Abstract
Background
The phenotypic variability in Beckwith–Wiedemann syndrome (BWS) reflects the genetic heterogeneity of the mechanism which by default leads to the deregulation of genes located at 11p15.5. Genotype–phenotype correlation studies have demonstrated an association between omphalocoele and CDKN1C/p57 mutations or hypermethylation. Paternal uniparental disomy 11 (pUPD11) has been described only in the mosaic condition with both uniparental and biparental cell lines, and no association with omphalocoele has been pointed out.
Methods
Two cases are presented here, in which a paternal segmental UPD11 was detected by molecular investigation of amniotic fluid cell cultures after the presence of apparently isolated omphalocoele was revealed in the fetuses by ultrasound scan. Further studies were performed on additional autoptic feto‐placental tissues to characterise the distribution of the uniparental cell line and to unmask any biparental lineage in order to document in more detail the as yet unreported association between omphalocoele and pUPD11.
Results
Results on the UPD distribution profile showed that the abdominal organs have a predominant uniparental constitution. This condition could mimic the effect of CDKN1C/p57 inactivation, causing the omphalocoele.
Conclusion
New genotype–phenotype correlations emerge from the investigated cases, suggesting that molecular analysis be extended to all cases with fetal omphalocoele in order to establish the incidence of pUPD11 in complete BWS and in monosymptomatic/mild forms.
Beckwith–Wiedeman Syndrome (BWS; OMIM 130650) is a complex overgrowth disorder with an incidence of about 1:13 700 live births, characterised by an increased susceptibility to childhood embryonal malignancies such as Wilms' tumour. Most cases are sporadic and only 15% show an autosomal dominant inheritance. The diagnosis of BWS is usually defined by the presence of at least three clinical findings (two major and one minor). Major signs include macrosomia, macroglossia, anterior linear ear lobe creases/posterior helical ear pits, omphalocoele, umbilical hernia, visceromegaly, embryonal tumours, hemihyperplasia, adrenocortical cytomegaly and renal abnormalities. Minor findings associated with BWS are polyhydramnios, prematurity, neonatal hypoglycaemia, facial nevus flammeus, haemangioma, characteristic facies, cardiac anomalies, diastasis recti and advanced bone age.1,2
An accurate prenatal diagnosis is crucial for accurate genetic counselling, decision of pregnancy termination or a proper mode of delivery and neonatal management. To address this issue, a review of 21 prenatal cases of BWS has recently been published, proposing a guideline for a clinically reliable prenatal diagnosis of this syndrome.3
The genetic heterogeneity of BWS is based on different mechanisms that might deregulate genes located at 11p15.5, a chromosomal region containing two clusters of imprinted genes, which have a role in cell cycle and growth regulation.4 The most telomeric imprinted region (BWSIC1) includes the insulin‐like growth factor 2 (IGF2) and H19; the centromeric cluster (BWSIC2) contains several genes: cyclin‐dependent kinase inhibitor 1C (CDKN1C/p57), potassium voltage‐gated channel, subfamily Q, member 1 (KCNQ1) and KCNQ1‐overlapping transcript 1 (KCNQ1OT1 or LIT1).
The most frequent genetic alteration occurring in patients with BWS is loss of imprinting of the maternally derived allele at the differentially methylated imprinting centre 2: BWSIC2 (∼50%), which is associated with reduction of CDKN1C/p57 expression. Less frequently, the epigenetic alteration occurs at the maternal differentially methylated region 1 leading to gain of methylation and consequent biallelic IGF2 expression: BWSIC1 (2–7%). Other mechanisms responsible for BWS are chromosome rearrangements (1–2% of BWS cases) and CDKN1C/p57 mutations (BWSMUTCDKN1C), accounting for 5–10% of sporadic cases and for 40% of familial cases.2 A further category is represented by mosaicism for paternal isodisomy of 11p15: BWSUPD11 (about 20%). In the vast majority of cases with uniparental disomy (UPD), the uniparental contribution is limited to a short‐arm segment, which results from post‐zygotic interchromatid recombination, establishing a somatic mosaicism in which a cell population with segmental paternal isodisomy coexists with a normal biparental cell line.5 In the presence of low‐level mosaicism, or when the isodisomy is confined to tissues not available for analysis, the genetic defect may be undetected, making this condition underscored. The biparental cell line can mitigate the clinical signs causing a very mild BWS phenotype, which might not fit into the classical diagnostic criteria. Epigenotype/genotype–phenotype correlation studies demonstrated an association between a specific molecular abnormality and the phenotypic presentation of BWS such as hemihypertrophy and hypoglycaemia in cases with BWSUPD11 and abdominal wall defects in patients with CDKN1C/p57 germinal mutations or KCNQ1OT1/LIT1 hypomethylation (BWSIC2 defect). Omphalocoele was never described in cases with UPD reported so far.6,7,8,9,10
Herein, we present two cases in which a paternal segmental UPD11 was detected by molecular investigation of amniotic fluid (AF) cell cultures after the identification of apparently isolated fetal omphalocoele at ultrasound evaluation. Targeted molecular studies were performed on additional fresh and autoptic feto‐placental tissues to characterise the distribution of the uniparental cell line and to unmask the biparental cell line to document in detail the observed novel association of pUPD11 with omphalocoele.
