Abstract
Introduction
Rett syndrome (RTT) is an X‐linked dominant neurodevelopmental disorder that is usually associated with mutations in the MECP2 gene. The most common mutations in the gene are p.R168X and p.T158M. The influence of X‐chromosome inactivation (XCI) on clinical severity in patients with RTT with these mutations was investigated, taking into account the extent and direction of skewing.
Methods
Female patients and their parents were recruited from the UK and Australia. Clinical severity was measured by the Pineda Severity and Kerr profile scores. The degree of XCI and its direction relative to the X chromosome parent of origin were measured in DNA prepared from peripheral blood leucocytes, and allele‐specific polymerase chain reaction was used to determine the parental origin of mutation. Combining these, the percentage of cells expected to express the mutant allele was calculated.
Results
Linear regression analysis was undertaken for fully informative cases with p.R168X (n = 23) and p.T158M (n = 20) mutations. A statistically significant increase in clinical severity with increase in the proportion of active mutated allele was shown for both the p.R168X and p.T158M mutations.
Conclusions
XCI may vary in neurological and haematological tissues. However, these data are the first to show a relationship between the degree and direction of XCI in leucocytes and clinical severity in RTT, although the clinical utility of this in giving a prognosis for individual patients is unclear.
Rett syndrome (RTT) is an X‐linked dominant neurodevelopmental disorder, usually caused by mutations in the methyl‐CpG‐binding protein 2 (MECP2, OMIM#300005) gene. Mutations often arise at CpG hotspots,1 and the most common mutations in MECP2 found in RTT cases are p.T158M and p.R168X (RettBASE, http://mecp2.chw.edu.au/). RTT has a wide clinical variability in terms of its severity.2 Studies investigating the association between genotype and phenotype were originally quite inconsistent in their findings. However, with larger studies and increasing numbers of publications, evidence for definite relationships between genotype and phenotype is becoming clearer.3 Apparent differences in study results often occurred because of the use of different means of classifying and recording clinical severity. Additionally, the effects of X‐chromosome inactivation (XCI) status and other epigenetic influences on MECP2 function are likely to have a real influence on the variation in phenotype associated with specific mutations.
XCI occurs early in embryogenesis at the blastula stage, and is usually a random process.4,5 The inactive X chromosome is determined as cells become pluripotent; once this has happened, lineages derived from each of these cells will all have the same X chromosome inactivated through a process of methylation. Some studies have shown that there is an increased tendency for skewing of XCI in lymphocytes in RTT when compared with age‐matched controls, and that this usually confers a protective effect.6,7,8 The most striking clinical examples of the effects of XCI in RTT are seen in twins with disparate severity9,10 and in healthy carrier mothers with skewed XCI (presumed favourable) with affected daughters.11,12,13,14,15
In this study, we adopted a new approach to investigate genotype–phenotype relationships in RTT by exploring the association between clinical severity and the proportion of active mutated allele for the two common MECP2 mutations, p.R168X and p.T158M.
Methods
Data sources
Patients were recruited from three sources in two countries (table 1): UK cases from two RTT research studies (Cardiff and Glasgow) and Australian cases from the Australian Rett Syndrome Database (www.ichr.uwa.edu.au/rett/aussierett/).
Table 1 Distribution of sample sources and informative data for mutations p.T158M and p.R168X.
