Abstract
Background
Syndromic hearing loss that results from contiguous gene deletions is uncommon. Deafness‐infertility syndrome (DIS) is caused by large contiguous gene deletions at 15q15.3.
Methods
Three families with a novel syndrome characterised by deafness and infertility are described. These three families do not share a common ancestor and do not share identical deletions. Linkage was established by completing a genome‐wide scan and candidate genes in the linked region were screened by direct sequencing.
Results
The deleted region is about 100 kb long and involves four genes (KIAA0377, CKMT1B, STRC and CATSPER2), each of which has a telomeric duplicate. This genomic architecture underlies the mechanism by which these deletions occur. CATSPER2 and STRC are expressed in the sperm and inner ear, respectively, consistent with the phenotype in persons homozygous for this deletion. A deletion of this region has been reported in one other family segregating male infertility and sensorineural deafness, although congenital dyserythropoietic anaemia type I (CDAI) was also present, presumably due to a second deletion in another genomic region.
Conclusion
We have identified three families segregating an autosomal recessive contiguous gene deletion syndrome characterised by deafness and sperm dysmotility. This new syndrome is caused by the deletion of contiguous genes at 15q15.3.
Keywords: CATSPER2 , contiguous gene deletion, deafness‐infertility syndrome, DIS, STRC , syndromic hearing loss
In developed countries, genetic deafness is estimated to affect one in 2000 newborns.1 Syndromic and non‐syndromic subclassifications are recognised, the former being differentiated from the latter by the presence of additional traits that segregate with the deafness phenotype. The most common types of syndromic hearing loss include Waardenburg syndrome and branchio‐oto‐renal syndrome, both of which are autosomal dominant, and Pendred syndrome and Usher syndrome, both of which are autosomal recessive. Waardenburg syndrome and Usher syndrome are heterogeneous, with several genes causally related to various syndrome subtypes.2
Syndromic hearing loss can also result from contiguous gene deletions, although these are uncommon. Examples include infantile hyperinsulinism associated with enteropathy, deafness and renal tubulopathy caused by a contiguous gene deletion located on chromosome 11p,3 HDR syndrome (hyperparathyroidism, deafness and renal dysplasia) caused by a deletion distal to the DiGeorge syndrome region on 10p13–14,4 BRESEK/BRESHECK syndrome (brain anomalies, retardation of mentality and growth, ectodermal dysplasia, skeletal malformations, Hirschsprung disease, ear deformity and deafness, eye hypoplasia, cleft palate, cryptorchidism and kidney dysplasia/hypoplasia), due to a contiguous deletion on the X chromosome,5 and choroideraemia and deafness with stapes fixation, a contiguous gene deletion syndrome on Xq21.6
We have identified three families segregating an autosomal recessive contiguous gene deletion syndrome characterised by deafness and sperm dysmotility. This phenotype is similar to that described in a French family in which affected persons were deaf and, if male, had infertility secondary to asthenoteratozoospermia.7 Fine mapping in this family showed that the phenotype cosegregated with a deletion which included the STRC and CATSPER2 genes. In the families we report, affected males are deaf and have asthenoteratozoospermia associated with a deletion of chromosome 15q15. Using site‐specific nucleotide dosage analysis, we defined the deletion breakpoints in these families, all of which involve four duplicated genes: KIAA0377 (unknown function), CKMT1 (creatine kinase mitochondrial 1), STRC (stereocilin) and CATSPER2 (cation channel, sperm‐associated 2). Both copies of CKMT1 differ by only two nucleotides which predict synonymous mutations and are functional (CKMT1B and CKMT1A, telomeric), while the telomeric copies of KIAA0377, STRC and CATSPER2 are pseudo‐genes.