Methods
Patients and procedures
Over a period of 2 years, we received from different public and private centres in the north of Italy a cohort of six pregnancies complicated by omphalocoele detected at routine prenatal ultrasound (US) scan. In each case, AF was collected and at least two cultures were harvested using the “in situ” method for karyotype analysis.
Cytogenetic and fluorescence in situ hybridisation analyses
Karyotype analysis of AF was performed in all six cases, and at least 15 metaphases from >2 cultures were karyotyped. On the basis of the molecular/cytogenetics results, fluorescence in situ hybridization (FISH) analysis was carried out, using the specific probe for the 11p arm telomeric region in accordance with the manufacturer's recommendations (SpectrumGreen probe, Vysis, Downers Grove, Illinois, USA). Normal control samples were also hybridised and 50 metaphases were analysed.
Molecular analysis
The quantitative UPD11 investigation was performed by segregation analysis from parents to fetuses in cases with a normal karyotype.11 DNAs were extracted from maternal and paternal peripheral blood lymphocytes and from amniocytes (AF) using the QIAamp DNA Mini Kit (Qiagen, Chatsworth, California, USA). D11S4046, D11S1338, D11S4146, D11S1363 and D11S1760 short tandem repeat (STR) markers mapping to the BWS critical region were analysed in a cohort of normal subjects (n = 30) to calculate the reference interval of the area ratio between paternal and maternal alleles. The same analysis was performed on the six prenatal samples, and when the ratio values were not included in the reference interval a UPD11 condition was diagnosed. In these cases analysis was extended to additional feto‐placental tissues, by testing seven additional chromosome 11 polymorphisms mapping to the BWS region (D11S2071, D11S922, D11S4088, D11S1923, D11S2345, D11S988 and D11S4181) and seven STRs spanning along 11p and 11q (D11S995, D11S871, D11S986, D11S1303, D11S4191, D11S987 and D11S911) to define the extension of the uniparental segment. DNA obtained from different fresh and paraffin wax‐embedded autoptic tissues was extracted with the MagneSil Genomic Fixed Tissue System (Promega, Madison, USA). Primers specific for the STRs were obtained from Genome Database and/or National Center for Biotechnology Information, together with the PCR conditions. Twenty‐eight cycles of PCR products were run on the fluorescent capillary systems ABI PRISM 310 (ABI, Foster City, California, USA) or SMDX 96‐12 (SpectruMedix LLC, State College, Pennsylvania, USA). Data were analysed using GeneScan Software (ABI) and Genospectrum 2.08 (SMDX). In the case of pUPD, STRs for an additional chromosome were also analysed to assess paternity.
The percentage of the pUPD component was calculated using the formula reported by Bliek et al12: UPD% = [(p−m)/(p + m)]×100% (p and m are the area peaks of the paternal and maternal alleles, respectively).
No additional tests for BWS (ie, CDKN1C mutation analysis) were performed on AF of the fetuses.
Results
Clinical reports
The cohort we collected comprises cases referred for karyotyping by different public and private obstetrics centres in the north of Italy. Since we do not have a direct involvement in the clinical management of pregnancies, we do not have follow‐up data of cases with normal karyotype and without UPD11.
Case 1
A 21‐year‐old pregnant woman with negative medical, surgical and family history had a routine antenatal US scan at the 16th week of gestation (wg), which revealed a female fetus with an apparently isolated omphalocoele; the finding was confirmed at level II US, which failed to detect any associated anomalies. Following detection of a normal fetal karyotype on cultured AF cells, a quantitative UPD11 investigation, performed by segregation analysis from parents to fetus, showed the presence of paternal uniparental disomy of 11p15. The patient underwent extensive counselling, after which she ultimately decided the voluntary interruption of the pregnancy (20 + 4 wg) and gave her consent for an autopsy to be performed. Fresh specimens were obtained from four placental cotyledons (V1–4) and skin (Sk1–3); autoptic biopsy specimens from the fetal heart, lung, kidney, liver, thymus, central nervous system (CNS) and umbilical cord were fixed in formalin and embedded in paraffin wax.