| Source | Cases (n) | Informative and clinical data available |
|---|---|---|
| T158M | ||
| Cardiff | 18 | 8 |
| Scotland | 7 | 3 |
| Australia | 15 | 9 |
| Total | 40 | 20 |
| R168X | ||
| Cardiff | 28 | 15 |
| Scotland | 3 | 2 |
| Australia | 12 | 6 |
| Total | 43 | 23 |
| Total | 83 | 43 |
UK patients were recruited from those referred to Cardiff for molecular genetic analysis, or were selected from those participating in a large genotype–phenotype study.2 Cardiff provides one of the two main diagnostic molecular testing services within the UK. By July 2005, 245 patients had a MECP2 mutation identified and all were invited to participate in the Cardiff Rett Study. DNA samples were already available from the proband, and a blood sample was obtained from at least one parent where this was not already available. In total, samples were available from 18 of 25 Cardiff cases with p.T158M and 28 of 37 cases with p.R168X. An additional seven p.T158M cases and three p.R168X cases were obtained from Scotland (the majority identified in Glasgow).2
The Australian Rett Syndrome Database is an ongoing population‐based registry, established in 1993, of Australian RTT cases born since 1976. Data are collected from families and clinicians on enrolment and, in combination with molecular data, evaluated for compliance with the RTT diagnostic criteria.16 By July 2005, mutation testing had been carried out in 254 (88.2%) of the 288 cases, with a pathogenic MECP2 mutation detected in 186 (73.2%). DNA samples from probands that had at least one parental sample available were accessible for testing in 15 of 21 cases of p.T158M and in 12 of 21 cases of p.R168X.
XCI studies
Determination of degree and direction of X‐inactivation status
XCI was determined using a modified standard method.17 Separate aliquots of DNA were predigested with the methylation‐sensitive enzymes HpaII and McrBC (New England Biolabs, Hitchin, Hertfordshire, UK). The triplet repeat at the HUMARA locus was then amplified by polymerase chain reaction (PCR) using fluorescent primers, and analysed using an ABI 3100 automated sequencer and Genotyper software (Applied Biosystems, Foster City, California, USA). Allele peak areas were compared for HpaII digested, McrBC digested and undigested DNA to determine the degree of XCI. HpaII predigestion will prevent PCR amplification of the HUMARA allele on the unmethylated (active) X chromosome; conversely, McrBC predigestion will prevent PCR amplification of the allele on the methylated (inactive) X chromosome. This allows identification of the allele that is present on the more active X chromosome. DNA samples from the parents were genotyped to determine the parental origin of the alleles, and thereby from which parent the more active X chromosome was inherited.
Determination of the parental origin of the mutation
Intron 3 and the 3′ untranslated region (3′UTR) of MECP2 were sequenced in the patients to find informative single‐nucleotide polymorphisms (SNPs). When an SNP was found, allele‐specific PCR primers were designed so that the base on the 3′ end of each primer was complementary to only one allele of the SNP (table 2). SNPs are numbered relative to the position of the stop codon (+878C→G) or the end of exon 3 (+266C→T, +516C→T and +648A→G). All SNPs are available in dbSNP (http://www.ncbi.nlm.nih.gov/SNP): +878C→G = rs3027924, +266C→T = rs2075596, +516C→T = rs3027931 and 648A→G = rs3850326.
Table 2 Primers used for allele‐specific polymerase chain reaction.
| Primer location | Primer sequence |
|---|---|
| 3′UTR sequencing primers | |
| UTR‐F | 5′‐gcaaggagagcagccccaagg‐3′ |
| UTR‐R | 5′‐agttctcaacactgtcacatc‐3′ |
| 3′UTR allele‐specific primers (+878C→G; use with EX4‐F or IVS3‐2F primer) | |
| 878C | 5′‐ccctgtccactaagtcacag‐3′ |
| 878G | 5′‐ccctgtccactaagtcacac‐3′ |
| Intron 3 sequencing primers | |
| IVS3‐F | 5′‐aagttctcaaggtagcag‐3′ |
| IVS3‐R | 5′‐aggaacaattagaggctc‐3′ |
| Intron 3 allele‐specific primers (+266C→T) | |
| 266C | 5′‐gcagaggaacttgcagagcc‐3′ |
| 266T | 5′‐gcagaggaacttgcagagct‐3′ |
| Intron 3 allele‐specific primers (+516C→T) | |
| 516C | 5′‐atgggcctcagcctgacaac‐3′ |
| 516T | 5′‐atgggcctcagcctgacaat‐3′ |
| Intron 3 allele‐specific primers (+648A→G) | |
| 648A | 5′‐gcagtgtgactctcgttcaa‐3′ |
| 648G | 5′‐gcagtgtgactctcgttcag‐3′ |
| Common reverse primer for intron 3 ASP | |
| EX4‐R | 5′‐tgagtcttagctggctccttg‐3′ |
| Extra sequencing primers | |
| IVS3‐2F | 5′‐gagcctctaattgttccttg‐3′ |
| EX4‐F | 5′‐tccacccaggtcatggtgatc‐3′ |
Allele‐specific PCR was performed to amplify a fragment including the position of the mutation. The allele‐specific PCR products were sequenced to determine which haplotype contained the mutation. It was then possible to determine the parent of origin by genotyping the SNP in the parents. By combining the parent of origin with the XCI results, the percentage of cells expected to be expressing the mutant MECP2 was calculated. Where parent of origin could not be determined, but it was known from which parent the more active X chromosome was inherited, it was assumed that the mutation was of paternal origin.
