Abstract
Background
Noonan syndrome, cardio‐facio‐cutaneous syndrome (CFC) and Costello syndrome constitute a group of developmental disorders with an overlapping pattern of congenital anomalies. Each of these conditions can be caused by germline mutations in key components of the highly conserved Ras‐MAPK pathway, possibly reflecting a similar pathogenesis underlying the three disorders. Germline mutations in KRAS have recently been identified in a small number of patients with Noonan syndrome and CFC.
Methods and results
260 patients were screened for KRAS mutations by direct sequencing. Overall, we detected KRAS mutations in 12 patients, including three known and eight novel sequence alterations. All mutations are predicted to cause single amino acid substitutions. Remarkably, our cohort of individuals with KRAS mutations showed a high clinical variability, ranging from Noonan syndrome to CFC, and also included two patients who met the clinical criteria of Costello syndrome.
Conclusion
Our findings reinforce the picture of a clustered distribution of disease associated KRAS germline alterations. We further defined the phenotypic spectrum associated with KRAS missense mutations and provided the first evidence of clinical differences in patients with KRAS mutations compared with Noonan syndrome affected individuals with heterozygous PTPN11 mutations and CFC patients carrying a BRAF, MEK1 or MEK1 alteration, respectively. We speculate that the observed phenotypic variability may be related, at least in part, to specific genotypes and possibly reflects the central role of K‐Ras in a number of different signalling pathways.
Noonan syndrome (OMIM 163950), cardio‐facio‐cutaneous syndrome (CFC; OMIM 115150) and Costello syndrome (OMIM 218040) are distinct entities that share a common pattern of congenital anomalies, including typical heart defects, overlapping craniofacial dysmorphisms, short stature and a variable degree of mental retardation. Discrimination between the three conditions is based mainly on distinct clinical features such as dry hyperkeratotic skin and hair abnormalities in patients with CFC,1 and redundant and loose skin with deep palmar and plantar creases as well as a coarse facial appearance in those with Costello syndrome.2 In addition, mental development is more severely impaired in CFC and Costello syndrome whereas Noonan syndrome is usually associated with minor cognitive deficits or even normal intelligence.3 While patients with Noonan syndrome and CFC have no or only a slightly increased risk of tumour development, the incidence of tumours in Costello syndrome has been estimated to be 7–21%.4 Although various attempts have been undertaken to develop standardised diagnostic criteria for these entities,5,6 considerable overlap exists and in some instances a patient's phenotype cannot be clearly assigned to one of these conditions.
Missense mutations in PTPN11 were first identified in patients with Noonan syndrome7 and subsequently have been shown to account for almost 50% of cases.8,9PTPN11 encodes the protein tyrosine phosphatase SHP‐2 which relays growth signals from activated tyrosine kinase receptors to other signalling molecules, particularly Ras (reviewed by Neel et al10). Noonan syndrome causing PTPN11 mutations have been thought to result in gain‐of‐function of SHP‐2 and cause deregulation of Ras dependent signalling cascades.11 Heterozygous germline mutations in KRAS were reported to occur in a minority of patients with Noonan syndrome12 and CFC,12,13 shortly after mutations in HRAS had been detected in the majority of individuals with Costello syndrome.14 Moreover, mutations in BRAF, MEK1 and MEK2, encoding proteins involved in Ras downstream signalling, were shown to cause CFC syndrome.13,15 Taken together, the current data suggest that germline missense mutations in the aforementioned genes culminate in deregulated Ras‐MAPK signalling that most likely represents the common pathogenetic basis of this group of developmental disorders.16,17