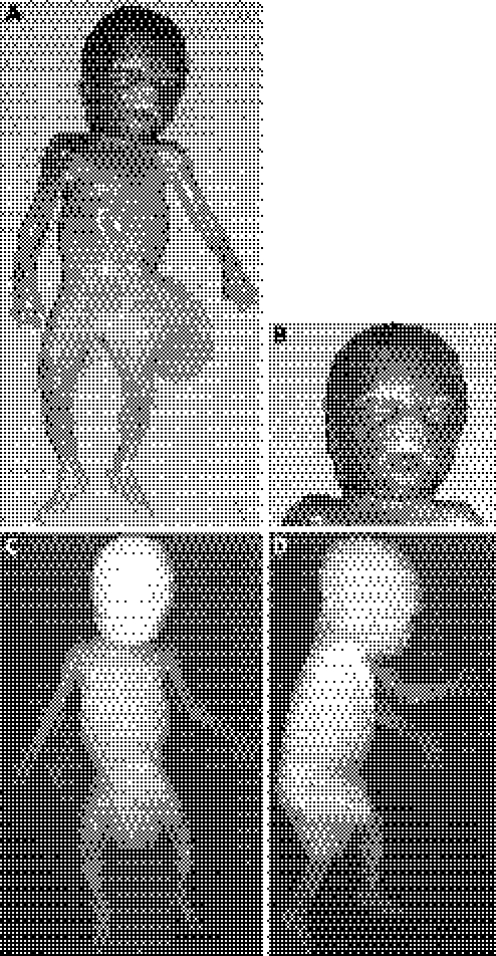
Physical examination revealed an overgrowth condition characterised by a body weight of 440 g (mean 250 g), a vertex–coccyx length of 18 cm (+2 SD = 15.3 cm) and a head circumference of 19.0 cm (mean 17.6 cm). The fetus had facial dysmorphisms (short nose with anteverted nostrils and a long, simple and prominent philtrum, symmetric macroglossia and earlobe creases), body asymmetry with hemihypertrophy of the right body side (right side length of 27.5 cm, left side length of 27.0 cm; right foot length of 4.0 cm, left foot length of 3.6 cm) and omphalocoele (fig 1A, B). At autopsy, cardiac biventricular hypertrophy, hepatomegaly, bilateral nephromegaly and adrenal gland hypertrophy (more evident on the right side) were observed. A histological inspection of the bilateral adrenal gland revealed a cytomegalic cortex with hyperchromatic nuclei and nuclear inclusions. Whole‐body radiography confirmed the skeletal asymmetry of the lower limbs without bone malformations (fig 1C,D). On the basis of such clinical findings, a diagnosis of BWS was confirmed.
Figure 1 Case 1 phenotype. (A, B) Frontal view showing body asymmetry with hemihypertrophy of the right body side, omphalocoele, short nose with anteverted nostrils and a long, simple and prominent philtrum, and macroglossia. (C, D) Frontal and lateral whole‐body radiographic images showing the skeletal asymmetry of the lower limbs without evident skeletal malformations. Parental/guardian informed consent was obtained for publication of this figure.
Case 2
A 28‐year‐old pregnant woman had a routine antenatal US scan at the 16th wg that revealed an omphalocoele. After genetic investigation and counselling, the parents decided to interrupt the pregnancy (18th wg) and gave their consent for an autopsy to be performed. Twenty‐four biopsy specimens taken from the lung (Lu1–2), central nervous system (CNS1–2), adrenal gland (S1–2), pancreas (Pa1–2), umbilical cord (C1–3), bowel (B1–2), uterus (U1–2), tongue (To1–2), heart (H) and placenta (V1–6) were fixed in formalin and embedded in paraffin wax.
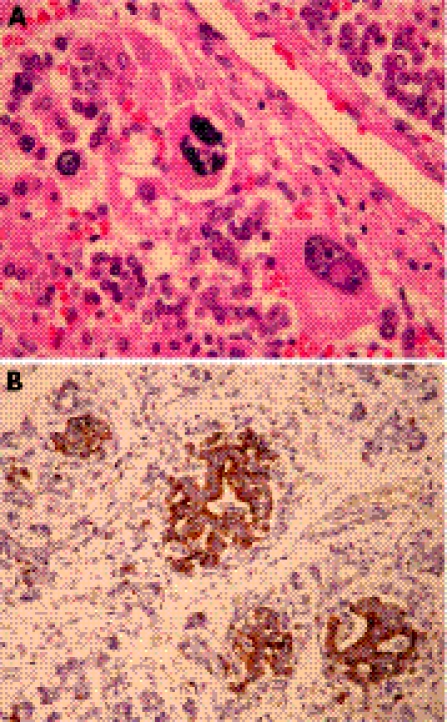
Post mortem examination revealed an overgrowth condition: the fetus weighted 298 g (mean 150 g); vertex–coccyx length 13.0 cm (+1.8 SD); crown–heel length 23.0 cm (+1.0 SD); head circumference 14.0 cm (mean 15.2 cm); chest circumference 14.0 cm (+1.0 SD); abdominal circumference 15.5 cm (>90th centile); humerus length 3.5 cm (95th centile 3.1 cm); femur length 5.0 cm (95th centile 3.2 cm); foot length 3.0 cm (+1.0 SD). At external examination, there was a severe omphalocoele (4.5 cm) containing some of the small‐bowel loops and the extremity of both lobes of the liver. The fetus had a short nose with anteverted nostrils, a long and prominent philtrum, macroglossia and low‐set ears (fig 2A–E). Limbs, genital and chest organs were normal (fig 2F). Placental weight was 94 g (5th centile 135 g), with cotyledons showing reduced growth without gross lesions. A histological inspection of the adrenal gland showed a high degree of cytomegaly of the cortex, and immunohistochemical analysis revealed pancreatic hyperplasia of the islets of Langherhans (fig 3A, B).