Phenotype scoring, data management and statistical analysis
Genotype–phenotype investigations by our groups have until now relied on scoring systems previously established by others. These scales use simple and objective measures that are not dependent on clinical skill or intuition. We refer to these scoring systems as the Kerr profile and the Pineda and Percy scores.18 As our previous work has shown that the Pineda score, which takes into account historical data as well as current clinical status, is the most sensitive to change, we used both the Kerr profile and Pineda Severity scores. These scores had already been generated for the Australian cases based on data collected in 2000.18,19 For the UK cases, when necessary, telephone interviews were arranged with families (conducted by HA) to obtain the necessary information and to supplement the details available from clinical sources.
Statistical analysis was undertaken using Stata V.9.0. For the 15 Australian cases the previously generated scores, where missing data were replaced with the average score for that item for the Rett classification type (ie, classic, mild atypical or early‐onset atypical), were used.18 For the 28 UK cases, an imputation routine, in which missing values for each scale were imputed on the basis of other values in that scale, was used to estimate missing values and to maximise the use of the scarce data.20 For this, imputation data that had been collected on a further 102 UK cases (either with p.T158M or p.R168X mutations but non‐informative XCI data, or with another mutation) were included. Linear regression was undertaken with the Kerr profile and Pineda scales as continuous variables and the percentage of the mutant allele as the independent variable. A case with both p.R168X and trisomy 21 was excluded from the analysis to avoid the potential confounding effects arising from coexisting Down's syndrome.21
Results
Molecular assay
Of the 83 cases recruited, XCI analysis failed in 17 cases due to poor DNA quality or insufficient DNA. Of the remaining 66 cases, 9 were homozygotic at the androgen receptor locus, and in 14 cases it was not possible to ascertain which parental allele was most active because of missing paternal samples or an approximate 50:50 ratio of XCI. Among these non‐contributory samples, two mutations were of maternal origin and three of paternal origin. In the 43 cases in which the most active allele was ascertained, 21 mutations were of paternal origin and none were of maternal origin. The normal allele was favoured in 18 of 21 (86%) cases. In the other 22 cases there were no informative SNPs to allow determination of parental origin.
Clinical data
For the 28 UK cases there were 29 missing individual items, equivalent to 5.2% of all items, for the Kerr profile and 26 missing items, equivalent to 9.3% of all items, for the Pineda scale. In the Kerr profile items relating to height, joint contractures, mood and sleep disturbance, and in the Pineda scale, items relating to age at sitting and acquisition and loss of ambulation, had the most missing items.
Correlation between proportion of mutated allele activity and clinical severity
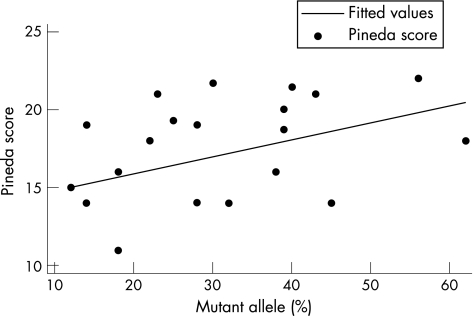
Among cases with the p.R168X mutation, clinical severity increased significantly as the proportion of cells with an active mutant allele increased (fig 1, table 3). This effect was observed with both the Pineda (p = 0.05) and Kerr (p = 0.014) scoring systems. When the data were adjusted for age, the effect with the Kerr profile became more significant (p = 0.005) and the Pineda score less so (p = 0.15).