Ras isoforms encoded by the three genes KRAS, HRAS and NRAS represent highly conserved signal transduction molecules. They act as molecular switches through cycling between an active GTP bound and an inactive GDP bound state,18 and in their active form they interact with a variety of downstream effector proteins.19RAS genes have long been known as proto‐oncogenes mutated in various types of human cancers (reviewed by Bos20). The majority of these oncogenic RAS mutations affect amino acid residues G12, G13 and Q61 and cause Ras to accumulate in the active GTP bound state by impairing intrinsic GTPase activity and conferring resistance to GTPase activating proteins (GAPs).20 Germline HRAS mutations associated with Costello syndrome almost exclusively affect codons 12 and 13 and are identical to somatic alterations identified in cancer,14 hence explaining the high risk of tumour development in Costello syndrome. In contrast, KRAS mutations described to date in patients with Noonan syndrome/CFC are distinct from those found in malignancies. Similar to the concept of activating PTPN11 mutations in Noonan syndrome and malignancies,11KRAS mutations associated with Noonan syndrome or CFC might give rise to mutant proteins with a relatively mild gain‐of‐function which are tolerated in the germline as well as during embryonic development. Specifically, KRAS mutations identified in Noonan syndrome patients include V14I and T58I whereas P34R and G60R were found in CFC patients.12,13 Mutations in KRAS exon 6, causing amino acid alterations in the C terminal portion of isoform B, such as D153V and V152G, were found to be associated with a severe Noonan syndrome or CFC phenotype.12,13,21 It has been proposed that all mutations lead to stabilisation of K‐Ras in the active conformation, most likely by different gain‐of‐function mechanisms.12,21
Here we report the results of KRAS mutation screening in a large cohort of patients with Noonan syndrome and related disorders.
Patients and methods
The original study population consisted of 333 individuals of predominantly European origin with a clinical diagnosis of Noonan syndrome, 37 with CFC and 30 with Costello syndrome. Patients were clinically assessed by experienced clinical geneticists and classified according to general diagnostic criteria.3,5,6 Ninety seven patients with Noonan syndrome (29%) were previously tested positive for a PTPN11 mutation, a proportion that is comparable with that published by Musante et al22 but lower than in other studies.8,9 These variations probably reflect differences in the selection of the referred cases. Twenty seven of 30 patients with Costello syndrome (90%) were found to have an HRAS mutation, a rate that has been consistently reported in previous studies.14,23,24,25 In 12 (32%) of the 37 CFC cases, a BRAF mutation was detected, a similar proportion to that reported by Niihori et al.13 Two of our patients with CFC (5%) had a mutation of MEK1 and two (5%) a mutation of MEK2 (M Zenker and K Kutsche, unpublished data). The remaining 236 PTPN11 negative patients with Noonan syndrome, 21 individuals with CFC and three patients with Costello syndrome were investigated for mutations in KRAS. Informed consent was obtained for genetic analyses from all patients or their legal guardians. Ethics approval for the study was obtained from the ethics committee of the University of Erlangen‐Nuremberg. Mutation screening of the entire coding sequence of both KRAS isoforms was carried out as described previously.12 Primer pairs and polymerase chain reaction conditions are available on request. Sequence analysis was performed by bidirectional sequencing using the ABI BigDye Terminator Sequencing Kit (Applied Biosystems, Weiterstadt, Germany) and automated capillary sequencers (Applied Biosystems). Where mutations were shown to have arisen de novo, we verified declared relationships by genotyping of both parents and the patient at 10 microsatellite loci.
Results
Spectrum of KRAS mutations
We discovered 11 different heterozygous KRAS mutations in 12 of the patients in our study population (table 1). All patients carrying a KRAS mutation were sporadic cases. In 10 individuals we could demonstrate that the mutation occurred de novo (including confirmation of paternity). DNA from both parents was not available in the remaining two cases (Nos 4 and 7; table 1).