The phenotype associated with the deletion of STRC and CATSPER2 is consistent with our knowledge of these genes. Stereocilin is expressed in both the testis and inner ear and mutations in STRC are associated with deafness at the DFNB16 locus.8CATSPER2 is a member of the CATSPER gene family (CATSPER1–4) and is reportedly expressed exclusively in sperm.9
Methods
Patients and family
Family D_SM, family L705 and family L1014 were ascertained through a genetics clinic at The Welfare Science and Rehabilitation University in Iran. Informed consent was obtained from all participants. Each individual underwent audiologic testing to quantitate the degree of hearing loss, and a complete physical examination by both an otolaryngologist and a clinical geneticist to exclude clinical features consistent with syndromic hearing loss. A sample (10 ml) of whole blood was obtained as a DNA source. Sperm analyses were also carried out in consenting males. The Human Research Institutional Review Boards at the Welfare Science and Rehabilitation University and the Iran University of Medical Sciences, Tehran, Iran, and the University of Iowa, Iowa City, Iowa, USA approved all procedures.
Linkage mapping
Linkage was established by completing a genome‐wide scan using 400 fluorescence dye‐labelled microsatellite markers with an average spacing of 10 cM across the 22 autosomes and chromosome X (Prism Linkage Mapping Set, version 2.5; Applied Biosystems, Foster City, CA). Microsatellite markers were amplified by polymerase chain reaction (PCR) and analysed on an ABI Prism 3100 Genetic Analyzer. Alleles were assigned using GeneMapper 3.0 software (Applied Biosystems). Haplotype reconstruction was determined with custom‐made GeneScan software.
Mutation screening
Candidate genes in the linked region were screened by direct sequencing. Primers for PCR amplification were selected using Primer3 (http://frodo.wi.mit.edu/cgi‐bin/primer3/primer3_www. cgi). PCR was performed in a volume of 25 or 50 µl containing 0.2 µM each of forward and reverse primers, 0.2 mM dNTPs, 1.5 mM MgCl2, 1× PCR buffer, 30 ng genomic DNA and 1–2 U Taq DNA polymerase. Cycling parameters were an initial denaturation at 94°C for 4 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50–55°C for 45 s and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. PCR products were visualised on agarose gels (Qiagen, Valencia, CA) and gel extracted for sequencing using ABI BigDye terminator chemistry and an ABI 3730 DNA Analyzer (Applied Biosystems).
Results
Clinical phenotype
Clinical findings in families D_SM, L705 and L1014 were consistent with a diagnosis of autosomal recessive non‐syndromic deafness (figs 1 and 2). In each family, affected individuals had prelingual auditory impairment with normal vestibular function as evidenced by age‐appropriate developmental motor milestones and physical tests of balance. No deaf person had evidence of syndromic features. However, based on the genotypic data, sperm motility was assessed in a consenting male in family D_SM. Although sperm counts were normal (78×106/ml) with normal volume (∼4 ml) and colour, malformed sperm (>88%) were observed (mainly thin heads, micro‐ and irregular acrosomes). About 30% of sperm had short, coiled flagella. Less than 5% of sperm had full swimming capacities after liquidation. The reduction in sperm motility and viability was consistent with the diagnosis of asthenoteratozoospermia (fig 1A, table 1).
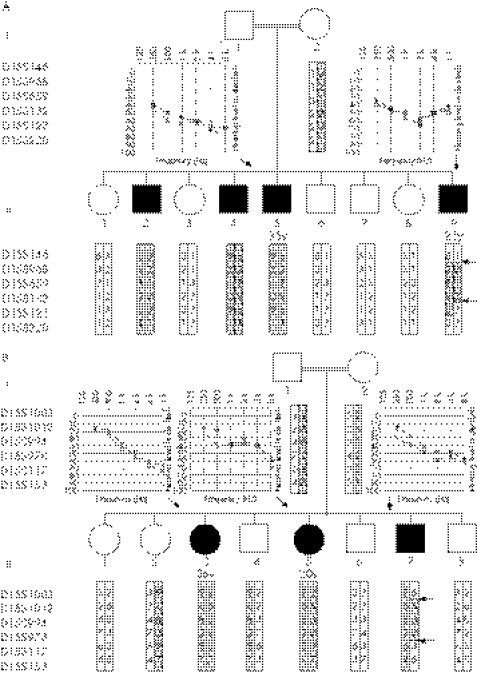
Figure 1 Pedigree of family D_SM (A) and family L705 (B). Haplotypes for markers on 15q15.3 are given. Regions shared by all affected individuals are shaded. Audiograms show moderate‐to‐severe hearing impairment at all frequencies. Normal auditory thresholds are above 25 dB (□, male; О, female; ▪, affected male; diagonal line, deceased; X, left ear; O, right ear).