Figure1 Correlation between proportion of R168X allele active and clinical severity. R2 = 17%.
Table 3 Relationship between phenotype severity scores and percentage of mutant allele active for mutations p.T158M and p.R168X.
| Mutation | Scoring system | Cases (n) | Mean (SD) | Minimum value | Maximum value | r | *Reg. coefft. | p Value |
|---|---|---|---|---|---|---|---|---|
| p.R168X | Pineda scale unadjusted | 23 | 16.87 (3.29) | 10.93 | 21.69 | 0.42 | 0.11 | 0.05 |
| Pineda scale age adjusted | 0.08 | 0.15 | ||||||
| p.R168X | Kerr profile unadjusted | 23 | 22.65 (4.40) | 14.25 | 30 | 0.49 | 0.16 | 0.02 |
| Kerr profile age adjusted | 0.21 | 0.005 | ||||||
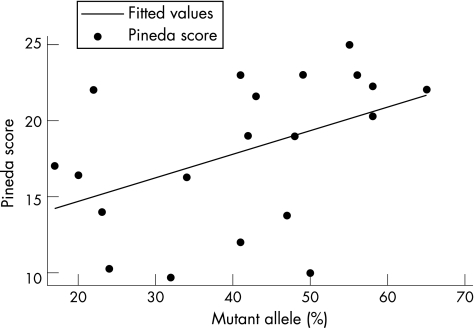
| p.T158M | Pineda scale unadjusted | 20 | 17.64 (4.78) | 9.68 | 25 | 0.45 | 0.16 | 0.05 |
| Pineda scale age adjusted | 0.17 | 0.04 | ||||||
| p.T158M | Kerr profile unadjusted | 20 | 24.90 (6.93) | 10 | 33 | 0.35 | 0.17 | 0.13 |
| Kerr profile age adjusted | 0.20 | 0.09 | ||||||
| p.T158M and p.R168X | Pineda scale unadjusted | 43 | 17.48 (4.17) | 9.68 | 25 | 0.44 | 0.13 | 0.003 |
| Pineda scale age adjusted | 0.12 | 0.005 | ||||||
| p.T158M and p.R168X | Kerr profile unadjusted | 43 | 23.70 (5.76) | 10 | 33 | 0.44 | 0.17 | 0.003 |
| Kerr profile age adjusted | 0.20 | 0.001 |
*Coefficient of the regression of severity score on percentage of mutant allele active—represents the increase in severity score for each percentage point increase.
As the proportion of cells with an active mutant allele increased in cases with the p.T158M mutation, the Pineda score increased significantly (p = 0.04), again indicating greater severity (fig 2, table 3), but the correlation was not significant in the case of the Kerr profile score. The magnitude of the association was the same as with the p.R168X mutation, both with and without age adjustment.
Figure 2 Correlation between proportion of T158M allele active and clinical severity. R2 = 20%.
For both mutations, repeating the analysis using the logistic transformation of the percentage of the mutant allele active had minimal effect on the results. In our dataset, these two mutations have very similar Kerr and Pineda scores, indicating that on average they are of similar severity. Therefore, we also assessed the effect of X inactivation on the two groups combined. This has shown that the increasing severity indicated by both the Pineda and the Kerr scales was associated with the effects of X inactivation both when not adjusted (p = 0.003 for either scale) and when adjusted for age (p = 0.005 for Pineda and p = 0.001 for Kerr), as well as with the direction of XCI skewing. The R2 (square of the correlation coefficient) was 19% for both; the direction of XCI has therefore been shown to account for 19% of the variance in phenotypic outcome, a substantial effect.