Table 1 Clinical features of patients with KRAS mutations.
| Patient No 1 | Patient No 2 | Patient No 3 | Patient No 4 | Patient No 5 | Patient No 6 | |
|---|---|---|---|---|---|---|
| KRAS mutation* | K5N (c.15A→T) | V14I (c.40G→A) | Q22E (c.64C→G) | Q22R (c.65A→G) | P34L (c.101C→T) | P34Q (c.101C→A) |
| Age at last follow up | 7.5 months | 16 years | 2 years | 3.3 years | 17 years | 3.4 years |
| Sex | Male | Male | Male | Female | Male | Male |
| Birth weight (g)† | 3195 | 4000 | 3400 | 2820 | 4470 | 3060 |
| Congenital heart defect | Pst | No | Pst | Pst | No | Pst |
| Facial features | Hypertelorism, downslanting palpebral fissures, coarse face, anteverted nostrils, low‐set dysplastic ears | Hypertelorism, low‐set ears | Hypertelorism, downslanting palpebral fissures, anteverted nostrils, low‐set ears | Hypertelorism, downslanting palpebral fissures, low‐set ears | Hypertelorism, downslanting palpebral fissures | Hypertelorism, low set ears, flat nasal bridge, posteriorly rotated ears |
| Stature | <1st centile | 1st centile | 15th centile | 3rd–10th centile | 1st centile | <1st centile |
| Relative macrocephaly | Yes | Yes | No | Yes | Yes | No |
| Short/webbed neck | Webbed neck | Short neck | Short and webbed neck | Short and webbed neck | Short neck | Webbed neck |
| Thorax deformity | Pectus carinatum | Mild pectus excavatum | No | Broad chest, mild pectus excavatum | Pectus excavatum (op) | No |
| Cryptorchidism | No | No | No | – | No | No |
| Ophthalmological problems | Ptosis, strabismus, nystagmus | Ptosis (op), strabismus | Ptosis | Ptosis, strabismus | Strabismus | Mild ptosis |
| Developmental delay/mental retardation | Moderate | Mild | Moderate | Moderate | Mild | Mild |
| Abnormal hair | Sparse | No, sparse hair in early childhood | Thin hair, sparse eyebrows | No | No | No |
| Skin abnormalities | Redundant skin, deep palmar and plantar creases | No | Deep palmar and plantar creases | No | No | No |
| Other | Macroglossia, severe feeding difficulties and failure to thrive | Seizures, mild hydrocephalus internus, cubitus valgus | Unilateral pyelectasia, failure to thrive | – | Received GH treatment | Failure to thrive and feeding problems in infancy |
| Clinical diagnosis | CS | NS | CFC | Severe NS | NS | NS |
| Patient No 7 | Patient No 8 | Patient No 9 | Patient No 10 | Patient No 11 | Patient No 12 | |
|---|---|---|---|---|---|---|
| KRAS mutation* | I36M (c.108A→G) | G60R (c.178G→C) | D153V (isoform B) (c.460A→T) | D153V (isoform B) (c.460A→T) | F156I (isoform B) (c.466T→A) | F156L (isoform B) (c.468C→G) |
| Age at last follow‐up | 17 years | 5 years | 20 years | 5.5 years | 8.5 years | 14 months‡ |
| Sex | Female | Female | Male | Male | Male | Male |
| Birth weight (g)† | 3000 | 3350 | 3210 | 2820 | 4850 | 3090 |
| Congenital heart defect | ASD | HOCM | No | Pst | No | HCM, Pst, ASD |
| Facial features | Hypertelorism, downslanting palpebral fissures, posteriorly rotated ears | Hypertelorism, downslanting palpebral fissures, coarse face, low‐set ears | Hypertelorism, downslanting palpebral fissures, posteriorly rotated ears | Hypertelorism, anteverted nostrils, low‐set ears | Hypertelorism, prominent forehead, bitemporal narrowing, epicanthic folds, downslanting palpebral fissures, ptosis, widely spaced teeth, low‐set ears | Hypertelorism, coarse face, anteverted nostrils, low‐set dysplastic ears |
| Stature | 3rd centile | <1st centile | 10th centile | <1st centile | 3rd centile | <3rd centile |
| Relative macrocephaly | Yes | No | Yes | Yes | Yes | Yes |
