Abstract
Introduction
Myopathy, lactic acidosis and sideroblastic anaemia (MLASA) is a rare condition that combines early‐onset myopathy with lactic acidosis and sideroblastic anaemia. MLASA has been associated with a missense mutation in pseudouridylate synthase 1 (PUS1), an enzyme located in both nucleus and mitochondria, which converts uridine into pseudouridine in several cytosolic and mitochondrial tRNA positions and increases the efficiency of protein synthesis in both compartments.
Subjects and methods
We have identified two Italian brothers, offspring of distantly related parents, both of whom are affected by MLASA. The six exons of the PUS1 gene were analysed by automated sequencing.
Results
We found combined defects in mitochondrial respiratory chain complexes in muscle and fibroblast homogenates of both patients, and low levels of mtDNA translation products in fibroblast mitochondria. A novel, homozygous stop mutation was present in PUS1 (E220X). We have investigated the structural and mechanistic aspects of the double localisation of PUS1, demonstrating that the isoform located in the nucleus contains an N‐terminal extension which is absent in the mature mitochondrial isoform.
Conclusions
The stop mutation in PUS1 is likely to determine the loss of function of the protein, since it predicts the synthesis of a protein missing 208/427 amino acid residues on the C terminus, and was associated with low mtDNA translation. The structural differences in nuclear versus mitochondrial isoforms of PUS1 may be implicated in the variability of the clinical presentations in MLASA.
Keywords: lactic acidosis, mitochondrial disorders, mtDNA translation, myopathy, pseudouridylation, PUS1, respiratory chain, sideroblastic anaemia, tRNA
Mitochondrial disorders are characterised by primary defects in the mitochondrial respiratory chain (RC). Biochemically, these disorders can affect single enzymatic activities or present as a combination of multiple RC defects. The latter, in particular combined defects in complex I and complex IV, account for approximately one third of all the biochemically‐defined mitochondrial disorders in humans. Genetically, defects in single enzymatic activities can be due to mutations in genes encoding individual subunits of each complex, or specific factors involved in their assembly and turnover.1 In adults, multiple defects in RC activities are often caused by mutations in the mtDNA‐encoded RNA products, mainly tRNAs, involved in mtDNA protein translation. These mutations are less frequently found in children. Conditions characterised by profound reduction in mtDNA (mtDNA depletion syndromes) account for a small fraction of infantile cases in which multiple defects in RC complexes are present in single or multiple tissues, such as muscle, liver or brain.2 Mutations in factors involved in mtDNA replication have been identified in some mtDNA depletion syndromes. However, in many cases the genetic and molecular bases of multiple RC defects remain undiagnosed. Nuclear‐encoded protein components of the mtDNA translation machinery are other likely candidates.3 In a cohort of seven children with multiple defects in the RC complexes, which were detectable in both muscle and fibroblasts, we identified two brothers with a homozygous stop mutation in the gene encoding pseudouridylate synthase 1 (PUS1). Similar to the reported cluster of Persian‐Jewish families in which the first PUS1 mutation was identified,4 our patients were affected by an infantile myopathy with lactic acidosis and sideroblastic anaemia (MLASA; MIM 600462). Myopathic changes were typical of a mitochondrial disorder, although the severity of the syndrome varied markedly in the two brothers. PUS1 is part of the truA family of tRNA pseudouridine synthases5,6 and converts uridine into pseudouridine in several tRNA positions encoded by either nuclear or mitochondrial genes,7,8 thereby acting in both cellular compartments. Pseudouridylation is the most frequently found modification in tRNAs and seems to increase the protein translation efficiency.9 The double localisation of PUS1, and its effects on two spatially and functionally separated translational machineries, may account, at least in part, for the variability in the clinical presentations associated with PUS1 mutations.
