M F, Kris M G, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 20052e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi S, Boggon T J, Dayaram T, Janne P A, Kocher O, Meyerson M, Johnson B E, Eck M J, Tenen D G, Halmos B. EGFR mutation and resistance of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2005352786–792. [DOI] [PubMed] [Google Scholar]
- 54.Bell D W, Gore I, Okimoto R A, Godin‐Heymann N, Sordella R, Mulloy R, Sharma S V, Brannigan B W, Mohapatra G, Settleman J, Haber D A. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005371315–1316. [DOI] [PubMed] [Google Scholar]
- 55.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T.cit-tl>Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications.Cancer Res 2004648919–8923. [DOI] [PubMed] [Google Scholar]
- 56.Soh J, Toyooka S, Aoe K, Asano H, Ichihara S, Katayama H, Hiraki A, Kiura K, Aoe M, Sano Y, Sugi K, Shimizu N, Date H. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer 20061192353–2358. [DOI] [PubMed] [Google Scholar]
- 57.Asano H, Toyooka S, Tokumo M, Ichimura K, Aoe K, Ito S, Tsukuda K, Ouchida M, Aoe M, Katayama H, Hiraki A, Sugi K, Kiura K, Date H, Shimizu N. Detection of EGFR gene mutation in lung cancer by mutant‐enriched polymerase chain reaction assay. Clin Cancer Res 20061243–48. [DOI] [PubMed] [Google Scholar]
- 58.Fan X, Furnari F B, Cavenee W K, Castresana J S. Non‐isotopic silver‐stained SSCP is more sensitive than automated direct sequencing for the detection of PTEN mutations in a mixture of DNA extracted from normal and tumor cells. Int J Oncol 2001181023–1026. [DOI] [PubMed] [Google Scholar]
- 59.Bosari S, Marchetti A, Buttitta F, Graziani D, Borsani G, Loda M, Bevilacqua G, Coggi G. Detection of p53 mutations by single‐strand conformation polymorphisms (SSCP) gel electrophoresis. A comparative study of radioactive and nonradioactive silver‐stained SSCP analysis. Diagn Mol Pathol 19954249–255. [DOI] [PubMed] [Google Scholar]
- 60.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction‐based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn 20057396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, Fukuyama S, Yokote A, Kobayashi K, Kanazawa M, Hagiwara K. Genetic heterogeneity of the epidermal growth factor receptor in non‐small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid‐locked nucleic acid PCR clamp. Cancer Res 2005657276–7282. [DOI] [PubMed] [Google Scholar]
- 62.Sugio K, Uramoto H, Ono K, Oyama T, Hanagiri T, Sugaya M, Ichiki Y, So T, Nakata S, Morita M, Yasumoto K. Mutations within the tyrosine kinase domain of EGFR gene specifically occur in lung adenocarcinoma patients with a low exposure of tobacco smoking. Br J Cancer 200694896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang X, Shigematsu H, Bekele B N, Roth J A, Minna J D, Hong W K, Gazdar A F, Wistuba EGFR tyrosine kinase domain mutations are detected in histologically normal respiratory epithelium in lung cancer patients. Cancer Res 2005657568–7572. [DOI] [PubMed] [Google Scholar]
- 64.Greulich H, Chen T H, Feng W, Janne P A, Alvarez J V, Zappaterra M, Bulmer S E, Frank D A, Hahn W C, Sellers W R, Meyerson M. Oncogenic transformation by inhibitor‐sensitive and ‐resistant EGFR mutants. PLoS Med 20052e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sordella R, Bell D W, Haber D A, Settleman J. Gefitinib‐sensitizing EGFR mutations in lung cancer activate anti‐apoptotic pathways. Science 20043051163–1167. [DOI] [PubMed] [Google Scholar]
- 66.Tracy S, Mukohara T, Hansen M, Meyerson M, Johnson B E, Janne P A. Gefitinib induces apoptosis in the EGFRL858R non‐small‐cell lung cancer cell line H3255. Cancer Res 2004647241–7244. [DOI] [PubMed] [Google Scholar]
- 67.Cappuzzo F, Hirsch F R, Rossi E, Bartolini S, Ceresoli G L, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin W A, Crino L, Bunn P A, Jr, Varella‐Garcia M. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non‐small‐cell lung cancer. J Natl Cancer Inst 200597643–655. [DOI] [PubMed] [Google Scholar]
- 68.Hirsch F R, Varella‐Garcia M, McCoy J, West H, Xavier A C, Gumerlock P, Bunn P A, Jr, Franklin W A, Crowley J, Gandara D R. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol 2005236838–6845. [DOI] [PubMed] [Google Scholar]
- 69.Reinmuth N, Brandt B, Kunze W P, Junker K, Thomas M, Achatzy R, Scheld H H, Semik M. Ploidy, expression of erbB1, erbB2, P53 and amplification of erbB1, erbB2 and erbB3 in non‐small cell lung cancer. Eur Respir J 200016991–996. [DOI] [PubMed] [Google Scholar]