| Short/webbed neck | Short and webbed neck | Short neck | Short and webbed neck | Short neck | Short and webbed neck | Short and webbed neck |
| Thorax deformity | Pectus excavatum | No | Pectus excavatum | Pectus excavatum | Pectus excavatum | No |
| Cryptorchidism | – | – | Yes | No | No | Yes |
| Ophthalmological problems | Mild ptosis | No | Ptosis | Mild ptosis, strabismus | Ptosis, nystagmus, strabismus | Ptosis |
| Developmental delay/mental retardation | Mild | Moderate | Mild | Mild | Moderate | Moderate |
| Abnormal hair | Thin hair | Thin and sparse hair | No | No | Low posterior hair line, sparse hair at early age | Sparse |
| Skin abnormalities | Loose/redundant skin in early childhood | No | No | No | Wrinkled skin of palms and soles | Loose/redundant and soft skin with deep palmar and plantar creases |
| Other | Received GH treatment, facial asymmetry, bleeding diathesis | Feeding difficulties and failure to thrive | – | Haemangioma of lower lip, vascular malformation of the brain, brachydactyly, GH treatment | Hearing loss, widely spaced nipples, inguinal hernia, muscular hypotonia | Dandy‐Walker malformation, laryngomalacia, conductive hearing deficits, severe feeding difficulties and failure to thrive |
| Clinical diagnosis | NS | CFC | NS | NS | NS/CFC | CS |
ASD, atrial septal defect; CFC, cardio‐facio‐cutaneous syndrome; CS, Costello syndrome; GH, growth hormone; H(O)CM, hypertrophic (obstructive) cardiomyopathy; MR, mental retardation; nd, not documented; NS, Noonan syndrome; op, operated; Pst, pulmonic stenosis.
*Novel mutations are in bold.
†All children were born at term.
‡Died from unexplained sudden death at 14 months of age.
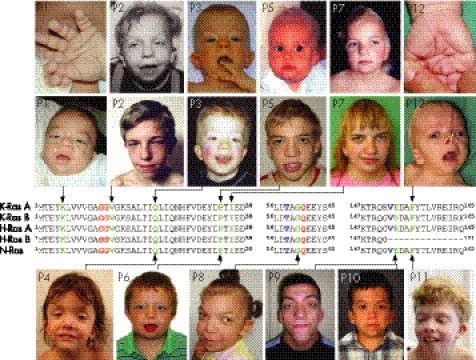
Three mutations, V14I, G60R and D153V, have been described previously12,13 whereas eight are novel (table 1). All newly identified mutations are predicted to cause single amino acid substitutions at highly conserved positions of K‐Ras (data not shown). All but three of the disease associated amino acid changes affect both isoforms of K‐Ras while the nucleotide substitutions leading to the missense mutations D153V, F156I and F156L were found in the alternatively spliced exon 6, thus being present only in isoform B (fig 1).
Figure 1 Novel mutations of KRAS in patients with Noonan syndrome spectrum disorders. Partial alignment of human K‐RasA, K‐RasB, N‐Ras, H‐RasA and H‐RasB is shown. Mutated amino acid residues presented in this study are indicated in green, those altered in previously reported patients (but not present in our study cohort) in blue and residues G12, G13 and Q61, which are most commonly found to be mutated in tumours, in red. Clinical photographs document the variable craniofacial phenotype of patients with heterozygous KRAS mutations (second panel from top and bottom panel). Arrows point from the patients' photographs to the respective residues mutated in K‐Ras. The patient's number (according to table 1) is given in the upper left corner of each photograph. Additional photographs documenting deep palms of patient Nos 1 and 12 as well as earlier photographs of patient Nos 2, 3, 5 and 7 are shown in the top row. We obtained written consent from the patient or his/her legal guardian for publication of the images.
Five of the novel mutations, P34L, P34Q, I36M, F156I and F156L, are located at or close to the known hotspots for KRAS germline mutations whereas missense mutations affecting codons 5 and 22, respectively, represent novel mutation hotspots (K5N, Q22R and Q22E). Notably, mutations at these positions have previously been reported to occur as somatic alterations in cancer (COSMIC database).