Case reports
The probands were two brothers with reportedly unrelated parents. However, investigation through birth registration records demonstrated that their parents are in fact sixth cousins. The family history was completely negative for MLASA, haematological or neurological genetic disease, including mental retardation. Informed consent was obtained from the parents of the patients, according to the Ethics Committee of the National Institute of Neurology and the guidelines of the Italian Public Ministry of Health. Patient 1 was a boy born at term after an uneventful pregnancy. The Apgar score was 9 at 1 min, and 10 at 5 min. Body weight was 2610 g, length was 45.5 cm and head circumference was 32.2 cm. Body growth was consistently below the 3rd centile and an arginine test failed to show increased secretion of growth hormone (GH). However, the child walked at 18 months and language and mental development were both normal, with good proficiency at school. At age 6 months generalised hypotonia was noted, together with joint laxity, pseudoepicanthus and hypertelorism. At 5 years of age the patient was diagnosed with severe sideroblastic anaemia, unresponsive to vitamin B6 supplementation (Hb 5 g%, Ht 16%, MCV 100 µm3, BRC 1.6×106/mm3), requiring periodic blood transfusions with associated iron‐chelating treatment with desferoxamine. He also received GH supplementation for the correction of hypopituitarism and severe growth failure. Physical examination at 10 years of age revealed marked growth retardation: body weight was 18.7 kg (<<3rd centile), height was 125 cm (<3rd centile) and head circumference was 49 cm (−2 SD). Flat nose, hypertelorism and prominent cheek bones were attributed to bone marrow hyperplasia. The child had profound, generalised muscle hypotrophy and weakness, more marked in the hands, winging scapulae, hyperlordosis of the trunk and anserine gait with a frank Gower's manoeuvre. No cerebellar or pyramidal signs were present. Extrinsic ocular motility was normal, with mild weakness of the upper eyelids. IQ was 120. Blood lactate at rest was 6.1 mM (normal value 0.5–2.2), blood pyruvate was 0.64 mg% (normal value 0.36–0.59) and serum CK was normal. ECG and ultrasound examination of the heart were both normal. The child had severe restrictive ventilatory syndrome, which was attributed to failure of the respiratory muscles, including the diaphragm. EMG, ENG, brain MRI and abdominal ultrasound examination were normal. The patient also developed milk protein and gluten intolerance, and non‐progressive pigmentary retinopathy.
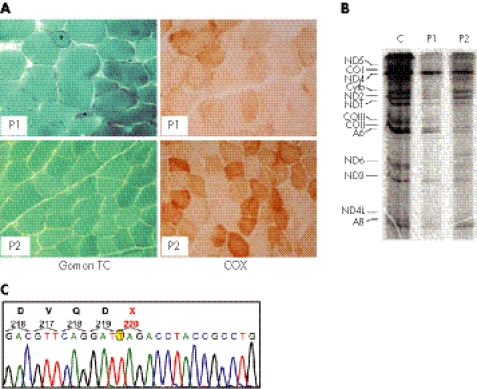
A muscle biopsy from the left quadriceps showed myopathic changes and the presence of ragged‐red and COX‐negative fibres (fig 1A). The activity of the RC complexes showed a profound deficiency of complex IV and less severe deficiency of complex I in both muscle homogenate and cultured skin fibroblasts (table 1). Progressive worsening of the muscle weakness and sideroblastic anaemia led to the child's death at 12 years of age from respiratory failure.
Figure 1 Morphological, biochemical and molecular findings in patients 1 and 2. (A) Gomori trichrome (TC) and cytochrome c oxidase (COX) staining of muscle biopsies in patient 1 (P1) and patient 2 (P2). Patient 1 shows more severe abnormalities than patient 2; note the presence of ragged‐red and atrophic fibres (asterisks) and diffuse reduction in COX reactivity. (B) mtDNA translation in cell culture. The bands in the autoradiograph correspond to the 13 polypeptides encoded by mtDNA genes. A, ATP synthase (complex V); CO, cytochrome c oxidase (complex IV); Cytb, cytochrome b (complex III); ND, polypeptides of NADH‐CoQ reductase (complex I). (C) Electropherogram of the PUS1 automated sequence analysis in patient 1. A mutant T, which replaces a wild‐type G at position 658 of the cDNA sequence, is encircled in yellow.
Table 1 RC activity.
| Complex I* (NADH‐CoQ1 red) | Complex II* (succ‐CoQ1 red) | Complex III* (DBH2‐cit C red) | Complex IV* (COX) | Complex V* (ATPase) | Citrate synthase† | ||
|---|---|---|---|---|---|---|---|
| Patient 1 | Muscle | 2.6 | 14.8 | 15 | 8.8 | 61 | 274 |
| Fibroblasts | 9.7 | 6.9 | 73 | 58 | 40 | 321 | |
| Patient 2 | Muscle | 3.7 | 16.9 | 26 | 22 | 70 | 249 |
| Fibroblasts | 6.7 | 14.4 | 110.2 | 12 | 60.6 | 136.5 | |
| Reference | Muscle | 15–31 | 20–40 | 80–160 | 80–180 | 100–220 | 90–200 |
| values | Fibroblasts | 13–34 | 10–20 | 95–155 | 70–170 | 60–110 | 100–250 |
*Values refer to citrate synthase activity; †nmol min−1 mg protein−1.