Discussion
In this collaborative international study, our data show a relationship between the extent and direction of skewing of XCI and clinical severity for cases with the p.T158M or p.R168X mutation in the MECP2 gene. Both p.R168X and p.T158M are typically associated with a severe presentation,2,7,19 but both have also been reported in mildly affected cases with the preserved speech variant of RTT.22,23,24 This suggests that the clinical variability associated with these mutations is broad, and that this may be due to the role of epigenetic influences, including XCI. This contrasts with those with the p.R133C mutation (usually mildly affected)25 or those with the p.R255X mutation (usually more severely affected),26 where there is less variability in terms of severity. The sensitivity of the available severity scales may not be great enough to detect subtle differences caused by the effects of XCI status.
An RNA‐based approach to determine the parent of origin would have resulted in a higher ascertainment, but was not possible in this study as fresh patient samples were not available for RNA extraction. The method used to determine XCI status and parental origin of the mutation in this study was straightforward and did not require further fresh blood samples from patients. However, there were losses at each stage of the testing process. The androgen receptor assay is usually uninformative in approximately 20–30% of cases,7,27 although in our experience this was true for 9 of 66 (15%) cases with DNA of suitable quantity and quality for analysis. Further cases (14/66, 21%) in this study were lost because of difficulty in ascertaining the most active parental allele, usually in cases where only one parental sample was available. In samples where we could determine the parental origin of the MECP2 mutation, 92% (24/26) were of paternal origin, as found in previous studies.28,29 Paternal origin of mutation was assumed in 51% (22/43) of cases included in the investigation, so it is likely that a small number of these cases had the incorrect allele assigned as the active one. It is interesting that so few cases were identified in which >60% of the mutant MECP2 allele was active. There might be a selective advantage in growth and cell division for those haematopoietic cells using the X chromosome with the intact MECP2 locus, but this is not confirmed. There may also be some selection against the long‐term survival of females, with >60% of their cells using the altered copy of the MECP2 locus, or their phenotype may be less typical of classic RTT and may more closely resemble the encephalopathy of males, with MECP2 mutations associated in females with classic RTT. In either case, they might be under‐represented in our clinically selected group. An additional part of the explanation may lie in the fact that, where there is skewing of XCI, it is usually the paternal X chromosome that is preferentially inactivated, and, combined with the fact that most mutations are paternal, this leads to the conclusion that the cases that are more heavily skewed are less likely to express the mutant copy of MECP2.8
In this study, we found that XCI almost always favours the normal allele, as has been reported in mouse models of RTT,30,31 and this is the pattern commonly seen in other X‐linked diseases.32 Although making assumptions based on XCI status in lymphocytes might be considered a compromise in this study, it was neither realistic nor ethical to obtain neurological tissues from patients to study the effects of direction of skewing. Although we have shown that there is a relationship between severity and XCI by mutation, it should not be surprising that there was still variability in those with the same mutation and the same proportion of active mutant allele. In this situation, each individual might be expected to have differences in inter‐tissue and intra‐tissue XCI status, as is sometimes observed in mouse models.30,33 This confirms the finding that milder cases of RTT have a greater degree of XCI skewing than more severe cases, although the parent of origin of the mutation was not determined in that study.8 The variability in the pattern of XCI is likely to be of particular importance in the brain, where abnormal expression of MECP2 in a neurone will affect the expression of MECP2 in the surrounding neurones, even if they are expressing the normal allele.34 Although XCI clearly affects clinical severity in RTT, assessment of XCI status is unlikely to be useful as a prognosticator for individual cases, with the possible exception of those with extreme skewing.
There are now a small number of other examples where influences other than XCI may be modulating phenotype in RTT. We might expect any male patient to represent the most severe phenotype associated with a particular mutation. There is a published report of a male patient with an early truncating mutation, which would be expected to be associated with a severe phenotype, yet he was relatively mildly affected at 11 years.35 There was no evidence for somatic mosaicism, although this could not be completely ruled out. This example suggests that there may be other background genetic and epigenetic influences that contribute significantly to the clinical manifestations of RTT, but the exact nature of these additional factors is yet to be determined.
Despite the limitations, we were able to show in DNA extracted from peripheral leucocytes that there was a correlation between the proportion of the mutated allele used by an individual and the clinical severity of a predominantly neurodevelopmental phenotype. Combining the efforts of three countries was essential for this study, but wider collaborative efforts will be required to study the phenotype associated with other common mutations in a similar way. We had hoped to look at the effects of the parent of origin based on the suggestion that this may also modulate the phenotype.36 As mutations of maternal origin are infrequent, substantially greater numbers of patients would be required to deal with this question.
