Abstract
Background
Fanconi anaemia is a heterogeneous genetic disease, where 12 complementation groups have been already described. Identifying the complementation group in patients with Fanconi anaemia constitutes a direct procedure to confirm the diagnosis of the disease and is required for the recruitment of these patients in gene therapy trials.
Objective
To determine the subtype of Fanconi anaemia patients in Spain, a Mediterranean country with a relatively high population (23%) of Fanconi anaemia patients belonging to the gypsy race.
Methods
Most patients could be subtyped by retroviral complementation approaches in peripheral blood T cells, although some mosaic patients were subtyped in cultured skin fibroblasts. Other approaches, mainly based on western blot analysis and generation of nuclear RAD51 and FANCJ foci, were required for the subtyping of a minor number of patients.
Results and conclusions
From a total of 125 patients included in the Registry of Fanconi Anaemia, samples from 102 patients were available for subtyping analyses. In 89 cases the subtype could be determined and in 8 cases exclusions of common complementation groups were made. Compared with other international studies, a skewed distribution of complementation groups was observed in Spain, where 80% of the families belonged to the Fanconi anaemia group A (FA‐A) complementation group. The high proportion of gypsy patients, all of them FA‐A, and the absence of patients with FA‐C account for this characteristic distribution of complementation groups.
Fanconi anaemia is a rare hereditary recessive disease characterised by developmental abnormalities, bone marrow failure and predisposition to cancer, mainly acute myeloid leukaemia.1 To date, 12 complementation groups have been reported (FA‐A, B, C, D1, D2, E, F, G, I, J, L and M) and 11 associated genes have already been identified: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG/XRCC9, BRIP1/FANCJ, FANCL and FANCM/Hef.2,3,4,5,6,7,8,9,10,11,12,13,14 In dividing cells or in cells exposed to DNA damage, eight Fanconi anaemia proteins (FANCA/B/C/E/F/G/L/M) form a Fanconi anaemia core complex, necessary for the monoubiquitination of FANCD2.2,5,15 In contrast with these Fanconi anaemia proteins, FANCD1 and FANCJ are not involved in FANCD2 monoubiquitination, indicating that these proteins participate downstream of FANCD2 in the FA/BRCA pathway.2,7
Because of the overlap in the phenotype and molecular pathways between the different chromosome fragility syndromes, Fanconi anaemia subtyping constitutes an invaluable approach to confirm the diagnosis of the disease.16,17 Additionally, in the case of patients with FAD1, subtyping analysis allows the identification of BRCA2 mutation carriers, characterised by an increased risk of developing breast, ovarian and other types of cancers.18 Fanconi anaemia subtyping also facilitates mutation screening studies and therefore the identification of mutations with particular pathogenic effects. In addition to the above‐mentioned applications, subtyping is essential before enrolling a patient with Fanconi anaemia in a gene therapy trial.
Progress in the cloning of Fanconi anaemia genes enabled the identification of mutations in specific Fanconi anaemia genes by means of DNA sequencing approaches or other methods.19 The large number and complexity of some Fanconi anaemia genes and their mutations, together with the necessity of verifying the pathogenicity of each new mutation, implies that subtyping of patients with Fanconi anaemia by mutational analysis is often time consuming and laborious. The possibility of reverting the phenotype of Fanconi anaemia cells by the transfer of functional Fanconi anaemia genes has been recently proposed as an efficient approach for identifying the pathogenic genes that account for the disease in patients with Fanconi anaemia.20,21 A different Fanconi anaemia subtyping approach is based on the western blot analysis of FANCD2.22 By means of the observation of the ubiquitinated (FANCD2‐L) and non‐ubiquitinated (FANCD2‐S) forms of the protein FANCD2, it is possible to predict pathogenic mutations in proteins upstream or downstream of FANCD2.23 In the case of patients belonging to rare complementation groups such as FAD1 or FA‐J, approaches based on the formation of RAD51 or BRIP1 nuclear foci are also highly informative in identifying their complementation group.24
With the purpose of determining the prevalence of the different Fanconi anaemia complementation groups in Spain, we conducted an extensive subtyping study of Fanconi anaemia in this Mediterranean country. In addition to a predominantly caucasian population, a relatively large population of about 500 000 gypsies also live in Spain. In this population, the incidence of recessive syndromes is high, owing to the high rates of consanguinity.25 This study will allow us to identify potential differences in the distribution of Fanconi anaemia subtypes due to geographical and ethnic characteristics, and will also allow us to conduct further mutation studies within the population of patients subtyped for Fanconi anaemia. Additionally, our subtyping study will facilitate the enrolling of patients with Fanconi anaemia in clinical gene therapy trials aimed at the genetic correction of their haematopoietic stem cells.
Methods
Patients, chromosome breakage tests, lymphoblast cell lines and skin fibroblasts
The national registry of patients with Fanconi anaemia from Spain was created in 1998. Currently the registry includes 125 patients. Patients were coded to protect their confidentiality, and informed consent was obtained from the patients or their relatives. Patients with Fanconi anaemia were diagnosed on the basis of clinical symptoms and positive results from chromosome breakage tests using a DNA cross‐linker drug.26 Fresh peripheral blood lymphocytes were stimulated with phytohaemagglutinin for 24 h and further incubated with or without diepoxybutane (DEB) for 48 h. Aberrant metaphases were defined by the presence of chromosomal breakages, gaps or radial chromosomes.26 Epstein–Barr virus‐transformed lymphoblast cell lines (LCLs) were generated from peripheral blood cells of healthy donors and patients with Fanconi anaemia, and maintained in RPMI medium (Gibco, Grand Island, NY, USA) supplemented with 20% fetal bovine serum (FBS; Gibco) and antibiotics. To generate fibroblast cells, a skin biopsy specimen was obtained, and then mechanically fragmented and incubated in Dulbecco's modified Eagle's medium (DMEM; Gibco) with collagenase 0.25% (Sigma, St Louis, MO, USA) for 12 h. Cells were centrifuged and cultured for 2–3 weeks in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% FBS (Gibco) at 37°C and 5% CO2.