The clinical course of patient 2 was much milder than that of his older brother. His weight at birth was 3280 g, and body growth has consistently remained at the lower normal limit. Motor and language milestones were reached at the appropriate age. However, his IQ (Leiter scale) was 85 at 6 years of age, and the visual‐perception and visual‐motor test results were <1st percentile, with impaired general visual perception and visual‐motor integration. Neurological examination showed mild generalised muscle hypotrophy, more severe in both hands, very mild weakness, and hypotonia, mainly in the lower limbs, but no Gower's manoeuvre. The patient has mild sideroblastic anaemia (Hb 11.8–9.8 g%), which has never required transfusion, and moderate elevation of blood lactate (3.9 mM) and pyruvate (1.31 mg%). The patient is currently 13 years old. He displays mild exercise intolerance and an initial ventilatory insufficiency. His haematological conditions are relatively compensated for and stable. He has medium mental insufficiency: total IQ is now 53, verbal IQ 59, performance IQ 51 using WISC‐R, Raven test 12/60, <5th centile. He has no pigmentary retinopathy.
Muscle homogenate and cultured fibroblasts showed combined deficiency of complex IV and complex I. The morphological features of the muscle biopsy were similar, although less severe, than those found in patient 1 (fig 1A).
Methods
Generation and cloning of human PUS1 variants
The full‐length human IMAGE cDNA clones for Homo sapiens pseudouridylate synthase 1, transcript variant 1 (accession number BC019320) and transcript variant 3 (accession number BC002901) were obtained from the German Resource Center for Genome Research, RZPD (clones IRAUp969C0377D and IRALp962M1515Q). The HA tagged cDNA fragments were generated by PCR using a chimeric oligonucleotide reverse primer containing the 3′ end of the human PUS1 cDNA ORF (open reading frame) in frame with the HA encoding sequence and a forward primer containing the Kozak sequence and the 5′ end of the human PUS1‐1 or PUS1‐3. The 5′ modified PUS1‐1 cDNA containing the Kozak sequence was also generated by amplification using the same forward primer and a reverse primer containing the natural 3′ end of the human PUS1 cDNA. All these PCR products were cloned into the pcDNA3.2/V5/GW/D‐TOPO vector (Invitrogen, Carlsbad, CA). The recombinant plasmids encoding the HA tagged proteins were transfected by electroporation into Cos7 cells for transient expression.
In a second set of experiments, the PCR products corresponding to nucleotides 188 to 573 of the PUS1‐1 cDNA (BC019320) and 75 to 358 of the PUS1‐3 cDNA (BC002901) were cloned into the pcDNA3.1/CT‐GFP‐TOPO vector (Invitrogen) in frame with the Cycle 3 GFP ORF already contained in the plasmid. The recombinant plasmids were transfected by electroporation into Cos7 cells for transient expression. The pcDNA3.1/CT‐GFP vector was used as an expression control in the Cos7 cells.
In organello import assay
In vitro transcription and translation of PUS1‐1 was performed with the TNT T7 Quick Coupled Transcription/Translation System (Promega, Madison, WI) in the presence of 20 mCi [35S]‐l‐methionine (Amersham, Little Chalfont, UK) using the 5′ modified PUS1‐1 cDNA cloned into pcDNA3.2/V5/GW/D‐TOPO plasmid as the template. The translation product of PUS1‐3 was obtained using the TNT SP6 Quick Coupled Transcription/Translation System (Promega) in the presence of 20 mCi [35S]‐l‐methionine (Amersham) and by adding the IRALp962M1515Q plasmid as the template. Fresh mitochondria were prepared starting from approximately 2×107 HeLa cells as described previously10 and the final fraction was resuspended in incubation buffer to a final concentration of 2 mg/ml. The in organello import assay was then performed as described.11 Samples were electrophoresed using a 12% SDS‐polyacrylamide gel. After fixation, the gel was washed for 30 min in Amplify (Amersham), dried and exposed using a phosphorimaging screen (Bio‐Rad, Hercules, CA).
Antibodies
The mouse monoclonal antibody to the haemagglutinin epitope of the influenza virus (anti‐HA) was from Roche (Indianapolis, IN). Polyclonal antibodies against human mitochondrial single‐stranded DNA binding protein were raised and characterised as described.12 Fluorescent anti‐mouse and anti‐rabbit IgG antibody conjugates were from Molecular Probes (Invitrogen).
Cell cultures and immunofluorescence studies
Mammalian cells were cultured in DMEM supplemented with 10% fetal calf serum at 37°C in 5% CO2 atmosphere. For immunofluorescence assays, transfected cells were plated on coverslips and the following day cells were fixed, permeabilised and incubated with primary and fluorescent dye‐conjugated secondary antibodies, as described.12 The cells transfected with the green fluorescent protein (GFP) fusion proteins were plated on coverslips and the following day the samples were stained with 30 nM MitoTracker Red dye (CMXRos; Molecular Probes) for 30 min at 37°C. Fluorescence patterns were visualised with a confocal microscope (Bio‐Rad).