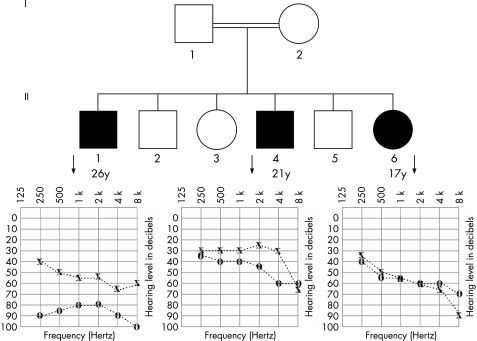
Figure 2 Pedigree of family L1014. Audiograms show moderate‐to‐severe hearing impairment at all frequencies. Normal auditory thresholds are above 25 dB (□, male; О, female; ▪, affected male; diagonal line, deceased; X, left ear; O, right ear).
Table 1 Semen analyses.
| Family D_SM | Family L1014 | Family L1014 | |
|---|---|---|---|
| II9 | II1 | II4 | |
| Age | 23 | 26 | 21 |
| Volume | 4 ml | 2.2 ml | 1 ml |
| Appearance | Normal | Normal | Normal |
| Colour | Normal | Normal | Normal |
| Viscosity | Normal | 3+ | Normal |
| Liquefaction | 30 min | 30 min | 30 min |
| Germinal cell | 1–2 | 3–4 | 3–4 |
| Concentration | 78×106/ml | 73×106/ml | 60×106/ml |
| Normal morphology10 | 11% | 7% | 11% |
| Motility (liquefaction) | 5% | 10% | 15% |
| Abnormal morphology observed | Thin head; irregular micro acrosome; short, coiled tail | Thin irregular head; irregular acrosome; short abnormal tail | Macrohead; irregular and double head; irregular acrosome; short, abnormal tail |
In family L705 (fig 1B), sperm motility was not tested since the only affected male was too young, while in family L1014 sperm analyses were completed on two deaf males based on genotypic data. Both II1 and II4 had normal sperm counts (73×106/ml and 60×106/ml, respectively), although each person had a many malformed sperm with poor motility (table 1), consistent with asthenoteratozoospermia.
These findings are suggestive of male infertility and are supported by the fact that the three affected males in family D_SM (II2, II4 and II5) were unable to father children without intracytoplasmic sperm injection. In family L1014, affected male II1 has been married for >1.5 years and although the couple is not using birth control, his wife has been unable to conceive. We do not know the fertility status for the other affected males (family D_SM, II9; family L1014, II4; family 705, II7), but based on mutation similarity we presume that while they are infertile, intracytoplasmic sperm injection is an option should they wish to father children.
Linkage mapping
In both families D_SM and L705, a genome‐wide screen using short tandem repeat polymorphic (STRP) marker analysis identified a single region of homozygosity by descent in affected persons on 15q15.1–15.3. This region was flanked by D15S968 and D15S132 in family D_SM, defining a 7‐cM region that includes the DFNB16 interval and the gene causally implicated in deafness at this locus, STRC (fig 1A). In family L705, the region of homozygosity by descent was on 15q14–21.1 and flanked by markers D15S1012 and D15S978 (fig 1B).