Clinical presentation of patients with KRAS mutations
Heterozygous KRAS mutations were identified in seven patients with Noonan syndrome, two individuals with CFC and one patient with an overlapping phenotype between Noonan syndrome and CFC (table 1), thus confirming that sequence alterations in KRAS account for approximately 2% (7/333) of cases with Noonan syndrome and less than 10% (2/37) with CFC.12,13 Moreover, we also detected mutations in two patients (Nos 1 and 12) with a clinical diagnosis of Costello syndrome who were 7.5 and 14 months old, respectively (table 1).
Clinical details of patients carrying KRAS mutations are listed in table 1. All patients with KRAS mutations exhibited the typical craniofacial features of the Noonan syndrome–CFC–Costello syndrome spectrum (fig 1). The majority had short stature, relative macrocephaly, short/webbed neck, thorax deformity and typical ophthalmological anomalies. It is noteworthy that all individuals with KRAS mutations had mild to moderate mental retardation. Three patients presented with additional cerebral abnormalities which are uncommon in Noonan syndrome and CFC, including mild hydrocephalus, intracranial vascular malformation and Dandy‐Walker malformation (table 1). In addition, CFC patients with a KRAS mutation had sparse and thin hair but none had hyperkeratotic or ichthyotic skin lesions (table 1).
Discussion
We identified three known and eight novel KRAS germline mutations in patients with variable phenotypes of the Noonan syndrome–CFC–Costello syndrome spectrum. The assumption that these novel sequence variations indeed represent pathogenic mutations is justified by the following findings: (i) all amino acid substitutions affect highly conserved residues of K‐Ras, (ii) in the majority of cases we demonstrated that the mutation had arisen de novo and (iii) all mutations are located at or near known hotspots for germline and somatic KRAS mutations, respectively. Our findings reinforce the picture of clustered germline mutations in KRAS that strikingly parallels but hardly overlaps the distribution of somatic mutations found in cancer. However, two exceptions exist, K5N and Q22R, representing germline mutations that have also been reported as somatic events in tumours. K5N was described as the probable causative mutation in gastric cancer26 while the Q22R mutation was concurrently found with the oncogenic G12S mutation on the same KRAS allele, suggesting that the transforming potential of this allele is related to G12S rather than Q22R.27 None the less, these findings indicate that at least some oncogenic KRAS mutations may be compatible with life when occurring in the germline and are associated with Noonan syndrome spectrum disorders. Patients carrying either of these KRAS mutations may have an increased tumour risk, similar to patients with Costello syndrome.4
Based on data obtained by functional experiments for the mutations T58I and V14I,12 as well as the structural analysis of both V152G and D153V,21 we hypothesise that the novel KRAS mutations most likely also confer gain‐of‐function effects. In line with this assumption, previous in vitro mutational studies showed that substitutions of I36 and P34, respectively, caused severe impairment of GAP stimulated GTP hydrolysis, thus causing mutant Ras proteins to accumulate in the active state.28,29 Moreover, Ras F156L mutant protein exhibited an extremely rapid off rate for both GDP and GTP in vitro and showed increased levels of its GTP bound form in vivo.30 These findings, together with data from our recent detailed structural analysis, suggest that KRAS mutations cause accumulation of Ras mutant proteins in the active GTP bound state, most likely by different gain‐of‐function mechanisms (R Dvorsky, M Zenker, K Kutsche, MR Ahmadian, personal communication, August 2006).