Western‐blot analysis
Approximately 2×106 cells were trypsinised, pelleted, sonicated and solubilised as previously described. SDS‐polyacrylamide gel with 100–200 µg protein/lane, and Western blot analyses were performed as described13 using the ECL‐chemiluminescence kit (Amersham).
Biochemical assays
Specific activities of individual RC complexes were measured on cell and muscle homogenates.14 Protein concentration was measured by the Folin‐Ciocalteau method.15 Specific activities of each complex were normalised to that of citrate synthase,14 an indicator of the number of mitochondria.
Results
Genetic and biochemical studies
Albeit characterised by wide variability, the essential clinical features of both patients resembled MLASA. MLASA is a rare condition which has recently been associated with a missense mutation in the gene encoding PUS1 in two, presumably related, families of Persian‐Jewish origin.4 The mutation predicts a R116W amino acid change in isoform 2 (PUS1‐2) of the corresponding protein (see below). We screened the six exons of the PUS1 gene and found a homozygous G>T transversion in DNA samples from both patients. The parents were both heterozygous for the same mutation. Considering the A of the first ATG of the PUS1‐1 ORF as nucleotide position +1, the mutation affects the G nucleotide at position 658 (658G>T). The 658G>T mutation causes the replacement of the codon for the amino acid residue E220 of isoform 1 (PUS1‐1) with a stop codon (TAG). The position equivalent to the PUS1‐1 E220X is E192X in the PUS1‐2 isoform. For both isoforms, the mutation found in our patients predicts the synthesis of a shorter polypeptide, which encompasses approximately the N‐terminal half of the wild‐type protein, lacks the last 208 C‐terminal amino acid residues, and is likely to be inactive. Accordingly, mtDNA‐specific protein translation was reduced in fibroblasts from both patients (fig 1), which would explain the combined deficiency of complex I and complex IV found in these cells (table 1).
Immunofluorescence studies
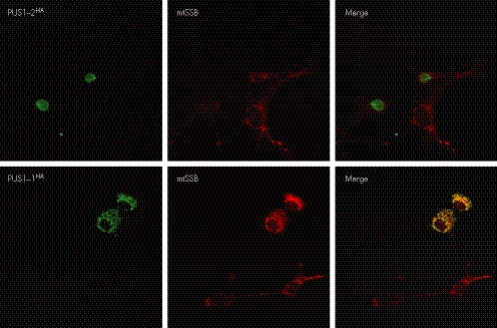
The same PUS1 gene encodes at least three transcription variants. Transcript variant 1 (accession number NM_025215) is translated into a longer protein isoform (isoform 1, PUS1‐1) that contains an N‐terminal extension, which has the features typical of a mitochondrial leader peptide. PUS1‐1 is believed to target, and be imported into, mitochondria, where it participates in the maturation of the 22 mtDNA‐encoded tRNAs involved in the in situ translation of the 13 mtDNA‐specific mRNA transcripts. A shorter isoform (PUS1‐2) is encoded by both transcript variants 2 (accession number NM_001002019) and 3 (NM_001002020), which differ from each other in the 5′ untranslated sequence but contain the same ORF sequence. The first ATG of PUS1‐2 corresponds to an internal methionine, which is 29 codons downstream from, and in‐frame with, the first ATG in transcript variant 1 (PUS1‐1). The sequence of PUS1‐2 is otherwise identical to the longer form, PUS1‐1. However, two additional variants consequent to differential splicing of exons 3 and 4 have been reported in the mouse, but their existence has not been proved in humans.16 The putative localisation of PUS1‐2 is in the nucleus, where it promotes the maturation of tRNAs serving cytosolic protein translation. Using HA‐tagged recombinant eukaryotic expression vectors, we demonstrated that each of the two isoforms of PUS1 has a distinct subcellular localisation. The immunofluorescence pattern specific for PUS1‐1HA was identical to that obtained using an antibody specific for the mitochondrial single‐stranded DNA binding protein (mtSSB) (fig 2), while no immunoreaction was present in the nucleus. By contrast, the immunofluorescence pattern specific for PUS1‐2HA was exclusively nuclear (fig 2).
Figure 2 Immunofluorescence‐based localisation of PUS1‐1 and PUS1‐2 isoforms in transiently transfected COS‐7 cells. The immunofluorescence pattern obtained by expressing recombinant PUS1‐1HA and PUS1‐2HA polypeptides was compared with the immunofluorescence pattern of mitochondrial single‐stranded DNA binding protein (mtSSB), a mitochondrion‐specific protein.