Figure 2 Case 2 phenotype. (A–C) Frontal and lateral views showing severe omphalocoele and the dysmorphic features (short nose with anteverted nostrils, a long and prominent philtrum, macroglossia and low‐set ears). (D, E) Omphalocoele containing some of the small‐bowel loops and the extremity of both lobes of the liver. (F) Whole‐body radiography showing the absence of evident skeletal malformations. Parental/guardian informed consent was obtained for publication of this figure.
Figure 3 Histological inspection of the adrenal gland (A) showing a high degree of cytomegaly of the cortex, and immunohistochemical analysis of pancreas (B) revealing hyperplasia of the Langherans islets.
Retrospectively, a clinical diagnosis of BWS was established.
Cytogenetic results
Karyotype analysis demonstrated a normal chromosome complement in five of six cases analysed. In one instance, a 47,XY, +18 karyotype was found in a fetus with omphalocoele and hydrocephalus detected by US scan.
Molecular analysis
Quantitative fluorescent PCR analysis (QF‐PCR) on AF demonstrated an evident abnormal short–to–long (As/Al) peak area ratio in two cases (1 and 2), both with a normal karyotype.
Case 1
Cytogenetic investigation on 18 metaphases revealed a normal female chromosome complement.
FISH analysis using a specific 11p telomeric probe demonstrated a normal fluorescent signal pattern on both homologues (data not shown).
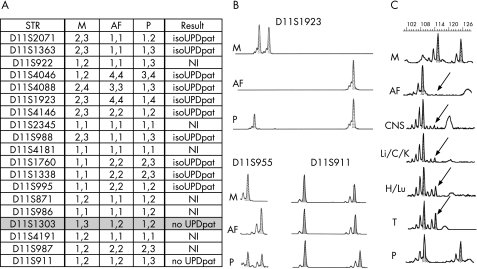
Figure 4A shows the segregation analysis of 12 STRs, mapping to the BWS critical region. The D11S2071, D11S1363, D11S4046, D11S4088, D11S1923, D11S988, D11S1760 and D11S1338 loci showed the presence of the paternal allele only, apparently without any maternal contribution. Among the seven STRs located outside the BWS critical region (11p13–11q23), D11S1303 (11p11.1) excluded the presence of paternal isodisomy and D11S911 showed the presence of a normal biparental contribution. Figure 4B shows the electropherograms of representative loci.
Figure 4 Schematic representation of short tandem repeat (STR) genotyping of case 1. (A) Allelic segregation of the tested polymorphic markers. (B) Electropherograms of D11S1923 and D11S995 STRs included in the uniparental region and of D11S911 mapped in 11q13 showing a normal biparental contribution. (C) Allelic segregation of D11S988 and quantification of the paternal uniparental disomy (pUPD) cell line in different fetal tissues and in alleles from amniotic fluid (AF). The biparental component is represented by the maternal allele (arrow). C, umbilical cord; CNS, central nervous system; H, heart; IsoUPDpat, paternal isodisomy; K, kidney; Li, liver; Lu, lung; M/P, maternal and paternal alleles; NI, not informative; T, thymus.
QF‐PCR analysis using D11S4046, D11S1338, D11S1760 and D11S1363 showed the presence of a variable maternal peak in fetal skin (Sk1–3) and placental (V1–4) DNAs. The normal short‐to‐long (As/Al) area peak ratio ranges (p = 0.05) of the three informative STRs are the following: D11S4046, 1.18 (±0.20); D11S1338 1.13 (±0.08); and D11S1363 1.18 (±0.21). The ratios obtained in the Sk1–3 specimens are all outside the expected range: 0.92–0.87–0.90 (D11S4046), 0.85–0.84–0.88 (D11S1338) and 1.43–1.40–1.46 (D11S1363).
The pUPD component was quantified as 4–30% using the informative STRs. Segregation analysis and area ratio values of V1–4 samples always showed a slight prevalence of the maternal component over the paternal one, suggesting maternal cell contamination (MCC), subsequently demonstrated using further STR mapping on other chromosomes (data not shown).
PCR amplification of AF DNA using a thermal profile with a higher (32–35) number of cycles unmasked the presence of a very low level of maternal/biparental lineage, which could be quantified to 0.8–6.29% using different informative STRs.