The phenotypic spectrum in our cohort of patients with KRAS mutations was remarkably broad and included two individuals (cases 1 and 12) with a clinical diagnosis of Costello syndrome which was based on their characteristic facial appearance, the presence of loose and redundant skin with deep palmar and plantar creases, heart abnormality, severe feeding difficulties in early infancy and failure to thrive (fig 1, table 1). Admittedly, none of these features is absolutely specific for Costello syndrome, and as both of these patients with Costello syndrome were very young at the time of clinical diagnosis we cannot exclude the fact that the phenotype may emerge to (severe) CFC later in life. None the less, some features of Costello syndrome and CFC have also been described in a recently reported patient with the KRAS V152G mutation.21 Bentires‐Alj et al proposed that each syndrome should be defined by the specific underlying genetic defect.16 In fact, it may be reasonable to reserve the diagnosis of Costello syndrome for patients who carry oncogenic HRAS mutations and hence must be considered to have an increased risk of tumour development. However, at this point the fact cannot be excluded that specific germline mutations of KRAS (this report) or BRAF31 may not only lead to clinical features of Costello syndrome but also have a similar oncogenic potential as activating HRAS mutations.
It is important to note that all patients, even those classified as Noonan syndrome, showed mild to moderate mental retardation (table 1) which is generally found in only one third of patients with Noonan syndrome.3 In patients with a PTPN11 mutation, this proportion is even lower compared with PTPN11 negative cases,9 and in one published cohort only 24% of patients were reported to have needed special education.8 The level of cognitive function is commonly regarded as one important feature facilitating discrimination between Noonan syndrome and CFC.1,5 Therefore, the presence of substantial mental retardation in patients with Noonan syndrome‐like features may shift the clinical diagnosis towards CFC and lead to classification of a severe Noonan syndrome or a Noonan syndrome–CFC overlapping phenotype in some patients with KRAS mutations. Similarly, Carta et al21 had difficulties in finding the correct clinical classification for their patients and suggested that they presented with a severe Noonan syndrome phenotype with some characteristics of CFC and Costello syndrome. The predominance of significant mental retardation in patients with Noonan syndrome and KRAS mutations could also explain the lack of familial cases.
Patients with CFC syndrome and BRAF mutations frequently show hyperkeratotic or ichthyotic skin lesions,13 which are typical findings in CFC patients.32 However, none of the CFC patients carrying a KRAS mutation—either previously reported13,21 or those presented here—showed at least one of these cutaneous features. Taken together, we propose that (i) patients with KRAS mutations display a particularly high phenotypic variability and that (ii) KRAS mutations are probably responsible for distinct phenotypic subclasses of both Noonan syndrome and CFC. These assumptions raise the question of whether or not the mutant K‐Ras proteins display a greater diversity of altered functions compared with other mutated signalling molecules implicated in the same pathway.
In summary, we have presented the largest cohort to date of patients with germline KRAS mutations and their association with a remarkably broad spectrum of clinical phenotypes. Clearly, the data underscore the central role of Ras in the pathogenesis of this group of related disorders, including Noonan syndrome, CFC and Costello syndrome. We speculate that the diversity of clinical phenotypes associated with different KRAS mutations is caused by the following two factors: (i) K‐Ras plays a key role in a number of downstream signalling cascades and (ii) various K‐Ras mutant proteins may have quantitatively and/or qualitatively different effects on these Ras dependent pathways. Furthermore, in vitro and clinical studies are necessary to elucidate the mechanisms leading to deregulated K‐Ras signalling by a specific mutation and its association with a defined disease phenotype.
Acknowledgements
We are grateful to the patients and families who participated in this study. We thank Inka Jantke, Angelika Diem, Randi Koll and Maren Steckel for skilful technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (GRK336 to KK) and the Deutsche José Carreras Leukämie Stiftung eV (DJCLS R02/10 JMML/MDS to CPK).
Abbreviations
CFC - cardio‐facio‐cutaneous syndrome
GAP - GTPase activating protein
Footnotes
Competing interests: None.
Electronic database information See Gene at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = gene for KRAS genomic (accession number NC_000012), cDNA (accession numbers NM_004985 and NM_033360) and K‐Ras amino acid (accession numbers NP_004976 and NP_203524) sequences; for N‐Ras (accession number NP_002515) and H‐Ras (accession numbers NP_005334 and NP_789765) amino acid sequences. See Catalogue of Somatic Mutations in Cancer (COSMIC) at http://www.sanger.ac.uk/genetics/CGP/cosmic/ for somatic mutations observed in KRAS.