In organello import
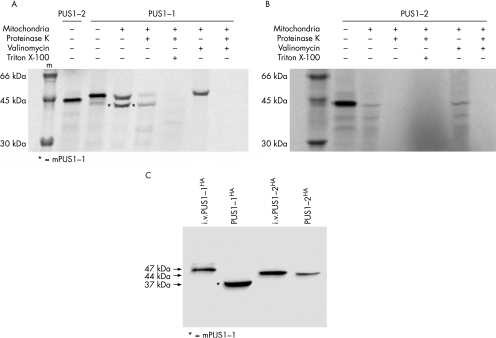
Using an in organello import assay,11 we demonstrated that PUS1‐1 is imported into mitochondria by the TOM/TIM protein translocation system, which is driven by the proton‐motive force (ΔΨ) provided by the mitochondrial membrane potential.17 As shown in fig 3A, when the in vitro translation product corresponding to the full‐length PUS1‐1 polypeptide was mixed with freshly prepared, energised mitochondria, a second, smaller band appeared (indicated by an asterisk in fig 3A), corresponding to a protein of lower molecular weight. The smaller band was protected from digestion with proteinase K (pK), while the band corresponding to full‐length PUS1‐1 was eliminated or markedly reduced. The smaller band corresponds to the mature PUS1‐1 (mPUS1‐1) mitochondrial protein, which is produced by cleavage of the N‐terminal targeting peptide by the mitochondrial matrix peptidase. Resistance to pK digestion indicates that this protein species was internalised within the inner compartment of mitochondria, which is impermeable to pK. Solubilisation of the inner mitochondrial membrane with Triton X‐100 before pK treatment made both bands disappear, due to the complete digestion of the corresponding proteins. Treatment of mitochondria with valinomycin, a mitochondrial ionophore that abolishes the ΔΨ, blocked the import process, thus preventing the generation of the shorter, mature form and allowing the PUS1‐1 precursor protein to be completely digested by pK. This result indicates that internalisation of PUS1‐1 into the inner compartment of mitochondria and, consequently, its maturation by cleavage of the mitochondrial leader peptide from the precursor, are dependent on the presence of an intact mitochondrial ΔΨ. A parallel in vitro import assay using PUS1‐2 confirmed that this protein is not imported into mitochondria (fig 3B).
Figure 3 In organello import and Western blot analysis. (A) In organello import assay of PUS1‐1. The in vitro translation product of PUS1‐2 is shown for comparison with the in vitro translation product corresponding to the PUS1‐1 precursor and with the mature PUS1‐1 (mPUS1‐1), generated by exposing the PUS1‐1 precursor to energised mitochondria. See text for details. (B) In organello import assay of PUS1‐2. In contrast with PUS1‐1, PUS1‐2 is not translocated into the inner compartment of mitochondria and is completely digested by exposure to proteinase K. See text for details. (C) Western blot analysis of PUS1‐1HA and PUS1‐2HA transfected in HeLa cells. i.v.PUS1‐1HA and i.v.PUS1‐2HA indicate in vitro translation products. The only protein species detected after transfection of PUS1‐1 is a shorter polypeptide corresponding to mPUS1‐1. See text for details.
Subcellular targeting of PUS1‐GFP chimaeric proteins
As also shown in fig 3A, the molecular weight (MW) of the precursor PUS1‐1 protein is approximately 47 kDa, while that of the PUS1‐2 protein is approximately 44 kDa. The mPUS1‐1 mitochondrial protein appears to be considerably smaller than that of PUS1‐2. The apparent MW of the mPUS1‐1 is approximately 37 kDa, which suggests that the mitochondrial targeting sequence of PUS1‐1 extends for approximately 100 amino acid residues.