Subtyping by complementation studies with retroviral vectors
Retroviral subtyping on blood T cells was based on the reversion of the mitomycin C (MMC) hypersensitivity of Fanconi anaemia cells mediated by the transfer of complementary Fanconi anaemia genes, using retroviral vectors as described previously.20 Mononuclear cells from heparinised peripheral blood were stimulated in plates coated (overnight at 4°C) with 30 μg/ml of purified anti‐human CD3 (OKT3 Ortho Biotech, Bridgewater, NJ, USA) and anti‐CD28 (Pharmingen, San Diego, California, US) monoclonal antibodies. After 5–6 days, proliferating T cells were placed in fibronectin‐coated wells (2 μg/cm2 CH‐296; Takara Shuzo, Otsu, Japan) pre‐loaded with retroviral supernatants. Two additional infections of 24 h in the presence of interleukin (IL)‐2 (100 IU/ml) were performed to achieve high percentages of cell transductions. After the second infection, cells were collected and exposed to increasing concentrations of MMC (0–1000 nmol/l) in fresh medium with IL‐2. Five days later, cells were resuspended in phophate‐buffered saline (PBS)–bovine serum albumin (BSA; 0.05%) containing 0.5 μg/ml propidium iodide (Sigma) and incubated for 10 min at 4°C. Cell viability was determined by flow cytometry (EPICS, Coulter Electronics, Hialeah, Florida, USA) based on the propidium iodide exclusion test, using a dot blot of forward scatter versus propidium iodide fluorescence. The following retroviral vectors were used: LFAPEG (FANCA+EGFP) and SFA (FANCA), LFCPEG (FANCC+EGFP), LFGPEG (FANCG+EGFP), S11FEIEG (FANCE+EGFP), LFFIEG (FANCF+EGFP), S11FD2IN (FANCD2+Neor) and a control LPEG (EGFP).20,27 Retroviral complementation was considered to have occurred if survivals to at least three different concentrations of MMC increased >20% as a result of the transduction of cells with one Fanconi anaemia vector, compared with viabilities obtained in cells treated under identical conditions, but transduced with the control vector. When EGFP‐retroviral vectors were used, the efficacy of transduction was also monitored by flow cytometry.
In mosaic Fanconi anaemia patients retroviral complementation was routinely conducted with a control retroviral vector expressing the neomicine phosphotransferase (Neor) gene (S11IN) and a vector coexpressing FANCA and Neor (S11FAIN) on skin fibroblasts subjected to G‐418 selection. In this case, the reversion in the G2/M arrest on exposure to MMC was used as the biological end point.21 Retroviral complementation was considered to have occurred if the percentage of MMC‐treated cells in G2/M was at least 15% lower in cells transduced with the complementing FA‐vector, compared with the G2/M value obtained in cells treated under identical conditions, but transduced with the control vector. The cell cycle was analysed by flow cytometry, and linear fluorescence emission of propidium iodide from 10 000 events was collected with doublet discrimination. Analysis of the cell cycle was performed using MODFIT‐LT (Verity Software House, Topsham, Maine, USA).
Western blot analyses
Western blot analyses were performed using extracts of proliferating LCLs. Two million cells per sample were collected by centrifugation, washed twice in PBS, lysed in 1× lysis buffer (50 nM TRIS‐HCl, 70 mM 2‐mercaptoethanol and 2% sodium dodecylsulphate), boiled for 5 min and subjected to 7.5% polyacrylamide sodium dodecylsulphate gel electrophoresis. After electrophoresis, proteins were transferred to a nitrocellulose membrane using a submerged transfer apparatus (BioRad, Hercules, CA, USA), filled with 25 mM TRIS Base, 200 mM glycine and 20% methanol. After blocking with 5% non‐fat dried milk in TRIS‐buffered saline Tween‐20 (50 mM Tris‐HCl (pH 8), 150 mM NaCl and 0.1% Tween 20), the membrane was incubated with primary antibodies (anti‐FANCD2, SantaCruz Biotechnology, Santa Cruz, California, USA; Ab‐1 anti‐BRCA2, Oncogene Research Products; or anti‐BRIP1, Novus Biologicals, Littleton, Colorado, USA) diluted in TRIS‐buffered saline Tween‐20, washed extensively and incubated with the appropriate horseradish peroxidase‐linked secondary antibody (Amersham Biosciences, Piscataway, NJ, USA). Detection was performed with the Western Breeze Immunodetection Kit (Invitrogen, Grand Island, NY, USA). When necessary, nuclear extracts were collected by the Nuclear Extraction Kit (Chemicon, Temecula, CA, USA). The concentration of protein was measured by the Bradford assay.