Genomic deletion
A tandem structure greater than 100 kb is present within the linkage region described above that includes the four genes KIAA0377, CKMT1B, STRC and CATSPER2, three of which have a highly homologous telomeric pseudo‐copy, ΨKIAA0377, ΨSTRC, and ΨCATSPER2. The copy of CKMT1B, CKMT1A, contains only two mismatches that predict synonymous mutations in the coding region in addition to two mismatches in the 3′ UTR. We observed these two synonymous mutations in RT‐PCR amplicons from fetal brain total RNA extracts (data not shown), suggesting that both copies of CKMT1 are functional.
D15S784, a marker present only in the centromeric half of the duplication region, lies between STRC and CATSPER2 and failed to amplify in any affected persons in families D_SM and L705 (data not shown). On that basis, we used D15S784 to screen probands from 300 additional consanguineous families segregating presumed autosomal recessive non‐syndromic deafness and identified a third family, L1014, segregating a deletion in this region (fig 2).
Haplotype analysis
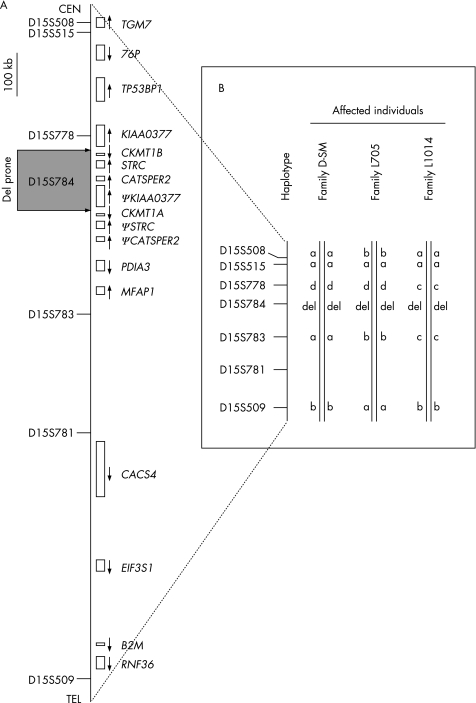
These three families have no apparent biological relationship to one another, although all families are Iranian. We therefore sought to determine whether the disease allele(s) were inherited from a common ancestor by genotyping six STRP markers closest to D15S784 (fig 3A) in each family. Results show that each family segregates a different haplotype, suggesting that this genomic region may be deletion prone (fig 3B).
Figure 3 The linked region showing genes and STRP markers (A) used to define the haplotypes segregating in each family (B).
Fine deletion mapping
To define the deletion breakpoints, we used site‐specific nucleotide dosage analysis. This approach capitalises on minor sequence mismatches, usually single nucleotide substitutions, between the centromeric and telomeric regions of the duplication. In normal controls, primer amplification and sequence analysis will demonstrate these mismatches, but in persons carrying a deletion in this region only one amplicon is generated.
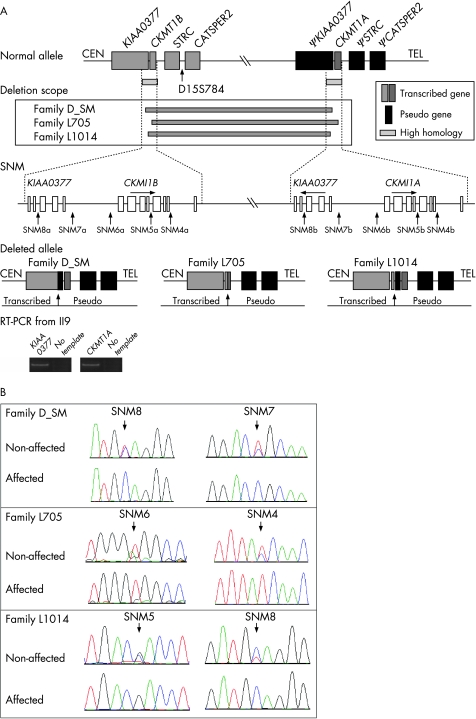
In family D_SM, we narrowed the breakpoints to a 2.5‐kb interval flanked by two critical mismatches, SNM7 and SNM8 (fig 4A, table 2). The absence of additional mismatches in this interval precluded us from narrowing the breakpoint further. As we expected, amplification across the breakpoint region generated PCR products in all persons in family D_SM and in all controls. However by sequencing, all controls demonstrated the presence of nucleotide sequence mismatches SNM7a, SNM7b and SNM8a, SNM8b (fig 4B), indicating the absence of a deletion. Affected persons from family D_SM, in contrast, failed to demonstrate nucleotide sequence mismatches (missing SNM7a and SNM8b; fig 4B) consistent with the presence of a deletion. Total RNA isolated from blood in all four affected individuals in this family showed that only the nucleotide mismatch of CKMT1A was present (fig 4A); in controls, both mismatches of CKMT1A and CKMT1B were expressed. Based on these data, affected individuals in family D_SM are homozygous for a approximately 100‐kb deletion with the proximal breakpoint at the 5′ flank of KIAA0377 (SNM8a‐SNM7a) and the distal breakpoint at the 5′ flank of ΨKIAA0377 (SNM8b‐SNM7b). Included in this deleted region are CATSPER2, STRC and CKMT1B (fig 4A).