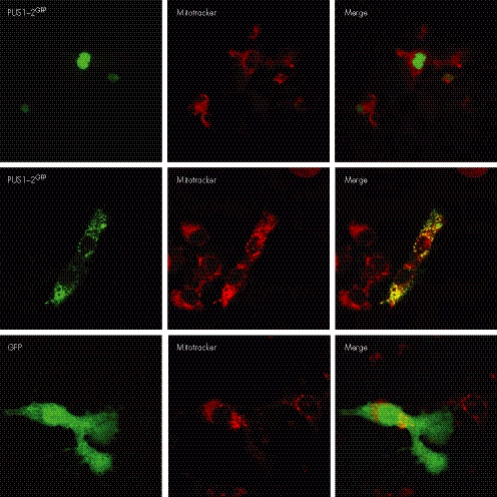
This result was confirmed by Western blot analysis on HeLa cells transfected with either the full‐length PUS1‐1HA or PUS1‐2HA. As shown in fig 3C, the MW of mPUS1‐1HA is not only lower than that of its in vitro translated precursor, but also of in vitro translated, or HeLa‐cell transfected, PUS1‐2HA. These findings are surprising because the maturation of the mitochondrial isoform into a functionally active species was thought to consist of the simple conversion of PUS1‐1 into a polypeptide identical to PUS1‐2. Ad hoc predictive software programs such as MitoProt II (http://ihg.gsf.de/ihg/mitoprot.html) and TargetP (http://www.cbs.dtu.dk/services/TargetP/), indeed indicate the existence of a potential mitochondrial matrix peptidase cleavage site of the PUS1‐1 sequence between M29 and A30 (MitoProt II), and also the existence of a second possible cleavage site between R102 and N103 (TargetP). We then reasoned that the sequence encompassing M29‐N103, which is present in PUS1‐2 but not in mPUS1‐1, must contain one or more nucleus‐localisation signals in order for PUS1‐2 to be targeted to the nucleus. There are two potential nucleus‐localisation signal motifs in the 29–103 PUS1 stretch, the first between amino acids 65 and 71 (PAKKLKS) and the second between amino acids 81 and 88 (PPKRKIVL) (fig 4). We decided to verify this hypothesis experimentally by constructing a reporter system based on the expression of two chimaeric proteins, PUS1‐1GFP and PUS1‐2GFP. PUS1‐1GFP contains the PUS1 M1‐E113 stretch on the N terminus, while PUS1‐2GFP contains the M29‐E113 stretch; each stretch is fused with the GFP sequence. As shown in fig 5, the PUS1‐2GFP fusion protein was specifically targeted to the nucleus of COS‐7 cells, while the PUS1‐1GFP fusion protein was targeted to mitochondria, since its pattern coincided with that of MitoTracker, a mitochondrion specific marker. GFP alone stained the cells diffusely.
Figure 4 PUS1 isoforms and post‐translation modifications. The putative mitochondrial targeting signal of PUS1‐1 is in blue and the sequence containing putative nuclear localisation signals for PUS1‐2 (underlined) is in red. The mature mitochondrial PUS1‐1 polypeptide (mPUS1‐1) is in black. The E220 residue which is mutated in patients 1 and 2 is encircled in yellow.
Figure 5 Subcellular localisation of PUS1/GFP chimeric proteins. The fluorescence patterns obtained by expressing PUS1‐1GFP, PUS1‐2GFP and GFP alone were compared with the fluorescence pattern of MitoTracker, a mitochondrial marker. See text for details.
Discussion
The haematological and neurological presentation varied in our patients, in spite of the common parental origin and identical PUS1 mutation. The first brother had a very severe mitochondrial myopathy with muscle wasting, severe sideroblastic anaemia and severe growth retardation, due to complete deficiency of GH secretion. However, he achieved the main psychomotor milestones normally, and his intelligence was above normal. He died in pre‐puberty. His brother, now 13 years old, is alive, and affected by a much milder neuromuscular syndrome with no muscle wasting. His sideroblastic anaemia never required the multiple transfusions that were needed by his older brother, but he displays obvious cognitive and behavioural abnormalities with mental retardation of medium severity, hyperactivity and panic attacks. The mitochondrial biochemical defect was equally severe in the two patients in both muscle and fibroblasts. The mutation affecting PUS1 in these patients predicts severe damage of the protein with the likely complete loss of enzymatic activity. Although circumstantial, strong evidence for such a conclusion is provided by the large reduction in mtDNA translation demonstrated in fibroblast mitochondria and by the defective biochemical profile of mitochondrial RC activities in muscle and cultured fibroblasts from both patients. Variability of clinical features has been reported in other MLASA families in the past, including the presence of additional symptoms such as microcephaly, micrognathia, dysthichiasis, high philtrum, high palate, and, as in our patient 2, mental retardation18,19 or, as in our patient 1, GH deficiency.20 Similar to our cases, wide clinical variability has previously been reported among affected members of a single family who carried the same homozygous PUS1 mutation.19,20
What is the molecular basis of this clinical heterogeneity? PUS1 has been shown to modify several tRNA positions of both nuclear and mitochondrial origin. The function of pseudouridylation on tRNAs is not completely clear. Pseudouridylation seems to stabilise base pairing in stems and base stacking in the anticodon loop; it may affect the interaction of the tRNA with its cognate aminoacyl tRNA synthetase and the fit of the aminoacyl‐tRNA in the P or A site of the ribosome. It may also affect the transport of tRNA out of the nucleus to the cytoplasm since the combined loss of PUS1 and PUS4 activity blocks this process in yeast.21 Whatever the mechanism, pseudouridylation increases the efficiency of protein translation in both cytoplasm and mitochondria, as shown by our own data on mtDNA translation products in fibroblast extracts. Some of the clinical features of MLASA that were found in our patients are clearly attributable to mitochondrial failure, including the myopathy with ragged red fibres, the lactic acidosis and the presence of multiple defects in RC enzymes. The latter defects also included partial reduction of complex II activity in fibroblasts of patient 1 (table 1). Complex II is not encoded by mtDNA, which suggests that its defect could be due to either abnormal cytosolic protein synthesis or to secondary destabilisation of the entire RC within the mitochondrial inner membrane, caused by severe damage of the mtDNA‐dependent complexes. Arrest in the maturation of erythroblasts and accumulation of iron deposits in mitochondria have occasionally been reported in some mitochondrial disorders and are the haematological hallmarks of Pearson's syndrome, a severe infantile disorder caused by multiorgan distribution of large scale rearrangements of mtDNA. The latter are typically characterised by the loss of several tRNA genes and are therefore associated with defective mitochondrial translation. Other abnormalities reported in some MLASA patients, such as psychiatric symptoms and facial dysmorphisms, are uncommon in mitochondrial disorders and are likely due to abnormalities in cytosolic protein synthesis. An interesting possibility stems from the recent observation that the pseudouridyne synthase activity of PUS1 is required for the activation of the steroid receptor RNA activator, which is in turn involved in regulation of nuclear receptor activity.22 As suggested by Patton et al,8 impairment of this important control of nuclear transcription could amplify the deleterious functional consequences of PUS1 mutations, and add to the pleiotropism and individual variability of the corresponding clinical phenotypes.