The area peak ratio values of all embedded specimens of fetuses were outside the normal range, and the quantification of the pUPD11 cell line at D11S988 identified two groups with different percentages of mosaicism (table 1 and fig 4C): a medium—high level group (76–85%) represented by the kidney, liver, umbilical cord and CNS, and a low‐level group (28–54%) composed of the thymus, heart and lung.
Table 1 Correlation between the percentages of pUDP cells in the panel of investigated fetal tissues and organomegaly.
| Case 1 | Case 2 | |||
|---|---|---|---|---|
| Enlarged | % UPD* | Enlarged | % UPD* | |
| Amniotic fluid | NA | 95 | NA | 85 |
| Placenta | — | MCC | reduced | MCC |
| Skin† | NA | 4–15–30 | NA | |
| Heart | + | 54 | — | 37 |
| Lung | – | 54 | — | 45–45 |
| Kidney | + | 76 | ND | |
| Liver | + | 78 | ND | |
| Thymus | – | 28 | ND | |
| CNS | NA | 85 | NA | 27–11 |
| Umbilical cord | NA | 78 | NA | 75–75–38 |
| Adrenal gland | + | + | 77–82 | |
| Bowel | ND | ND | 72–77 | |
| Tongue | + | + | 78 | |
| Uterus | ND | – | 48–37 | |
| Pancreas | ND | + | 70–70 | |
+, present; –, absent; CNS, central nervous system; MCC, maternal cell contamination; NA, not applicable; ND, not determined; UPD, uniparental disomy.
*Multiple values are reported in tissues for which more than one specimen was analysed.
†Not in figure 5.
Case 2
A normal 46,XX karyotype was found. FISH, by means of an 11p telomeric probe, evidenced the expected signals on both 11 homologues, without any additional signal at an atypical site.
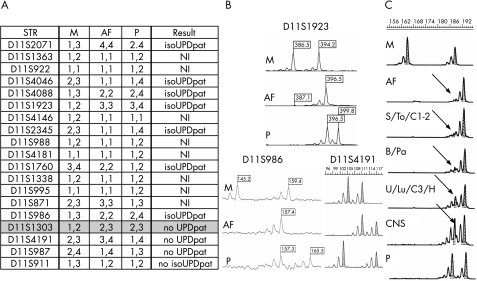
Figure 5A summarises the segregation analysis of 12 STRs mapping in the BWS‐specific region (11p15.5). The polymorphic markers D11S2071, D11S4046, D11S4088, D11S1923, D11S2345 and D11S1760 showed the presence of the paternal allele with a low level (15%) of the biparental component. Segregation analysis of the polymorphic loci located outside the BWS‐specific region and along the q arm showed the presence of a normal biparental contribution at the D11S1303 (11p11.1) and D11S4191 (11q12) loci. Figure 5B shows the representative electropherograms.
Figure 5 Schematic representation of case 2 short tandem repeat (STR) analysis. (A) Allelic segregation of the tested polymorphic markers. (B) Electropherograms of D11S1923 and D11S986 STRs included in the uniparental region and of D11S4191 mapped in 11q12 showing a normal biparental contribution. (C) Allelic segregation of D11S2071 and quantification of the biparental cell line in different fetal tissues and in alleles from amniotic fluid (AF). The paternal uniparental disomy (pUPD) component is represented by the maternal allele (arrow). B, bowel; C, umbilical cord; CNS, central nervous system; H, heart; isoUPDpat, paternal isodisomy; K, kidney; Lu, lung; M/P, maternal and paternal alleles; NI, not informative; Pa, pancreas; S, adrenal gland; To, tongue; U, uterus.
QF‐PCR analysis using the D11S4046 marker showed a maternal peak with variable ratios on DNAs from all 24 embedded tissue biopsy specimens. The area peak ratio values of all fetal specimens were outside the normal range (from 1.49 in CNS2 to 12.54 in S1 for D11S4046). Segregation analysis of parental alleles in the placental samples always showed borderline values owing to the presence of a low‐level MCC unmasked by the presence of the second maternal allele in the electropherogram (data not shown). Quantification of the parental alleles in the fetal tissues at D11S2071 and D11S4046 loci showed that the pUPD cell line ranged from a medium–high level (70–82%) in the pancreas (70% in both specimens), umbilical cord (75% in both specimens), bowel (72% and 77%), tongue (78%) and adrenal glands (77% and 82%) to a low level (27–48%) in the heart (37%), lung (45%), CNS (11% and 27%), umbilical cord (specimen 3: 38%) and uterus (37–48%; table 1 and fig 5C).