Figure 4 The genomic organisation of 15q15.3 includes a duplication with several single nucleotide mismatches, which were used to define the deletions in each family (A). Electropherograms show representative non‐deleted and deleted alleles (B).
Table 2 Single nucleotide mismatches position in non‐pseudo and pseudo region.
| SNM | Location on 15q | Nucleotide | Region |
|---|---|---|---|
| SNM4a | 41676103 | C | Centromeric |
| SNM4b | 41775935 | T | Telomeric |
| SNM5a | 41672406 | G | Centromeric |
| SNM5b | 41772158 | C | Telomeric |
| SNM6a | 41670757 | T | Centromeric |
| SNM6b | 41770509 | A | Telomeric |
| SNM7a | 41665337 | T | Centromeric |
| SNM7b | 41765172 | C | Telomeric |
| SNM8a | 41662746 | C | Centromeric |
| SNM8b | 41762581 | T | Telomeric |
In family L705, we narrowed the breakpoints to the interval flanked by the two mismatches, SNM6 and SNM4 (fig 4A, table 2), which is telomeric to the breakpoint interval in family D_SM. All non‐affected individuals in family L705 show mismatches at SNM6 and SNM4 by sequencing long‐PCR amplicons across this region, while the three affected persons are missing the mismatch of SNM6b and the mismatch of SNM4a (fig 4B). The deletion size in this family is also approximately 100 kb, and extends proximally from CKMT1B (5′ – intron 7; SNM6a‐SNM4a) to CKMT1A distally (5′ – intron 7; SNM6b‐SNM4b). Functional copies of STRC and CATSPER2 are deleted (fig 4A).
In family L1014, we observed the first loss of a single nucleotide mismatch at SNM5a and the last loss at SNM8b in all three patients but not in unaffected individuals (fig 4B) consistent with a approximately 90‐kB deletion starting in the mid portion of CKMT1B and ending 5′ of ΨKIAA0377. The deletion includes CATSPER2 and STRC (fig 4A).