From the above discussion, it is clear that an important source of variability depends on the dual spatial distribution of the PUS1 protein, and its activity on two physically and functionally separated translation machineries. Regulation of the differential expression of the PUS1 isoforms may in fact play a relevant role in controlling and coordinating cytosolic versus mitochondrial translation in normal and, possibly, disease conditions. Alternative splicing of the first exon accounts for the synthesis of the longer and shorter PUS1 isoforms in both mouse16 and humans. In order to clarify the mechanistic aspects of the post‐translation processing and sub‐cellular localisation of the PUS1 isoforms, we have set up in vitro and cellular assays using a series of recombinant PUS1 constructs. We have demonstrated that, in contrast with previous assumptions, the two PUS1 isoforms that are active in the two cell compartments, that is the mitochondrion and the nucleus, are structurally different. The mitochondrial enzyme is produced after the translocation of a 47‐kDa precursor protein, PUS1‐1, into the inner compartment of mitochondria. This process is carried out through the canonical ΔΨ‐driven TOM‐TIM import system, and is followed by cleavage of the mitochondrial targeting signal, to give rise to a mature form of approximately 37 kDa. The mitochondrial mature protein species is different from the protein imported into the nucleus, PUS1‐2, which is a 44‐kDa polypeptide. This result suggests that after translocation and maturation in the mitochondrial matrix, PUS1‐1 is not re‐exported from mitochondria into other cellular compartments, as has been shown to occur for some proteins which also display a double localisation (for example, fumarase).23 The first N‐terminal 72 amino acid residues of the PUS1‐2 protein are identical to residues 29–103 of the PUS1‐1 presequence but are not part of the mature mitochondrial PUS1‐1 polypeptide. Using a GFP‐based reporter system we have demonstrated that the PUS1‐2 specific sequence contains a nucleus‐targeting signal. An attractive hypothesis is that this sequence, in addition to determining the correct localisation of the nuclear enzyme, can also affect its kinetic properties, modifying them from those of the mitochondrial enzyme. An interesting possibility, consequent to our observations, is that deleterious mutations affecting the PUS1‐2 specific sequence could determine the onset of clinical presentations different from typical MLASA, due to defective translation in the cytoplasmic but not in the mitochondrial compartments. Conversely, mutations affecting the first 30 amino acid residues could in principle hamper mitochondrial import of PUS1‐1, leaving intact the nuclear activity, thereby causing an exclusively mitochondrial translation failure.
Electronic database information
The following URLs have been mentioned in this article: http://www.mitopedia.org (Center for the Study of Mitochondrial Disorders of Infancy and Childhood, National Institute of Neurology “C. Besta”), http://ihg.gsf.de/ihg/mitoprot.html (MitoProt software program) and http://www.cbs.dtu.dk/services/TargetP/ (TargetP software).
Acknowledgements
We thank Mr Franco Carrara and Dr Marilena Greco for skilful technical assistance.
Abbreviations
GH - growth hormone
GFP - green fluorescent protein
MLASA - myopathy, lactic acidosis and sideroblastic anaemia
mPUS1‐1 - mature PUS1‐1
MW - molecular weight
ORF - open reading frame
pK - proteinase K
PUS1 - pseudouridylate synthase 1
RC - respiratory chain
Footnotes
This study was supported by Fondazione Telethon‐Italy (grant no GGP030039), Fondazione Pierfranco e Luisa Mariani, MITOCIRCLE and EUMITOCOMBAT network grants from the European Union Framework Program 6.