Discussion
BWS is a complex disorder in which different molecular subgroups can be correlated with specific phenotypic presentations.6,7,8,9,10 In this paper, we describe two cases in which the antenatal US revealed the presence of an apparently isolated omphalocoele. This is one of the major symptoms of BWS; other possible prenatal findings are polydramnios, enlarged placenta, fetal overgrowth, distended abdomen, visceromegaly and macroglossia. Most of them become evident by ultrasound after 21 wg, as reported in the literature.3,13,14,15,16,17,18,19 Indeed, the only descriptions of prenatal diagnosis of BWS before 20 wg concerned familial cases.20,21 In our cases, none of the additional reported findings was revealed by ultrasound; nevertheless, owing to the early gestational week, this was not considered sufficient to rule out the suspected diagnosis confidently, and the presence of pUPD of the 11p15.5 region was primarily investigated on amniocyte DNA. Quantitative segregation analysis using a panel of BWS‐specific polymorphic loci demonstrated paternal isodisomy encompassing the BWS critical region in both cases.10
Unlike previous studies, the cases with BWSUPD11 described here demonstrate the atypical prenatal occurrence of omphalocoele, which is a recurrent clinical finding of postnatally diagnosed BWSMUTCDKN1C.6,7,8,9,10,22 Paternal isodisomy was homogeneous, apparently without a discernible biparental cell line in case 1. These findings induced us to investigate the extension of the uniparental chromosome segment and the presence and distribution of the pUPD cell line in order to document this peculiar phenotype–genotype association.
Only three cases with complete pUPD of chromosome 11 have been reported and their meiotic origin has been suggested.23,24,25 In line with a previous study investigating the extent of paternal disomy, segregation analysis of STRs mapping along chromosome 11 demonstrated the presence of segmental UPD extending onto the long arm in both our investigated cases, confirming that somatic recombination is the mechanism underlying the two reported BWSUPD11 cases.5,26
Samples from different fetal tissues were analysed to investigate the presence of the biparental cell line and assess its relative frequency. The results showed a peculiar distribution profile in which the abdominal organs (adrenal gland, bowel, pancreas, liver and kidney) displayed a predominance of uniparental constitution (70–82%), with respect to other anatomical sites. Interestingly, this distribution is superimposable on that of the organs mainly involved in the omphalocoele. In addition, other fetal tissues (tongue, CNS and umbilical cord) showed a high level (74–85%) of the pUPD11 cell line. To interpret these findings, we hypothesise that the mitotic recombination event might have occurred at a very early stage of embryonic development; as a consequence, the pUPD11 cell line colonised most fetal tissues owing to the selective proliferative advantage of the uniparental cell progenitor. Such a genetic constitution may justify the severe phenotype observed in both cases, characterised by a constellation of major and minor clinical findings. The reduction of the maternal H19 mRNA, combined with the increased dosage of the paternal IGF2 product, might account for the overgrowth, whereas mosaicism of normal and pUPD11 cells might explain the asymmetrical growth and hemihypertrophy.27
Itoh et al28 provided data on a correlation between the proportion of pUPD cells and enlarged organs. In accordance with their data, we found that a high proportion of isodisomic cells correlates with organomegaly: hyperplastic kidney and liver (case 1), adrenal gland, bowel, tongue and pancreas (case 2) all had a medium–high level of the pUPD component (⩾70%); in contrast, organs with normal growth (thymus of case 1 and heart, lung and uterus of case 2) demonstrated a lower proportion of UPD cells (<50%). In case 1, organ enlargement and hemihypertrophy of the right side of the body did not coincide (nephromegaly was bilateral). A possible explanation for the different levels of isodisomic cells in the different organs might include a combination of events such as the timing and the cell progenitor that is involved in the somatic interchromatid recombination and the proliferative selection of pUPD cells within a single organ.
Lam et al12 pointed out a contradiction in the phenotypic expression in BWSUPD11cases: in the presence of such a molecular condition, a reduction in CDKN1C expression is expected that consequently leads to the associated omphalocoele. Consistent with this prediction, the presence of the paternal isodisomic cell line at high level in the abdominal organs could have mimicked the effect of CDKN1C/p57 inactivating mutations or hypermethylation causing the omphalocoele. Similarly, it has been demonstrated that somatic mosaicism for 11p15 limited to pancreas is the mechanism causing persistent hyperinsulinaemic hypoglycaemia.29
In our two cases, segregation analysis on amniocytes demonstrated an evident and definite UPD11 condition, possibly because the presence of omphalocoele in the fetus might have enriched the AF with cells derived from abdominal organs. In addition, during the AF culture the pUPD11 cell line might have a proliferative advantage due to the autocrine effect of IGF2 getting the upper hand over the normal biparental cell line, which is found at a very low level, and leading to the unmasking of the UPD cell line. This hypothesis can explain the higher percentage of isodisomic cells detected in AF than in the different abdominal organs. On this basis, other tissues such as chorionic villi are not similarly suitable for prenatal diagnosis purposes. This view is also supported by the fact that the only case of pUPD11 previously reported in the literature was detected in cultured AF from a fetus with normal karyotype and BWS phenotype (not omphalocoele).19 In addition, allele area ratio values in chorionic villi could be included in the normal range, possibly owing to the presence of MCC and/or to the confinement of the UPD11 cell line in fetal tissues without placental involvement. This observation suggests that AF cell cultures could be the preferential prenatal sample for BWSUPD11 testing.