Discussion
Rather than being random events, many rearrangements characterised by non‐allelic homologous recombination reflect genome architecture.11 Abundant repeat elements, for example, lead to large deletions of MECP2 in approximately 25% of cases of classic Rett syndrome. These deletions are facilitated by a region in intron 2 that is highly enriched for Alu repeats.12 Relatively large non‐contiguous duplications also lead to deletions, as in the case of the FVIII related gene A (F8A), which is found in intron 22 of the factor VIII gene and is transcribed in the opposite direction to FVIII. Two further copies of F8A that are 99.9% identical over 8 kb13 are found approximately 500 kb upstream (telomeric) of the FVIII gene and are transcribed in the same direction as FVIII. Intrachromosomal recombination between F8A and either of these copies interrupts the factor VIII gene and underlies about half of all cases of severe haemophilia A.14,15
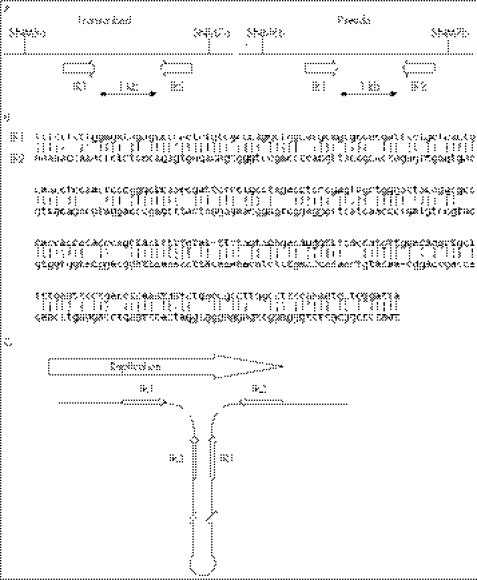
In the region we describe on chromosome 15q15.3, another architectural feature, a large tandem repeat, is present that is prone to rearrangement. A duplicated four‐gene array, which includes KIAA0377 (recently named by HUGO as HISPPD2A, histidine acid phosphatase domain containing 2A), CKMT1B, STRC and CATSPER2 and spans 83 kb, is separated by a 10 kb of intermediate unique sequence which contains a 6‐kb LINE sequence.7 These four genes are syntenically conserved among higher mammalian species, including cow, rat, dog, mouse and chimpanzee (based on Genome BLAST, data not shown), however only the human genome includes a duplication, placing its appearance as a recent evolutionary event. Lower vertebrates also do not have STRC and CATSPER2. A comparison of the two duplicated portions shows a 30‐kb region of high sequence homology (99.97% identical) from KIAA0377 to CKMT1B (30 kb centromeric) and from pseudo‐KIAA0377 to CKMT1A (30 kb telomeric). Using site‐specific nucleotide dosage mapping, we identified a 100‐kb deletion in family D_SM that extended 5′ of centromeric KIAA0377 to 5′ of telomeric pseudo KIAA0377, deleting all of CKMT1B, STRC and CATSPER2 (fig 4A). Within the breakpoints lies a pair of imperfect inverted repeats (fig 5A) which are known to promote secondary structure formation.16 This finding suggests that this region is prone to deletions triggered concordantly by inverted repeat pairing and misalignment due to replication slippage between tandem sequences. As shown in fig 5C, the intervening sequence between the transcribed region and the pseudo region forms a loop during replication, allowing the second inverted repeat element in the transcribed region and the first inverted repeat element in the pseudo region to Watson‐Crick base pair with 76% homology (fig 5B).
Figure 5 A schematic representation of the breakpoint regions in family D_SM (A) showing the predicted alignments of the two inverted repeats, IR1 and IR2, with 76% base pair matching as predicted by EMBOSS (B), and the resultant secondary DNA structure (C).
The disease phenotype associated with these deletions is characterised by deafness and infertility. Deafness‐infertility syndrome (DIS) was described first by Avidan and colleagues7 in a consanguineous family in which three male siblings had deafness (40‐dB hearing loss involving all frequencies) and infertility (asthenoteratozoospermia). In addition to DIS, congenital dyserythropoietic anaemia type I (CDAI) was also recognised in this family due to a second deletion in another genomic region. The DIS phenotype was attributed to a 70‐kb deletion of chromosome 15q15.3 that included the 5′ region of KIAA0377 (exons 1–24), CKMT1B, STRC and the 3′ region of CATSPER2 (exons 12 and 13). In the three families we studied, the phenotypes in affected persons segregated with deletions of chromosome 15q15. One STRP marker in the deleted interval, D15S784, failed to amplify in all families and may be a useful marker to screen for DIS.