Immunofluorescence studies of RAD51 and BRIP1 foci
For immunofluorescence studies, cells were fixed with 3.7% paraformaldehyde in PBS for 15 min followed by permeabilisation with 0.5% Triton X‐100 in PBS for 5 min. After blocking for 30 min in blocking buffer (10% FBS, 0.1% NP‐40 in PBS), cells were incubated with either polyclonal rabbit anti‐RAD51 antibody (Calbiochem, La Jolla, California, USA) or polyclonal rabbit anti‐BRIP1 antibody (Sigma‐Aldrich, St Louis, Missouri, USA). Cells were subsequently washed three times in TBS and incubated with anti‐rabbit Alexa 488 nm and Alexa 594 (Molecular Probes, Leiden, The Netherlands) as secondary antibodies. Alternatively, anti‐rabbit Texas red‐conjugated antibody (Jackson Immunoresearch Laboratories, Cambridgeshire, California, USA) was used. After 45 min, cells were washed three times with TBS and the slides were mounted in Vectashield (Vector Laboratories, Burlingame, California, USA) with 4,6‐diamidino‐2phenylindole.
Results and discussion
Response of peripheral blood T cells from healthy donors and patients with Fanconi anaemia to DEB and MMC
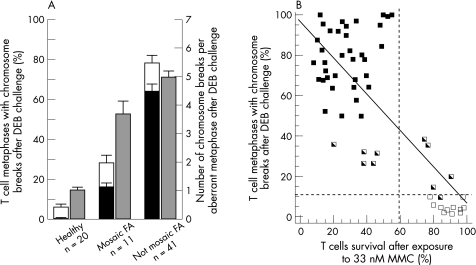
Cytogenetic analyses allowed us to determine the proportion of metaphases with single and multiple chromosome breaks in a population of 72 patients with Fanconi anaemia and healthy donors whose T cells were exposed to DEB (fig 1A). In contrast with results obtained from healthy donors, a high proportion of metaphases from patients with typical (non‐mosaic) Fanconi anaemia contained single or multiple breaks after DEB challenge (mean (SD): 6.40% (1.13%) vs 77.8% (2.4%), respectively). As observed in previous studies,23,28,29,30 samples from a relatively reduced number of patients showed intermediate proportions of T cell metaphases with chromosome aberrations after DEB challenge, thus complicating the diagnosis of these patients. In these cases, the presence of metaphases with ⩾2 breaks was most helpful for the diagnosis. Although only 0.70% (0.32%) of metaphases from healthy donors showed more than one break, 16.3% (2.1%) and 63.9% (3.1%) metaphases from mosaic and non‐mosaic patients, respectively, had more than one break. In addition to this parameter, the number of DEB‐induced chromosome breaks per aberrant metaphase constituted a powerful parameter for the diagnosis of mosaic patients (fig 1A). Although DEB‐treated cells from healthy donors contained a mean (SD) of 1.04 (0.18) breaks per aberrant cell, this parameter increased to 3.75 (0.42) and 5.02 (0.3) in the case of mosaic and non‐mosaic Fanconi anaemia patients. On the basis of these observations, initial suspects of somatic mosaicism in T cells are routinely considered when samples treated with DEB contain 5–50% of metaphases with chromosome breaks and intermediate or no sensitivity to MMC in the retroviral subtyping study. T cell mosaicism is more evident when the proportions of metaphases with multiple breaks range between 10–60% (additionally, LCLs from all these samples were resistant to MMC). When relatively low percentages of metaphases with single or multiple breaks are observed, the quantification of the mean number of breaks per aberrant metaphase is highly useful in identifying mosaic patients, since mean values >2 breaks per aberrant metaphase are highly indicative of the presence of Fanconi anaemia cells. In some instances, however, to exclude a negative diagnosis of a patient with Fanconi anaemia with a high level of mosaicism in peripheral blood, cytogenetic and cell cycle analyses of skin fibroblasts are required.
Figure 1 Analysis of the response of peripheral blood T cells from healthy donors and patients with Fanconi anaemia to diepoxybutane (DEB) and mitomycin C (MMC). (A) Proportion of T cells with single (white bars) and multiple (black bars) chromosome aberrations, as well as the number of chromosome breaks per aberrant cell after DEB treatment (grey bars). Each bar represents the mean (SD) of individual data. (B) Relationship between the proportion of peripheral blood T cells with aberrant metaphases after DEB challenge and the viability of these cells after exposure to 33 nM MMC. Individual points correspond to samples from healthy donors (□) and from mosaic patients ( ) and non‐mosaic Fanconi anaemia patients (▪).
) and non‐mosaic Fanconi anaemia patients (▪).
For the subtyping of patients with Fanconi anaemia, the retroviral complementation assay was routinely used as a first approach.20 This assay is based on the analysis of the MMC sensitivity of peripheral blood T cells, followed by the correction of the MMC hypersensitivity conferred by the complementing Fanconi anaemia gene. The retrospective analysis of 57 peripheral blood samples from healthy donors and from mosaic and non‐mosaic patients showed a good relationship—fitting to a linear regression model (p<0.01)—between the proportion of T cell metaphases with DEB‐induced aberrant chromosomes and the sensitivity of T cells to 33 nM MMC—a dose that showed more marked differences in viability between healthy cells and Fanconi anaemia cells (fig 1B). Although survivals >60% after an exposure to 33 nM MMC corresponded either to healthy donors (n = 11) or to mosaic patients (n = 5), T cell survivals below this threshold always corresponded to patients with Fanconi anaemia (36 non‐mosaic and 5 mosaic). This simple analysis can be conducted by any laboratory with experience in cell culture and flow cytometry, and would have diagnostic value in all cases where a high sensitivity to MMC is observed.