Competing interests: None declared.
Further information: Parental informed consent was obtained for publication of the patients' details in this report.
References
- 1.DiMauro S, Hirano M. Mitochondrial encephalomyopathies: an update. Neuromuscul Disord 200515276–286. [DOI] [PubMed] [Google Scholar]
- 2.Spinazzola A, Zeviani M. Disorders of nuclear‐mitochondrial intergenomic signaling. Gene 2005354162–168. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs H T, Turnbull D M. Nuclear genes and mitochondrial translation: a new class of genetic disease. Trends Genet 200521312–314. [DOI] [PubMed] [Google Scholar]
- 4.Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel‐Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 2004741303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J 1996152270–2284. [PMC free article] [PubMed] [Google Scholar]
- 6.Lecointe F, Simos G, Sauer A, Hurt E C, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J Biol Chem 19982731316–1323. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Patton J R. Cloning and characterization of a mammalian pseudouridine synthase. RNA 19995409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patton J R, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel‐Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J Biol Chem 200528019823–19828. [DOI] [PubMed] [Google Scholar]
- 9.Lecointe F, Namy O, Hatin I, Simos G, Rousset J P, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem 200227730445–30453. [DOI] [PubMed] [Google Scholar]
- 10.Fernández‐Vizarra E, López‐Perez M J, Enriquez J A. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods 200226292–297. [DOI] [PubMed] [Google Scholar]
- 11.Petruzzella V, Tiranti V, Fernandez P, Ianna P, Carrozzo R, Zeviani M. Identification and characterization of human cDNAs specific to BCS1, PET112, SCO1, COX15, and COX11, five genes involved in the formation and function of the mitochondrial respiratory chain. Genomics 199854494–504. [DOI] [PubMed] [Google Scholar]
- 12.Tiranti V, D'Adamo P, Briem E, Ferrari G, Mineri R, Lamantea E, Mandel H, Balestri P, Garcia‐Silva M T, Vollmer B, Rinaldo P, Hahn S H, Leonard J, Rahman S, Dionisi‐Vici C, Garavaglia B, Gasparini P, Zeviani M. Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am J Hum Genet 200474239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini M P, Zeviani M. Characterization of SURF‐1 expression and Surf‐1p function in normal and disease conditions. Hum Mol Genet 199982533–2540. [DOI] [PubMed] [Google Scholar]
- 14.Bugiani M, Invernizzi F, Alberio S, Briem E, Lamantea E, Carrara F, Moroni I, Farina L, Spada M, Donati M A, Uziel G, Zeviani M. Clinical and molecular findings in children with complex I deficiency. Biochim Biophys Acta 20041659136–147. [DOI] [PubMed] [Google Scholar]
- 15.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem 1951193265–275. [PubMed] [Google Scholar]
- 16.Chen J, Patton J R. Mouse pseudouridine synthase 1: gene structure and alternative splicing of pre‐mRNA. Biochem J 2000352465–473. [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedemann N, Frazier A E, Pfanner N. The protein import machinery of mitochondria. J Biol Chem 200427914473–14476. [DOI] [PubMed] [Google Scholar]
- 18.Inbal A, Avissar N, Shaklai M, Kuritzky A, Schejter A, Ben‐David E, Shanske S, Garty B Z. Myopathy, lactic acidosis, and sideroblastic anemia: a new syndrome. Am J Med Genet 199555372–378. [DOI] [PubMed] [Google Scholar]
- 19.Zeharia A, Fischel‐Ghodsian N, Casas K, Bykhovskaya Y, Tamari H, Lev D, Mimouni M, Lerman‐Sagie T. Mitochondrial myopathy, sideroblastic anemia, and lactic acidosis: an autosomal recessive syndrome in Persian Jews caused by a mutation in the PUS1 gene. J Child Neurol 200520449–452. [DOI] [PubMed] [Google Scholar]
- 20.Casas K A, Fischel‐Ghodsian N. Mitochondrial myopathy and sideroblastic anemia. Am J Med Genet A 2004125201–204. [DOI] [PubMed] [Google Scholar]
- 21.Grosshans H, Lecointe F, Grosjean H, Hurt E, Simos G. Pus1p‐dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J Biol Chem 200127646333–46339. [DOI] [PubMed] [Google Scholar]
- 22.Zhao X, Patton J R, Davis S L, Florence B, Ames S J, Spanjaard R A. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 200415549–558. [DOI] [PubMed] [Google Scholar]
- 23.Singh B, Gupta R S. Mitochondrial import of human and yeast fumarase in live mammalian cells: retrograde translocation of the yeast enzyme is mainly caused by its poor targeting sequence. Biochem Biophys Res Commun 2006346911–918. [DOI] [PubMed] [Google Scholar]