Using site‐specific nucleotide dosage mapping, we found that each family segregated a unique deletion, thus implicating four different deletions with DIS. The hearing loss phenotype is similar by audioprofiling to DFNB16‐related hearing loss, suggesting that deletion of STRC is causally related to the deafness in DIS, while deletion of CATSPER2 appears to be responsible for the male infertility. Consistent with this hypothesis, the CatSper2‐/‐ mouse mutant is completely infertile in spite of having a normal sperm count and only slightly decreased swimming capacity because sperm cannot transition to the hyperactivated state required for penetration of the zona pellucida.9
The two other genes in the deletion interval, CKMT1 (creatine mitochondrial kinase 1) and KIAA0377, are both ubiquitously expressed. CKMT1 is present as two transcribed copies, CKMT1A and CKMT1B, which encode the isoenzyme of mitochondrial creatine kinase that is responsible for the transfer of high energy phosphate from mitochondria to creatine in tissues with large fluctuating energy demands. Another isoenzyme in this family is encoded by CKMT2, the expression of which is limited to sarcomeric tissues of heart and skeletal muscle.17 Deleting only two of four copies of CKMT1 does not give an obvious phenotype, presumably due to functional redundancy.
KIAA0377 contains a single histidine acid phosphatase domain with unknown function.7 The large deletions in families L705 and L1014 do not affect the expression of KIAA0377. However in family D_SM, the 5′ portion of KIAA0377 (5′UTR to exon 3) is replaced by its counterpart in ΨKIAA0377. Over the replacement interval, KIAA0377 and ΨKIAA0377 differ by only one nucleotide (intron 2, SMN8), and hence the expression of KIAA0377 would be predicted to be unaltered. We confirmed this prediction by demonstrating KIAA0377 mRNA transcripts in all affected individuals by RT‐PCR amplification and direct sequencing (the primers target exon 3 and exon 5) (fig 4A).
In summary, we have identified three families segregating an autosomal recessive contiguous gene deletion syndrome characterised by deafness and sperm dysmotility. This new syndrome is caused by the deletion of contiguous genes at 15q15.3. The region contains a pair of imperfect inverted repeats, which are known to promote secondary structure formation, and suggests that the deletion is triggered concordantly by inverted repeat pairing and misalignment due to replication slippage between tandem sequences.
Electronic‐database information
The Primer3 website is at http://frodo.wi.mit.edu/ cgi‐bin/primer3/primer3_www.cgi
Acknowledgements
We wish to thank our patients and their families for their participation in this study.
Abbreviations
CDAI - congenital dyserythropoietic anaemia type I
DIS - deafness‐infertility syndrome
PCR - polymerase chain reaction
STRP marker - short tandem repeat polymorphic marker
Footnotes
This work was supported by the Iranian Deputy of Research and Technology, Ministry of Health and Medical Education Grant (P 6193, HN) and the National Institutes of Health (R01‐DC02842, RJHS).
Competing interests: None declared.
References
- 1.Petit C. Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 199614(4)385–391. [DOI] [PubMed] [Google Scholar]
- 2.Smith R J, Bale J F, Jr, White K R. Sensorineural hearing loss in children. Lancet 2005365(9462)879–890. [DOI] [PubMed] [Google Scholar]
- 3.Hussain K, Bitner‐Glindzicz M, Blaydon D, Lindley K J, Thompson D A, Kriss T, Rajput K, Ramadan D G, Al‐Mazidi Z, Cosgrove K E, Dunne M J, Aynsley‐Green A. Infantile hyperinsulinism associated with enteropathy, deafness and renal tubulopathy: clinical manifestations of a syndrome caused by a contiguous gene deletion located on chromosome 11p. J Pediatr Endocrinol Metab 200417(12)1613–1621. [DOI] [PubMed] [Google Scholar]