Subtyping analysis of non‐gypsy patients with Fanconi anaemia
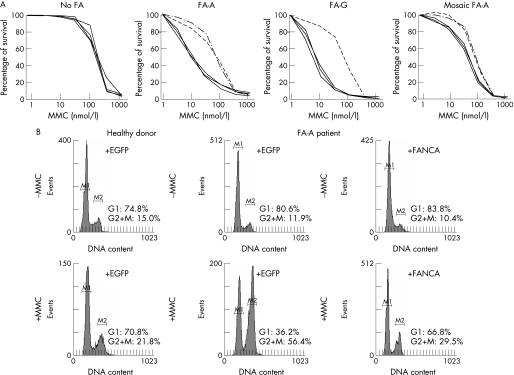
Once the diagnosis of Fanconi anaemia was confirmed, patients were subtyped by transducing their T cells with retroviral vectors expressing three different Fanconi anaemia genes: FANCA (two vectors), FANCC and FANCG, and one negative control (EGFP). In some instances, samples that were not complemented with these vectors were subtyped with additional retroviral vectors carrying three further Fanconi anaemia genes (FANCD2, FANCE and FANCF) and the control EGFP vector. The MMC hypersensitivity of T cells from patients assigned to the FA‐A subgroup was clearly reversed by the transfer of FANCA, using either the LFAPEG or SFA retroviral vectors. Similar results were obtained in samples corresponding to other complementation groups after transduction with the corresponding FANC gene (for a representative analysis of a patient with FA‐G see fig 2A). Of 14 mosaic patients, six could be subtyped using the same approach, since their T cells were sensitive enough to MMC (fig 2A). In other instances, however, T cells from mosaic patient were resistant to MMC, and those patients were subtyped in skin fibroblasts, where somatic mosaicism has not been reported so far. In this case, the reversion of the MMC‐induced G2/M arrest produced after the transduction with the complementing Fanconi anaemia gene was chosen as a biological end point (fig 2B).30
Figure 2 Genetic complementation of cells from patients with Fanconi anaemia (FA), with retroviral vectors expressing functional FANC genes. Panel A shows representative reversions in the mitomycin C (MMC) hypersensitivity of T cells after retroviral‐mediated gene transfer of different Fanconi anaemia genes. Since FA‐A subtype is the most frequently represented group, two different FANCA vectors were routinely used (LFAPEG and SFA), together with one vector encoding FANCC (LFCPEG), FANCG (LFGPEG) and a control EGFP vector (LPEG). Data corresponding to a healthy donor, to two non‐mosaic patients (FA‐A and FA‐G), and a mosaic FA‐A patient (with partial sensitivity to MMC) are shown. Broken lines represent complementation of MMC‐hypersensitivity. Panel B shows a representative correction of the MMC‐induced G2 phase accumulation in fibroblasts from a mosaic FA‐A patient after retroviral‐mediated gene transfer of FANCA. For subtyping in skin fibroblasts, a retroviral vector coexpressing FANCA and the neomycin phosphotransferase gene was used (S11FAIN).
Taken together, the retroviral complementation approach allowed us to subtype 57 of the 73 caucasian patients with Fanconi anaemia who entered the study. A total of 50 patients (45 families) were FA‐A, three patients were FAD2 (FA78 and siblings FA70 and FA71), two patients were FA‐E (FA202 and FA281) and two patients were FA‐G (FA59 and FA116). Regarding the other 16 patients, five patients could not be further studied (mosaics with MMC‐resistant T cells; skin samples not available) and 11 patients were classified as non‐FA‐A/C/G.
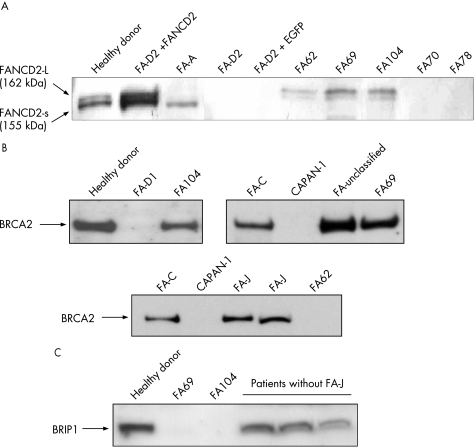
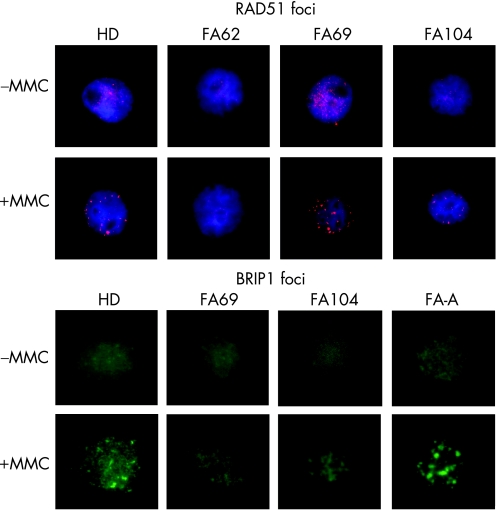
In five patients (FA62, 69, 104, 177 and 287), further subtyping studies could be conducted using complementary strategies to retroviral subtyping. Additionally, the subtype of patient FA72 was deduced from analyses of her sibling (FA69). Western blot analyses of FANCD2 were conducted with these samples to determine whether the mutated gene was upstream or downstream of the FANCD2 monoubiquitination pathway (fig 3A). Consistent with retroviral complementation analyses, FANCD2 was not detected in samples from FA70 and FA78. Cells from FA287 (not shown) only showed the small FANCD2 band, suggesting that this patient must be deficient in a FANC protein upstream of FANCD2, except for FANCA, FANCC and FANCG (ruled out by retroviral complementation). Cells from three unassigned patients (FA62, 69 and 104) showed both the non‐ubiquitinated and monoubiquitinated FANCD2 isoforms, indicating an intact Fanconi anaemia complex upstream of FANCD2 in these cells. Western blot analysis of BRCA2 showed a band of the expected size in FA69 and FA104 cells, whereas no band was detected in cells from patient FA62 (fig 3B). As BRCA2 is required for generating RAD51 foci after exposure to MMC, we investigated the formation of RAD51 foci in cells from these patients after an MMC challenge.24 Consistent with the lack of the BRCA2 band in the western blot analysis, RAD51 foci were not induced by MMC in FA62 cells, but were evident in FA69 and FA104 cells (fig 4), indicating that FA62 is a FAD1 patient. This finding has been recently confirmed by BRCA2 mutational analysis (data not shown).