Acknowledgements
We thank all the families and clinicians who have contributed to the study, the Rett Syndrome Association UK, Dr Alison Kerr and all other clinicians, who recruited patients from the UK, and the Australian Paediatric Surveillance Unit and the Rett Syndrome Association of Australia, who facilitated case ascertainment in Australia. We also acknowledge the molecular work undertaken by Dr Mark Davis.
Abbreviations
PCR - polymerase chain reaction
RTT - Rett syndrome
SNP - single‐nucleotide polymorphism
3′UTR - 3′ untranslated region
XCI - X‐chromosome inactivation
Footnotes
Funding: The work in the UK was supported by The Health Foundation, Jeans for Genes and the Rett Syndrome Association UK, Rett Syndrome Research Foundation, International Rett Syndrome Association, Rett Syndrome Association Scotland, Scottish Hospital Endowments Research Trust (SHERT/Cruden scholarship to MESB), and Institute of Biomedical and Life Sciences Research Committee, University of Glasgow. The Australian Rett Syndrome research was funded by the National Institutes of Health (1 R01 HD43100‐01A1). HL and LC are currently funded by NHMRC program grant 353514, and JC by NHMRC project grants 185202 and 346603, the Rett Syndrome Australian Research Fund, the International Rett Syndrome Association and the Rett Syndrome Research Foundation.
Competing interests: None declared.
References
- 1.Amir R E, Van dV I, Wan M, Tran C Q, Francke U, Zoghbi H Y. Rett syndrome is caused by mutations in X‐linked MECP2, encoding methyl‐CpG‐binding protein 2. Nat Genet 199923185–188. [DOI] [PubMed] [Google Scholar]
- 2.Charman T, Neilson T C, Mash V, Archer H, Gardiner M T, Knudsen G P, McDonnell A, Perry J, Whatley S D, Bunyan D J, Ravn K, Mount R H, Hastings R P, Hulten M, Orstavik K H, Reilly S, Cass H, Clarke A, Kerr A M, Bailey M E S. Dimensional phenotypic analysis and functional categorisation of mutations reveal novel genotype‐phenotype associations in Rett syndrome. Eur J Hum Genet 2005131121–1130. [DOI] [PubMed] [Google Scholar]
- 3.Ham A L, Kumar A, Deeter R, Schanen N C. Does genotype predict phenotype in Rett syndrome? J Child Neurol 200520768–778. [DOI] [PubMed] [Google Scholar]
- 4.Gartler S M, Dyer K A, Graves J A, Rocchi M. A two step model for mammalian X‐chromosome inactivation. Prog Clin Biol Res 1985198223–235. [PubMed] [Google Scholar]
- 5.Lyon M F. X‐chromosome inactivation as a system of gene dosage compensation to regulate gene expression. Prog Nucleic Acid Res Mol Biol 198936119–130. [DOI] [PubMed] [Google Scholar]
- 6.Weaving L S, Williamson S L, Bennetts B, Davis M, Ellaway C J, Leonard H, Thong M K, Delatycki M, Thompson E M, Laing N, Christodoulou J. Effects of MECP2 mutation type, location and X‐inactivation in modulating Rett syndrome phenotype. Am J Med Genet 2003118A103–114. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda T, Yamashita Y, Nagamitsu S, Miyamoto K, Jin J J, Ohmori I, Ohtsuka Y, Kuwajima K, Endo S, Iwai T, Yamagata H, Tabara Y, Miki T, Matsuishi T, Kondo I. Methyl‐CpG binding protein 2 gene (MECP2) variations in Japanese patients with Rett syndrome: pathological mutations and polymorphisms. Brain Dev 200527211–217. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen G P S, Neilson T C S, Pedersen J, Kerr A, Schwartz M, Hulten M, Bailey M E S, Orstavik K H. Increased skewing of X chromosome inactivation in Rett syndrome patients and their mothers. Eur J Hum Genet 200614(11)1189–1194. [DOI] [PubMed] [Google Scholar]