- 70.Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, Yamamoto S, Nokihara H, Yamamoto N, Sekine I, Kunitoh H, Shibata T, Sakiyama T, Yoshida T, Tamura T. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non‐small‐cell lung cancer. J Clin Oncol 2005236829–6837. [DOI] [PubMed] [Google Scholar]
- 71.Amann J, Kalyankrishna S, Massion P P, Ohm J E, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart Salmon J, Kim Y H, Pollack J R, Yanagisawa K, Gazdar A, Minna J D, Kurie J M, Carbone D P. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res 200565226–235. [PubMed] [Google Scholar]
- 72.Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, Tabata M, Ueoka H, Tanimoto M, Date H, Gazdar A F, Shimizu N. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non‐small cell lung cancers. Clin Cancer Res 2005111167–1173. [PubMed] [Google Scholar]
- 73.Shigematsu H, Gazdar A F. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer 2006118257–262. [DOI] [PubMed] [Google Scholar]
- 74.Groves C, Lamlum H, Crabtree M, Williamson J, Taylor C, Bass S, Cuthbert‐Heavens D, Hodgson S, Phillips R, Tomlinson I. Mutation cluster region, association between germline and somatic mutations and genotype‐phenotype correlation in upper gastrointestinal familial adenomatous polyposis. Am J Pathol 20021602055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phillips D H, Hewer A, Martin C N, Garner R C, King M M. Correlation of DNA adduct levels in human lung with cigarette smoking. Nature 1988336790–792. [DOI] [PubMed] [Google Scholar]
- 76.Brownson R C, Alavanja M C, Caporaso N, Simoes E J, Chang J C. Epidemiology and prevention of lung cancer in nonsmokers. Epidemiol Rev 199820218–236. [DOI] [PubMed] [Google Scholar]
- 77.Gartner L P, Hiatt J L.Color text book of histology. Philadelphia: WB Saunders, 2001
- 78.Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. Prospective phase II study of gefitinib for chemotherapy‐naive patients with advanced non‐small‐cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol 2006243340–3346. [DOI] [PubMed] [Google Scholar]
- 79.Asahina H, Yamazaki K, Kinoshita I, Sukoh N, Harada M, Yokouchi H, Ishida T, Ogura S, Kojima T, Okamoto Y, Fujita Y, Dosaka‐Akita H, Isobe H, Nishimura M. A phase II trial of gefitinib as first‐line therapy for advanced non‐small cell lung cancer with epidermal growth factor receptor mutations. Br J Cancer 200695998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jackman D M, Yeap B Y, Sequist L V, Lindeman N, Holmes A J, Joshi V A, Bell D W, Huberman M S, Halmos B, Rabin M S, Haber D A, Lynch T J, Meyerson M, Johnson B E, Janne P A. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non‐small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 2006123908–3914. [DOI] [PubMed] [Google Scholar]
- 81.Choong N W, Dietrich S, Seiwert T Y, Tretiakova M S, Nallasura V, Davies G C, Lipkowitz S, Husain A N, Salgia R, Ma P C. Gefitinib response of erlotinib‐refractory lung cancer involving meninges—role of EGFR mutation. Nat Clin Pract Oncol 2006350–7 quiz 1 p following 57. [DOI] [PubMed] [Google Scholar]
- 82.Engelman J A, Mukohara T, Zejnullahu K, Lifshits E, Borras A M, Gale C M, Naumov G N, Yeap B Y, Jarrell E, Sun J, Tracy S, Zhao X, Heymach J V, Johnson B E, Cantley L C, Janne P A. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR‐amplified lung cancer. J Clin Invest 20061162695–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Antonescu C R, Besmer P, Guo T, Arkun K, Hom G, Koryotowski B, Leversha M A, Jeffrey P D, Desantis D, Singer S, Brennan M F, Maki R G, DeMatteo R P. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res 2005114182–4190. [DOI] [PubMed] [Google Scholar]
- 84.Shah N P, Nicoll J M, Nagar B, Gorre M E, Paquette R L, Kuriyan J, Sawyers C L. Multiple BCR‐ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 20022117–125. [DOI] [PubMed] [Google Scholar]
- 85.Keohavong P, DeMichele M A, Melacrinos A C, Landreneau R J, Weyant R J, Siegfried J M. Detection of K‐ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res 19962411–418. [PubMed] [Google Scholar]
- 86.Siegfried J M, Gillespie A T, Mera R, Casey T J, Keohavong P, Testa J R, Hunt J D. Prognostic value of specific KRAS mutations in lung adenocarcinomas. Cancer Epidemiol Biomarkers Prev 19976841–847. [PubMed] [Google Scholar]
- 87.Toyooka S, Tsuda T, Gazdar A F. The TP53 gene, tobacco exposure, and lung cancer. Hum Mutat 200321229–239. [DOI] [PubMed] [Google Scholar]