Figure 3 Western blot analyses of the FANCD2, BRCA2 and BRIP1 proteins in cells from patients with Fanconi anaemia (FA) not complemented with FANCA, FANCC and FANCG retroviral vectors. Protein extracts from proliferating lymphoblast cell line were transferred to a nitrocellulose membrane, incubated with the corresponding antibodies, and then revealed as described in the Methods section. Western blot analysis of FANCD2, showing the ubiquitinated (L‐FANCD2) and non‐ubiquitinated (S‐FANCD2) forms of FANCD2. (B and C) Western blot analyses of BRCA2 and BRIP1 proteins, respectively. CAPAN‐1: negative control consisting of cells not expressing BRCA2.7,2
Figure 4 Mitomycin C (MMC) induction of RAD51 and BRIP1 foci in Fanconi anaemia (FA) samples not complemented with FANCA, FANCC and FANCG retroviral vectors. For immunofluorescence analysis of RAD51 and BRIP1 foci, untreated and MMC‐treated cells were stained with anti‐RAD51 or anti‐BRIP1 antibodies, and labelled with an anti‐rabbit Alexa antibody, as indicated in the Methods section. RAD51 foci were not observed in cells from patient FA62. BRIP1 foci were not observed in cells from patient FA69 and were reduced in cells from patient FA104 compared with cells from a healthy donor (HD) or a patient with FA‐A patient.
On the basis of the recent discovery that mutations in BRIP1 account for the FA‐J complementation group, western blot analyses of BRIP1 as well as analyses of BRIP1 foci in cells from patients FA69 and FA104 were conducted.12,13 FA69 cells did not generate any band of the expected size in the BRIP1 western blot (fig 3C), and neither generated BRIP1 foci after exposure to MMC (fig 4). This indicates that FA69 and her sibling FA72 are FA‐J patients. Regarding patient FA104, the analysis of the BRIP1 band was not evident in extracts from these cells (fig 3C), although nuclear extracts showed a tiny putative BRIP1 band (not shown). Analyses of BRIP1 foci in FA104 cells were not conclusive, as BRIP1 foci were observed in 10% of these cells after exposure to MMC, thus indicating the necessity of confirming the complementation group of this patient.
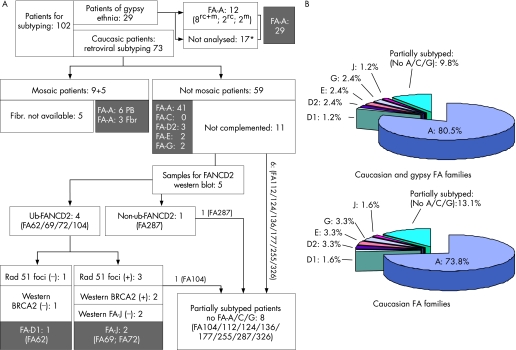
Figure 5 shows a detailed description of the progress of the subtyping studies conducted with samples from the 73 Caucasian patients with Fanconi anaemia and the 29 gypsy patients with Fanconi anaemia.
Figure 5 Summarised subtyping studies conducted in patients with Fanconi anaemia from the Spanish registry. (A) Subtyping screening analyses conducted to determine the complementation groups in patients with Fanconi anaemia corresponding to the gypsies (n = 29) and the caucasians (n = 73). (B) Complementation groups distributed by unrelated families affected by Fanconi anaemia, both in the total (n = 82) and in the Caucasian (n = 61) populations. Symbols used in the Gypsy populations: mSamples analysed for the 295C→T mutation; rvSamples subjected to retroviral complementation; rv+mSamples subjected to both analyses. *Considered FA‐A patients; see the Results and Discussion section for details.
Subtyping of gypsy patients with Fanconi anaemia
A total of 29 gypsy patients with Fanconi anaemia have been identified, which constitute 23% of the 125 patients included in the Spanish registry of Fanconi anaemia. Initially, these patients were subtyped by the standard retroviral analysis, showing that all 10 analysed gypsy patients corresponded to the FA‐A complementation group. Thereafter, we observed that patients with Fanconi anaemia corresponding to the gypsy ethnic group shared a mutation in FANCA consisting of a novel homozygotic truncating mutation (295C→T) in the fourth exon, which leads to a truncation in FANCA.31 In fact, 10 out of 10 Spanish gypsy patients (8 of whom had also been analysed by the retroviral complementation method) shared this mutation. Additionally, eight of eight Spanish gypsy patients shared the same genetic haplotype based on four microsatellite markers flanking the FANCA gene, and every analysed obligate carrier was heterozygotic for this mutation. On the basis of these observations, patients with Fanconi anaemia from the gypsy race of Spanish origin are now routinely screened by the presence of this ancestral mutation using a simple PCR reaction. Although samples from 17 gypsy patients were not available for subtyping or mutation screening, the fact that all tested patients of the gypsy ethnic group shared the 295C→T ancestral mutation and an identical genetic haplotype allows us to presume that all our 29 gypsy patients belong to the FA‐A complementation group.