- 9.Ishii T, Makita Y, Ogawa A, Amamiya S, Yamamoto M, Miyamoto A, Oki J. The role of different X‐inactivation pattern on the variable clinical phenotype with Rett syndrome. Brain Dev 200123(Suppl 1)S161–S164. [DOI] [PubMed] [Google Scholar]
- 10.Gill H, Cheadle J P, Maynard J, Fleming N, Whatley S, Cranston T, Thompson E M, Leonard H, Davis M, Christodoulou J, Skjeldal O, Hanefeld F, Kerr A, Tandy A, Ravine D, Clarke A. Mutation analysis in the MECP2 gene and genetic counselling for Rett syndrome. J Med Genet 200340380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan M, Lee S S, Zhang X, Houwink‐Manville I, Song H R, Amir R E, Budden S, Naidu S, Pereira J L, Lo I F, Zoghbi H Y, Schanen N C, Francke U. Rett syndrome and beyond: recurrent spontaneous and familial MECP2 mutations at CpG hotspots. Am J Hum Genet 1999651520–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bienvenu T, Carrie A, De Roux N, Vinet M C, Jonveaux P, Couvert P, Villard L, Arzimanoglou A, Beldjord C, Fontes M, Tardieu M, Chelly J. MECP2 mutations account for most cases of typical forms of Rett syndrome. Hum Mol Genet 200091377–1384. [DOI] [PubMed] [Google Scholar]
- 13.Obata K, Matsuishi T, Yamashita Y, Fukuda T, Kuwajima K, Horiuchi I, Nagamitsu S, Iwanaga R, Kimura A, Omori I, Endo S, Mori K, Kondo I. Mutation analysis of the methyl‐CpG binding protein 2 gene (MECP2) in patients with Rett syndrome. J Med Genet 200037608–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffbuhr K C, Moses L M, Jerdonek M A, Naidu S, Hoffman E P. Associations between MeCP2 mutations, X‐chromosome inactivation, and phenotype. Ment Retard Dev Disabil Res Rev 2002899–105. [DOI] [PubMed] [Google Scholar]
- 15.Villard L, Levy N, Xiang F, Kpebe A, Labelle V, Chevillard C, Zhang Z, Schwartz C E, Tardieu M, Chelly J, Anvret M, Fontes M. Segregation of a totally skewed pattern of X chromosome inactivation in four familial cases of Rett syndrome without MECP2 mutation: implications for the disease. J Med Genet 200138435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurvick C, De Klerk N. Bower C, Christodoulou J, Ravine D, Ellaway C, Williamson S, Leonard H. Rett syndrome in Australia; a review of the epidemiology. J Pediatr 2006148347–352. [DOI] [PubMed] [Google Scholar]
- 17.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen‐receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992511229–1239. [PMC free article] [PubMed] [Google Scholar]
- 18.Colvin L, Fyfe S, Leonard S, Schiavello T, Ellaway C, de Klerk N, Christodoulou J, Msall M, Leonard H. Describing the phenotype in Rett syndrome using a population database. Arch Dis Child 20038838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colvin L, Leonard H, de Klerk N, Davis M, Weaving L, Williamson S, Christodoulou J. Refining the phenotype of common mutations in Rett syndrome. J Med Genet 20044125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little R J A, Rubin D B.Statistical analysis with missing data. 2nd edn. Hoboken, NJ: Wiley, 2002
- 21.Leonard H, Weaving L, Eastaugh P, Smith L, Delatycki M, Witt E I, Christodoulou J. Trisomy 21 and Rett syndrome: a double burden. J Paediatr Child Health 200440406–409. [DOI] [PubMed] [Google Scholar]
- 22.Huppke P, Laccone F, Kramer N, Engel W, Hanefeld F. Rett syndrome: analysis of MECP2 and clinical characterization of 31 patients. Hum Mol Genet 200091369–1375. [DOI] [PubMed] [Google Scholar]