References
- 1.Roberts A, Allanson J, Jadico S K, Kavamura M I, Noonan J, Opitz J M, Young T, Neri G. The cardiofaciocutaneous syndrome. J Med Genet 200643833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hennekam R C. Costello syndrome: an overview. Am J Med Genet C Semin Med Genet 200311742–48. [DOI] [PubMed] [Google Scholar]
- 3.Allanson J E. Noonan syndrome. J Med Genet 1987249–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gripp K W. Tumor predisposition in Costello syndrome. Am J Med Genet C Semin Med Genet 200513772–77. [DOI] [PubMed] [Google Scholar]
- 5.Kavamura M I, Peres C A, Alchorne M M, Brunoni D. CFC index for the diagnosis of cardiofaciocutaneous syndrome. Am J Med Genet 200211212–16. [DOI] [PubMed] [Google Scholar]
- 6.van der Burgt I, Berends E, Lommen E, van Beersum S, Hamel B, Mariman E. Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet 199453187–191. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M, Mehler E L, Goldberg R, Zampino G, Brunner H G, Kremer H, van der Burgt I, Crosby A H, Ion A, Jeffery S, Kalidas K, Patton M A, Kucherlapati R S, Gelb B D. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP‐2, cause Noonan syndrome. Nat Genet 200129465–468. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia M, Kalidas K, Shaw A, Song X, Musat D L, van der Burgt I, Brunner H G, Bertola D R, Crosby A, Ion A, Kucherlapati R S, Jeffery S, Patton M A, Gelb B D. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype‐phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet 2002701555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zenker M, Buheitel G, Rauch R, Koenig R, Bosse K, Kress W, Tietze H U, Doerr H G, Hofbeck M, Singer H, Reis A, Rauch A. Genotype–phenotype correlations in Noonan syndrome. J Pediatr 2004144368–374. [DOI] [PubMed] [Google Scholar]
- 10.Neel B G, Gu H, Pao L. The ‘Shp'ing news: SH2 domain‐containing tyrosine phosphatases in cell signaling. Trends Biochem Sci 200328284–293. [DOI] [PubMed] [Google Scholar]
- 11.Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, Zampino G, Burgt I, Palleschi A, Petrucci T C, Sorcini M, Schoch C, Foa R, Emanuel P D, Gelb B D. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet 200678279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubbert S, Zenker M, Rowe S L, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner L E, Nguyen H, West B, Zhang K Y, Sistermans E, Rauch A, Niemeyer C M, Shannon K, Kratz C P. Germline KRAS mutations cause Noonan syndrome. Nat Genet 200638331–336. [DOI] [PubMed] [Google Scholar]
- 13.Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam R C, Gillessen‐Kaesbach G, Wieczorek D, Kavamura M I, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio‐facio‐cutaneous syndrome. Nat Genet 200638294–296. [DOI] [PubMed] [Google Scholar]
- 14.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto‐oncogene cause Costello syndrome. Nat Genet 2005371038–1040. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez‐Viciana P, Tetsu O, Tidyman W E, Estep A L, Conger B A, Cruz M S, McCormick F, Rauen K A. Germline mutations in genes within the MAPK pathway cause cardio‐facio‐cutaneous syndrome. Science 20063111287–1290. [DOI] [PubMed] [Google Scholar]
- 16.Bentires‐Alj M, Kontaridis M I, Neel B G. Stops along the RAS pathway in human genetic disease. Nat Med 200612283–285. [DOI] [PubMed] [Google Scholar]
- 17.Gelb B D, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS‐mitogen activated protein kinase signal transduction. Hum Mol Genet 200615(Suppl 2)R220–R226. [DOI] [PubMed] [Google Scholar]