Distribution of Fanconi anaemia complementation groups in Spain
By means of the different subtyping approaches shown above, we can propose, for the first time, the distribution of complementation groups in Spain (fig 5B). From the 102 samples available from subtyping, 89 patients have been totally subtyped and eight patients partially subtyped. Since the registry of Spanish patients with Fanconi anaemia includes 125 patients, our subtyping study represents 82% of our registry. Figure 5 shows the distribution of the complementation groups in non‐related families with Fanconi anaemia. Since all gypsy patients are FA‐A, the distribution of the complementation groups in the Caucasian Fanconi anaemia population is slightly modified (fig 5B).
Our study shows the high prevalence of the FA‐A subgroup in Spain (80% including the gypsy patients). This observation is mainly a consequence of the absence of FA‐C patients and the low prevalence of FA‐G complementation groups in Spain, which in other series were around 19% and 15%, respectively.1,2 Also, the high proportion of Fanconi anaemia gypsies found in our country (23% of all the Spanish patients with Fanconi anaemia), all sharing an ancestral mutation in FANCA, accounts for the high prevalence of the FA‐A subtype in Spain.31 Also, in Italy and in Tunisia, two other Mediterranean countries, a very high proportion of FA‐A patients has been observed.32,33 Whether or not common mutations are present in patients with Fanconi anaemia from these countries is still unknown, although this might be the case, since we have detected an insertion in FANCA (c3558–3559 insG) in a Spanish patient previously detected in an Italian patient (data not shown).34
With regard to the frequency of mosaic patients anaemia, we have determined the presence of 14 Caucasian patients corresponding to the FA‐A complementation group, with clear evidences of somatic mosaicism in the peripheral blood. This represents 19% of the caucasian patients—proportion that is similar to the 15% value recently reported in other populations with Fanconi anaemia.23
A proposed approach for the subtyping of patients with Fanconi anaemia
On the basis of the different approaches used in this study, we propose the algorithm shown in fig 6 for the subtyping of patients with Fanconi anaemia. In addition to chromosomal instability studies, the analysis of the MMC sensitivity of peripheral blood T cells is proposed to evaluate whether the subtyping can be conducted in peripheral blood or, alternatively, whether it should be conducted in skin fibroblasts (step 1 in fig 6). Since most patients with Fanconi anaemia distributed along the world are FA‐A, FA‐C or FA‐G (85% on average), most patients with Fanconi anaemia could be subtyped by the transduction of peripheral blood T cells with only three retroviral vectors carrying FANCA, FANCC and FANCG (step 2 in fig 6). Moreover, in Mediterranean countries such as Spain or Italy, the use of a single vector encoding FANCA would facilitate the subtyping of a similar proportion of patients. In those cases in which the suspicion of Fanconi anaemia is high, steps 1 and 2 could be conducted in parallel, so that the subtyping of the patient could be obtained in only 2–3 weeks. It is expected that about 15% of the analysed samples would not be complemented by FANCA, FANCC and FANCG. In these patients, the analysis of FANCD2 by western blot (step 3 in fig 6) would indicate whether the mutated gene is FANCD2 itself or is upstream or downstream of the ubiquitination of this regulatory protein.22 According to previous studies, it is predicted that 20% of samples not complemented by FANCA, FANCC and FANCG will not present any FANCD2 band (estimated frequency of FAD2 patients: 3%).2 It is also expected that 40% of these samples will present both bands of FANCD2 (groups FAD1 and FA‐J; estimated frequencies: 2–4% each). Another 40% will only show a single non‐ubiquitinated FANCD2 band (groups FA‐B, E, F, I, L and M; total estimated frequency: 6%). According to the information provided by the FANCD2 western blot, further studies of retroviral complementation, RAD51 or BRIP1 foci formation, cell fusion, immunoprecipitation, mutation screening and so on could be conducted (step 4 in fig 6), aimed at determining the assignment of the patient to a rare complementation group or to a new unidentified group.
Figure 6 Proposed algorithm for the subtyping of patients with Fanconi anaemia (FA). A four‐step process is proposed for the subtyping of patients with Fanconi anaemia. Step 1 aims to confirm the diagnosis of Fanconi anaemia and somatic mosaicism. According to step 2, 85% of patients with Fanconi anaemia would be subtyped by retroviral complementation. The complementation groups of the remaining 15% of the patients could be discriminated by means of analyses shown in step 3. Finally, the subtype of rare complementation groups or the discovery of new complementation groups could be determined by means of different research approaches described in step 4. DEB, diepoxybutane; MMC, mitomycin C; PB, peripheral blood; Ub, ubiquitinated.
Our study constitutes the first systematic subtyping analysis of patients with Fanconi anaemia in Spain, a Mediterranean country with a significant population of gypsy patients. We have observed a skewed distribution of complementation groups in this country with respect to other international studies. Further analyses of mutational screening are in progress to investigate the presence of common mutations in patients from particular regions from this country and to facilitate the prenatal and pre‐implantation diagnosis of Fanconi anaemia. It is our hope that the subtyping of this large population of patients with Fanconi anaemia will improve their clinical management and also their recruitment into optimised gene therapy trials with clinical efficacy.