- 23.Zappella M, Meloni I, Longo I, Hayek G, Renieri A. Preserved speech variants of the Rett syndrome: molecular and clinical analysis. Am J Med Genet 200110414–22. [DOI] [PubMed] [Google Scholar]
- 24.Kerr A M, Archer H L, Evans J C, Prescott R J, Gibbon F. People with MECP2 mutation positive Rett disorder who converse. J Intellect Disabil Res 200650386–394. [DOI] [PubMed] [Google Scholar]
- 25.Leonard H, Colvin L, Christodoulou J, Schiavello T, Williamson S, Davis M, Ravine D, Fyfe S, de Klerk N, Matsuishi T, Kondo I, Clarke A, Hackwell S, Yamashita Y. Patients with the R133C mutation: is their phenotype different from patients with Rett syndrome with other mutations? J Med Genet 200340e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jian L, Archer H L, Ravine D, Kerr A, de Klerk N, Christodoulou J, Bailey M E, Laurvick C, Leonard H. p.R270X MECP2 mutation and mortality in Rett syndrome. Eur J Hum Genet 2005131235–1238. [DOI] [PubMed] [Google Scholar]
- 27.Amir R E, Van dV I, Schultz R, Malicki D M, Tran C Q, Dahle E J, Philippi A, Timar L, Percy A K, Motil K J, Lichtarge O, Smith E O, Glaze D G, Zoghbi H Y. Influence of mutation type and X chromosome inactivation on Rett syndrome phenotypes. Ann Neurol 200047670–679. [PubMed] [Google Scholar]
- 28.Trappe R, Laccone F, Cobilanschi J, Meins M, Huppke P, Hanefeld F, Engel W.MECP2 mutations in sporadic cases of Rett syndrome are almost exclusively of paternal origin. Am J Hum Genet 2001681093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard M, Couvert P, Carrie A, Tardieu M, Chelly J, Beldjord C, Bienvenu T. Parental origin of de novo MECP2 mutations in Rett syndrome. Eur J Hum Genet 20019231–236. [DOI] [PubMed] [Google Scholar]
- 30.Young J I, Zoghbi H Y. X‐chromosome inactivation patterns are unbalanced and affect the phenotypic outcome in a mouse model of Rett syndrome. Am J Hum Genet 200474511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson C M, Pelka G J, Radziewic T, Shahbazian M D, Christodoulou J, Williamson S L, Tam P P. Reduced proportion of Purkinje cells expressing paternally derived mutant Mecp2308 allele in female mouse cerebellum is not due to a skewed primary pattern of X‐chromosome inactivation. Hum Mol Genet 2005141851–1861. [DOI] [PubMed] [Google Scholar]
- 32.Azofeifa J, Voit T, Hubner C, Cremer M. X‐chromosome methylation in manifesting and healthy carriers of dystrophinopathies: concordance of activation ratios among first degree female relatives and skewed inactivation as cause of the affected phenotypes. Hum Genet 199596167–176. [DOI] [PubMed] [Google Scholar]
- 33.Gibson J H, Williamson S L, Arbuckle S, Christodoulou J. X chromosome inactivation patterns in brain in Rett syndrome: implications for the disease phenotype. Brain Dev 200527266–270. [DOI] [PubMed] [Google Scholar]
- 34.Braunschweig D, Simcox T, Samaco R C, LaSalle J M. X‐chromosome inactivation ratios affect wild‐type MeCP2 expression within mosaic Rett syndrome and Mecp2−/+ mouse brain. Hum Mol Genet 2004131275–1286. [DOI] [PubMed] [Google Scholar]
- 35.Ravn K, Nielsen J B, Uldall P, Hansen F J, Schwartz M. No correlation between phenotype and genotype in boys with a truncating MECP2 mutation. J Med Genet 200340e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelka G J, Watson C M, Christodoulou J, Tam P P. Distinct expression profiles of Mecp2 transcripts with different lengths of 3′UTR in the brain and visceral organs during mouse development. Genomics 200585441–452. [DOI] [PubMed] [Google Scholar]