In conclusion, on the basis of our genetic and pathological findings, we believe that omphalocoele could be associated with this specific BWS molecular subtype. Recently, a case of giant omphalocoele and “prune belly” sequence in a prenatal case of BWS was investigated only for KCNQ1OT1/LIT1 promoter methylation analysis (and not H19) in fetal liver DNA showing 10% of residual methylation.30 This finding does not exclude the presence of a somatic mosaic of UPD11 (90%) and normal (10%) cell lines, even in a pattern confined to a single organ. Investigations of UPD11 in abdominal tissues from patients with BWS with omphalocoele and negative for CDKN1C mutations might strengthen the importance of UPD11 tissue specificity in the BWS phenotype.
Abbreviations
AF - amniotic fluid
BWS - Beckwith–Wiedemann syndrome
CNS - central nervous system
CDKN1C - cyclin‐dependent kinase inhibitor 1C
FISH - fluorescence in situ hybridisation
KCNQ1 - potassium voltage‐gated channel, subfamily Q, member 1
IGF2 - insulin‐like growth factor 2
MCC - maternal cell contamination
pUPD - paternal uniparental disomy
QF‐PCR - quantitative fluorescent PCR
STR - short tandem repeat
US - ultrasound
wg - weeks of gestation
Footnotes
Competing interests: None declared.
Parental/guardian informed consent was obtained for publication of figures 1 and 2.
References
- 1.Elliott M, Maher E R. Beckwith‐Wiedemann syndrome. J Med Genet 199431560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weksberg R, Shuman C, Smith A C. Beckwith‐Wiedemann syndrome. Am J Med Genet C 200513712–23. [DOI] [PubMed] [Google Scholar]
- 3.Williams D H, Gauthier D W, Maizels M. Prenatal diagnosis of Beckwith‐Wiedemann syndrome. Prenat Diagn 200525879–884. [DOI] [PubMed] [Google Scholar]
- 4.Maher E R, Reik W. Beckwith‐Wiedemann syndrome: imprinting in clusters revisited. J Clin Invest 2000105247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henry I, Bonaiti‐Pellie C, Chehensse V, Beldjord C, Schwartz C, Utermann G, Junien C. Uniparental paternal disomy in a genetic cancer‐predisposing syndrome. Nature 1991351665–667. [DOI] [PubMed] [Google Scholar]
- 6.Cooper W N, Luharia A, Evans G A, Raza H, Haire A C, Grundy R, Bowdin S C, Riccio A, Sebastio G, Bliek J, Schofield P N, Reik W, Macdonald F, Maher E R. Molecular subtypes and phenotypic expression of Beckwith‐Wiedemann syndrome. Eur J Hum Genet 2005131025–1032. [DOI] [PubMed] [Google Scholar]
- 7.Bliek J, Gicquel C, Maas S, Gaston V, Le Bouc Y, Mannens M. Epigenotyping as a tool for the prediction of tumor risk and tumor type in patients with Beckwith‐Wiedemann syndrome (BWS). J Pediatr 2004145796–799. [DOI] [PubMed] [Google Scholar]
- 8.DeBaun M R, Niemitz E L, McNeil D E, Brandenburg S A, Lee M P, Feinberg A P. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith‐Wiedemann syndrome with cancer and birth defects. Am J Hum Genet 200270604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaston V, Le Bouc Y, Soupre V, Burglen L, Donadieu J, Oro H, Audry G, Vazquez M P, Gicquel C. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith‐Wiedemann syndrome. Eur J Hum Genet 20019409–418. [DOI] [PubMed] [Google Scholar]
- 10.Engel J R, Smallwood A, Harper A, Higgins M J, Oshimura M, Reik W, Schofield P N, Maher E R. Epigenotype‐phenotype correlations in Beckwith‐Wiedemann syndrome. J Med Genet 200037921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo S, Mencarelli M, Cavalleri F, Selicorni A, Cogliati F, Larizza L. A fluorescent method for detecting low‐grade 11patUPD mosaicism in Beckwith‐Wiedemann syndrome. Mol Cell Probes 200317295–299. [DOI] [PubMed] [Google Scholar]
- 12.Bliek J, Maas S M, Ruijter J M, Hennekam R C, Alders M, Westerveld A, Mannens M M. Increased tumour risk for BWS patients correlates with aberrant H19 and not KCNQ1OT1 methylation: occurrence of KCNQ1OT1 hypomethylation in familial cases of BWS. Hum Mol Genet 200110467–476. [DOI] [PubMed] [Google Scholar]
- 13.Mulik V, Wellesley D, Sawdy R, Howe D T. Unusual prenatal presentation of Beckwith‐Wiedemann syndrome. Prenat Diagn 200424501–503. [DOI] [PubMed] [Google Scholar]