Acknowledgements
This work was supported by a grant from the Spanish Ministry of Health for the Cooperative Network on Fanconi Anemia (G03/073). CIEMAT has also been supported by grants from the European Programme “Life Sciences, Genomics and Biotechnology for Health” (CONSERT; Ref 005242 and FI6R‐CT‐2003‐508842). Comisión Interministerial de Ciencia y Tecnología (SAF2002‐03234; 2003‐020328; SAF2004‐20372E; 2005‐00058), Fondo Investigación Sanitaria (PI051205) La Caixa Foundation and the Marcelino Botín Foundation.
We thank Sergio García for invaluable help in the subtyping of patients, and Gloria Umbert for cytogenetic analyses. We also thank Mrs Aurora de Lacal, Elena García and M Dolores López for coordination in the delivery of the samples. We are indebted to the Spanish patients and their families, always enthusiastic for promoting the research on Fanconi anaemia.
Abbreviations
DEB - diepoxybutane
FBS - fetal bovine serum
LCL - lymphoblast cell line
MMC - mitomycin C
PBS - phophate‐buffered saline
TBS - TRIS‐buffered saline
Footnotes
Competing interests: None declared.
References
- 1.Taniguchi T, D'Andrea A D. The molecular pathogenesis of Fanconi anemia: recent progress. Blood 20061074223–4233. [DOI] [PubMed] [Google Scholar]
- 2.Levitus M, Rooimans M A, Steltenpool J, Cool N F, Oostra A B, Mathew C G, Hoatlin M E, Waisfisz Q, Arwert F, de Winter J P, Joenje H. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood 20041032498–2503. [DOI] [PubMed] [Google Scholar]
- 3.Meetei A R, Medhurst A L, Ling C, Xue Y, Singh T R, Bier P, Steltenpool J, Stone S, Dokal I, Mathew C G, Hoatlin M, Joenje H, de Winter J P, Wang W. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 200537958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo Ten Foe J R, Rooimans M A, Bosnoyan‐Collins L, Alon N, Wijker M, Parker L, Lightfoot J, Carreau M, Callen D F, Savoia A, Cheng N C, van Berkel C G, Strunk M H, Gille J J, Pals G, Kruyt F A, Pronk J C, Arwert F, Buchwald M, Joenje H. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet 199614320–323. [DOI] [PubMed] [Google Scholar]
- 5.Meetei A R, Levitus M, Xue Y, Medhurst A L, Zwaan M, Ling C, Rooimans M A, Bier P, Hoatlin M, Pals G, de Winter J P, Wang W, Joenje H. X‐linked inheritance of Fanconi anemia complementation group B. Nat Genet 2004361219–1224. [DOI] [PubMed] [Google Scholar]
- 6.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature 1992356763–767. [DOI] [PubMed] [Google Scholar]
- 7.Howlett N G, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die‐Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox E A, D'Andrea A D. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002297606–609. [DOI] [PubMed] [Google Scholar]
- 8.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D'Andrea A D, Moses R, Grompe M. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 20017241–248. [DOI] [PubMed] [Google Scholar]
- 9.de Winter J P, Leveille F, van Berkel C G, Rooimans M A, van Der Weel L, Steltenpool J, Demuth I, Morgan N V, Alon N, Bosnoyan‐Collins L, Lightfoot J, Leegwater P A, Waisfisz Q, Komatsu K, Arwert F, Pronk J C, Mathew C G, Digweed M, Buchwald M, Joenje H. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am J Hum Genet 2000671306–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Winter J P, Rooimans M A, van Der Weel L, van Berkel C G, Alon N, Bosnoyan‐Collins L, de Groot J, Zhi Y, Waisfisz Q, Pronk J C, Arwert F, Mathew C G, Scheper R J, Hoatlin M E, Buchwald M, Joenje H. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet 20002415–16. [DOI] [PubMed] [Google Scholar]
- 11.de Winter J P, Waisfisz Q, Rooimans M A, van Berkel C G, Bosnoyan‐Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, Pronk J C, Arwert F, Hoehn H, Digweed M, Buchwald M, Joenje H. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet 199820281–283. [DOI] [PubMed] [Google Scholar]
- 12.Levran O, Attwooll C, Henry R T, Milton K L, Neveling K, Rio P, Batish S D, Kalb R, Velleuer E, Barral S, Ott J, Petrini J, Schindler D, Hanenberg H, Auerbach A D. The BRCA1‐interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet 200537931–933. [DOI] [PubMed] [Google Scholar]
- 13.Levitus M, Waisfisz Q, Godthelp B C, de Vries Y, Hussain S, Wiegant W W, Elghalbzouri‐Maghrani E, Steltenpool J, Rooimans M A, Pals G, Arwert F, Mathew C G, Zdzienicka M Z, Hiom K, De Winter J P, Joenje H. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet 200537934–935. [DOI] [PubMed] [Google Scholar]
- 14.Meetei A R, de Winter J P, Medhurst A L, Wallisch M, Waisfisz Q, van de Vrugt H J, Oostra A B, Yan Z, Ling C, Bishop C E, Hoatlin M E, Joenje H, Wang W. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet 200335165–170. [DOI] [PubMed] [Google Scholar]
- 15.Garcia‐Higuera I, Taniguchi T, Ganesan S, Meyn M S, Timmers C, Hejna J, Grompe M, D'Andrea A D. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 20017249–262. [DOI] [PubMed] [Google Scholar]