- 18.Vetter I R, Wittinghofer A. The guanine nucleotide‐binding switch in three dimensions. Science 20012941299–1304. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann C. Ras‐effector interactions: after one decade. Curr Opin Struct Biol 200313122–129. [DOI] [PubMed] [Google Scholar]
- 20.Bos J L. ras oncogenes in human cancer: a review. Cancer Res 1989494682–4689. [PubMed] [Google Scholar]
- 21.Carta C, Pantaleoni F, Bocchinfuso G. Germline missense mutations affecting KRAS isoform B are associated with a severe Noonan syndrome phenotype. Am J Hum Genet 200679129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musante L, Kehl H G, Majewski F, Meinecke P, Schweiger S, Gillessen‐Kaesbach G, Wieczorek D, Hinkel G K, Tinschert S, Hoeltzenbein M, Ropers H H, Kalscheuer V M. Spectrum of mutations in PTPN11 and genotype–phenotype correlation in 96 patients with Noonan syndrome and five patients with cardio‐facio‐cutaneous syndrome. Eur J Hum Genet 200311201–206. [DOI] [PubMed] [Google Scholar]
- 23.Estep A L, Tidyman W E, Teitell M A, Cotter P D, Rauen K A. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild‐type allele in malignancy. Am J Med Genet A 20061408–16. [DOI] [PubMed] [Google Scholar]
- 24.Gripp K W, Lin A E, Stabley D L, Nicholson L, Scott C I, Jr, Doyle D, Aoki Y, Matsubara Y, Zackai E H, Lapunzina P, Gonzalez‐Meneses A, Holbrook J, Agresta C A, Gonzalez I L, Sol‐Church K. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am J Med Genet A 20061401–7. [DOI] [PubMed] [Google Scholar]
- 25.Kerr B, Delrue M A, Sigaudy S, Perveen R, Marche M, Burgelin I, Stef M, Tang B, Eden O B, O'Sullivan J, De Sandre‐Giovannoli A, Reardon W, Brewer C, Bennett C, Quarell O, M'Cann E, Donnai D, Stewart F, Hennekam R, Cave H, Verloes A, Philip N, Lacombe D, Levy N, Arveiler B, Black G. Genotype–phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J Med Genet 200643401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S H, Lee J W, Soung Y H, Kim H S, Park W S, Kim S Y, Lee J H, Park J Y, Cho Y G, Kim C J, Nam S W, Kim S H, Lee J Y, Yoo N J. BRAF and KRAS mutations in stomach cancer. Oncogene 2003226942–6945. [DOI] [PubMed] [Google Scholar]
- 27.Miyakura Y, Sugano K, Fukayama N, Konishi F, Nagai H. Concurrent mutations of K‐ras oncogene at codons 12 and 22 in colon cancer. Jpn J Clin Oncol 200232219–221. [DOI] [PubMed] [Google Scholar]
- 28.Chung H H, Benson D R, Cornish V W, Schultz P G. Probing the role of loop 2 in Ras function with unnatural amino acids. Proc Natl Acad Sci U S A 19939010145–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stone J C, Colleton M, Bottorff D. Effector domain mutations dissociate p21ras effector function and GTPase‐activating protein interaction. Mol Cell Biol 1993137311–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quilliam L A, Zhong S, Rabun K M, Carpenter J W, South T L, Der C J, Campbell‐Burk S. Biological and structural characterization of a Ras transforming mutation at the phenylalanine‐156 residue, which is conserved in all members of the Ras superfamily. Proc Natl Acad Sci U S A 1995921272–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rauen K A. Distinguishing Costello versus cardio‐facio‐cutaneous syndrome: BRAF mutations in patients with a Costello phenotype. Am J Med Genet A 20061401681–1683. [DOI] [PubMed] [Google Scholar]
- 32.Wieczorek D, Majewski F, Gillessen‐Kaesbach G. Cardio‐facio‐cutaneous (CFC) syndrome—a distinct entity? Report of three patients demonstrating the diagnostic difficulties in delineation of CFC syndrome. Clin Genet 19975237–46. [DOI] [PubMed] [Google Scholar]