- 4.Lichtner P, Konig R, Hasegawa T, Van Esch H, Meitinger T, Schuffenhauer S. An HDR (hypoparathyroidism, deafness, renal dysplasia) syndrome locus maps distal to the DiGeorge syndrome region on 10p13/14. J Med Genet 200037(1)33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reish O, Gorlin R J, Hordinsky M, Rest E B, Burke B, Berry S A. Brain anomalies, retardation of mentality and growth, ectodermal dysplasia, skeletal malformations, Hirschsprung disease, ear deformity and deafness, eye hypoplasia, cleft palate, cryptorchidism, and kidney dysplasia/hypoplasia (BRESEK/BRESHECK): new X‐linked syndrome? Am J Med Genet 199768(4)386–390. [DOI] [PubMed] [Google Scholar]
- 6.Merry D E, Lesko J G, Sosnoski D M, Lewis R A, Lubinsky M, Trask B, van den Engh G, Collins F S, Nussbaum R L. Choroideremia and deafness with stapes fixation: a contiguous gene deletion syndrome in Xq21. Am J Hum Genet 198945(4)530–540. [PMC free article] [PubMed] [Google Scholar]
- 7.Avidan N, Tamary H, Dgany O, Cattan D, Pariente A, Thulliez M, Borot N, Moati L, Barthelme A, Shalmon L, Krasnov T, Ben‐Asher E, Olender T, Khen M, Yaniv I, Zaizov R, Shalev H, Delaunay J, Fellous M, Lancet D, Beckmann J S. CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur J Hum Genet 200311(7)497–502. [DOI] [PubMed] [Google Scholar]
- 8.Verpy E, Masmoudi S, Zwaenepoel I, Leibovici M, Hutchin T P, Del Castillo I, Nouaille S, Blanchard S, Laine S, Popot J L, Moreno F, Mueller R F, Petit C. Mutations in a new gene encoding a protein of the hair bundle cause non‐syndromic deafness at the DFNB16 locus. Nat Genet 200129(3)345–349. [DOI] [PubMed] [Google Scholar]
- 9.Quill T A, Ren D, Clapham D E, Garbers D L. A voltage‐gated ion channel expressed specifically in spermatozoa. Proc Natl Acad Sci U S A 200198(22)12527–12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruger T F, Menkveld R, Stander F S, Lombard C J, Van der Merwe J P, van Zyl J A, Smith K. Sperm morphologic features as a prognostic factor in in vitro fertilization. Fertil Steril 198646(6)1118–1123. [DOI] [PubMed] [Google Scholar]
- 11.Stankiewicz P, Inoue K, Bi W, Walz K, Park S S, Kurotaki N, Shaw C J, Fonseca P, Yan J, Lee J A, Khajavi M, Lupski J R. Genomic disorders: genome architecture results in susceptibility to DNA rearrangements causing common human traits. Cold Spring Harb Symp Quant Biol 200368445–454. [DOI] [PubMed] [Google Scholar]
- 12.Laccone F, Junemann I, Whatley S, Morgan R, Butler R, Huppke P, Ravine D. Large deletions of the MECP2 gene detected by gene dosage analysis in patients with Rett syndrome. Hum Mutat 200423(3)234–244. [DOI] [PubMed] [Google Scholar]
- 13.Naylor J A, Buck D, Green P, Williamson H, Bentley D, Giannelli F. Investigation of the factor VIII intron 22 repeated region (int22h) and the associated inversion junctions. Hum Mol Genet 19954(7)1217–1224. [DOI] [PubMed] [Google Scholar]
- 14.Goodeve A C, Preston F E, Peake I R. Factor VIII gene rearrangements in patients with severe haemophilia A. Lancet 1994343(8893)329–330. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins P V, Collins P W, Goldman E, McCraw A, Riddell A, Lee C A, Pasi K J. Analysis of intron 22 inversions of the factor VIII gene in severe hemophilia A: implications for genetic counseling. Blood 199484(7)2197–2201. [PubMed] [Google Scholar]
- 16.Lovett S T. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol Microbiol 200452(5)1243–1253. [DOI] [PubMed] [Google Scholar]
- 17.Payne R M, Strauss A W. Expression of the mitochondrial creatine kinase genes. Mol Cell Biochem 1994133–134235–243. [DOI] [PubMed] [Google Scholar]