- 16.Surralles J, Jackson S P, Jasin M, Kastan M B, West S C, Joenje H. Molecular cross‐talk among chromosome fragility syndromes. Genes Dev 2004181359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimamura A, D'Andrea A D. Subtyping of Fanconi anemia patients: implications for clinical management. Blood 20031023459. [DOI] [PubMed] [Google Scholar]
- 18.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995378789–792. [DOI] [PubMed] [Google Scholar]
- 19.Wijker M, Morgan N V, Herterich S, van Berkel C G, Tipping A J, Gross H J, Gille J J, Pals G, Savino M, Altay C, Mohan S, Dokal I, Cavenagh J, Marsh J, van Weel M, Ortega J J, Schuler D, Samochatova E, Karwacki M, Bekassy A N, Abecasis M, Ebell W, Kwee M L, de Ravel T, Gibson R A, Gluckman E, Arwert F, Joenje H, Savoia A, Hoehn H, Pronk J C, CG Mathew et al Heterogeneous spectrum of mutations in the Fanconi anaemia group A gene. Eur J Hum Genet 1999752–59. [DOI] [PubMed] [Google Scholar]
- 20.Hanenberg H, Batish S D, Pollok K E, Vieten L, Verlander P C, Leurs C, Cooper R J, Gottsche K, Haneline L, Clapp D W, Lobitz S, Williams D A, Auerbach A D. Phenotypic correction of primary Fanconi anemia T cells with retroviral vectors as a diagnostic tool. Exp Hematol 200230410–420. [DOI] [PubMed] [Google Scholar]
- 21.Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, Henry R, Milton K, Batish S D, Cancelas J A, Hanenberg H, Auerbach A D, Williams D A. A rapid method for retrovirus‐mediated identification of complementation groups in Fanconi anemia patients. Mol Ther 200512976–984. [DOI] [PubMed] [Google Scholar]
- 22.Shimamura A, Montes de Oca R, Svenson J L, Haining N, Moreau L A, Nathan D G, D'Andrea A D. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood 20021004649–4654. [DOI] [PubMed] [Google Scholar]
- 23.Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, Esperou H, Ferry C, Jubert C, Feugeas J P, Henri A, Toubert A, Socie G, Baruchel A, Sigaux F, D'Andrea A D, Gluckman E. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood 20051051329–1336. [DOI] [PubMed] [Google Scholar]
- 24.Godthelp B C, Wiegant W W, Waisfisz Q, Medhurst A L, Arwert F, Joenje H, Zdzienicka M Z. Inducibility of nuclear Rad51 foci after DNA damage distinguishes all Fanconi anemia complementation groups from D1/BRCA2. Mutat Res 200659439–48. [DOI] [PubMed] [Google Scholar]
- 25.Hajioff S, McKee M. The health of the Roma people: a review of the published literature. J Epidemiol Community Health 200054864–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auerbach A D, Rogatko A, Schroeder‐Kurth T M. International Fanconi Anemia Registry: relation of clinical symptoms to diepoxybutane sensitivity. Blood 198973391–396. [PubMed] [Google Scholar]
- 27.Jacome A, Navarro S, Casado J A, Rio P, Madero L, Estella J, Sevilla J, Badell I, Ortega J J, Olive T, Hanenberg H, Segovia J C, Bueren J A. A simplified approach to improve the efficiency and safety of ex vivo hematopoietic gene therapy in Fanconi anemia patients. Hum Gene Ther 200617245–250. [DOI] [PubMed] [Google Scholar]
- 28.Auerbach A D, Rogatko A, Schroeder‐Kurth T M. International Fanconi Anemia Registry (IFAR): first report. In: Schroeder‐Kurth TM, Auerbach AD, Obe G, eds. Fanconi anemia, clinical and experimental aspects. Heidelberg: Springer‐Verlag, 19893–17.
- 29.Gregory J J, Jr, Wagner J E, Verlander P C, Levran O, Batish S D, Eide C R, Steffenhagen A, Hirsch B, Auerbach A D. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci U S A 2001982532–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, Gruhn B, Schindler D, Hoehn H. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self‐correction. Cytogenet Genome Res 200298126–135. [DOI] [PubMed] [Google Scholar]
- 31.Callen E, Casado J A, Tischkowitz M D, Bueren J A, Creus A, Marcos R, Dasi A, Estella J M, Munoz A, Ortega J J, de Winter J, Joenje H, Schindler D, Hanenberg H, Hodgson S V, Mathew C G, Surralles J. A common founder mutation in FANCA underlies the world highest prevalence of Fanconi anemia in gypsy families from Spain. Blood 20051051946–1949. [DOI] [PubMed] [Google Scholar]
- 32.Savoia A, Zatterale A, Del Principe D, Joenje H. Fanconi anaemia in Italy: high prevalence of complementation group A in two geographic clusters. Hum Genet 199697599–603. [DOI] [PubMed] [Google Scholar]
- 33.Bouchlaka C, Abdelhak S, Amouri A, Ben Abid H, Hadiji S, Frikha M, Ben Othman T, Amri F, Ayadi H, Hachicha M, Rebai A, Saad A, Dellagi K, Tunisian Fanconi Anemia Study Group Fanconi anemia in Tunisia: high prevalence of group A and identification of new FANCA mutations. J Hum Genet 200348352–361. [DOI] [PubMed] [Google Scholar]
- 34.Savino M, Ianzano L, Strippoli P, Ramenghi U, Arslanian A, Bagnara G P, Joenje H, Zelante L, Savoia A. Mutations of the Fanconi anemia group A gene (FAA) in Italian patients. Am J Hum Genet 1997611246–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]