- 14.Lodeiro J G, Byers JW I I I, Chuipek S, Feinstein S J. Prenatal diagnosis and perinatal management of the Beckwith‐Wiedemann syndrome: a case and review. Am J Perinatol 19896446–449. [DOI] [PubMed] [Google Scholar]
- 15.Wieacker P, Wilhelm C, Greiner P, Schillinger H. Prenatal diagnosis of Wiedemann‐Beckwith syndrome. J Perinat Med 198917351–355. [DOI] [PubMed] [Google Scholar]
- 16.Cobellis G, Iannoto P, Stabile M, Lonardo F, Della Bruna M, Caliendo E, Ventruto V. Prenatal ultrasound diagnosis of macroglossia in the Wiedemann‐Beckwith syndrome. Prenat Diagn 1988879–81. [DOI] [PubMed] [Google Scholar]
- 17.Fremond B, Poulain P, Odent S, Milon J, Treguier C, Babut M J. Prenatal detection of a congenital pancreatic cyst and Beckwith‐Wiedemann syndrome. Prenat Diagn 199717276–280. [PubMed] [Google Scholar]
- 18.Nowotny T, Bollmann R, Pfeifer L, Windt E. Beckwith‐Wiedemann syndrome: difficulties with prenatal diagnosis. Fetal Diagn Ther 19949256–260. [DOI] [PubMed] [Google Scholar]
- 19.Reish O, Lerer I, Amiel A, Heyman E, Herman A, Dolfin T, Abeliovich D. Wiedemann‐Beckwith syndrome: further prenatal characterization of the condition. Am J Med Genet 2002107209–213. [DOI] [PubMed] [Google Scholar]
- 20.Winter S C, Curry C J, Smith J C, Kassel S, Miller L, Andrea J. Prenatal diagnosis of Beckwith‐Wiedemann syndrome. Am J Med Genet 198624137–141. [DOI] [PubMed] [Google Scholar]
- 21.Viljoen D L, Jaquire Z, Woods D L. Prenatal diagnosis in autosomal dominant Beckwith‐Wiedemann syndrome. Prenat Diagn 199111167–175. [DOI] [PubMed] [Google Scholar]
- 22.Lam W W, Hatada I, Ohishi S, Mukai T, Joyce J A, Cole T R, Donnai D, Reik W, Schofield P N, Maher E R. Analysis of germline CDKN1C (p57KIP2) mutations and sporadic Beckwith‐Wiedemann syndrome (BWS) provides a novel genotype‐phenotype correlation. J Med Genet 199936518–523. [PMC free article] [PubMed] [Google Scholar]
- 23.Grundy P, Telzerow P, Paterson M C, Haber D, Berman B, Li F, Garber J. Chromosome 11 uniparental isodisomy predisposing to embryonal neoplasm [letter]. Lancet 19913381079–1080. [DOI] [PubMed] [Google Scholar]
- 24.Webb A, Beard J, Wright C, Robson S, Wolstenholme J, Goodship J. A case of paternal uniparental disomy for chromosome 11. Prenat Diagn 199515773–777. [DOI] [PubMed] [Google Scholar]
- 25.Dutly F, Baumer A, Kayserili H, Yuksel‐Apak M, Zerova T, Hebisch G, Schinzel A. Seven cases of Beckwith‐Wiedemann syndrome, including the first reported case of mosaic paternal isodisomy along the whole chromosome 11. Am J Med Genet 199879347–353. [DOI] [PubMed] [Google Scholar]
- 26.Catchpoole D, Lam W W K, Valler D, Temple I K, Joyce J A, Reik W, Scholfield P N, Maher E R. Epigenetic modification and uniparental inheritance of H19 in Beckwith‐Wiedemann syndrome. J Med Genet 199734353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slater H, Shaw J H, Dawson G, Bankier A, Forrest S M. Mosaic uniparental disomy in Beckwith‐Wiedemann syndrome. J Med Genet 199431749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh N, Becroft D M, Reeve A E, Morison I M. Proportion of cells with paternal 11p15 uniparental disomy correlates with organ enlargement in Wiedemann‐Beckwith syndrome. Am J Med Genet 200092111–116. [PubMed] [Google Scholar]
- 29.de Lonlay P, Fournet J C, Rahier J, Gross‐Morand M S, Poggi‐Travert F, Foussier V, Bonnefont J P, Brusset M C, Brunelle F, Robert J J, Nihoul‐Fekete C, Saudubray J M, Junien C. Somatic deletion of the imprinted 11p15 region in sporadic persistent hyperinsulinemic hypoglycemia of infancy is specific of focal adenomatous hyperplasia and endorses partial pancreatectomy. J Clin Invest 1997100802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinico M, Touboul C, Haddad B, Encha‐Razavi F, Paniel J B, Gicquel C, Gerard‐Blanluet M. Giant omphalocoele and “prune belly” sequence as components of the Beckwith‐Wiedemann syndrome. Am J Med Genet A 2004129198–200. [DOI] [PubMed] [Google Scholar]