Abstract
Background
During a genetic study of autism, a female child who met diagnostic criteria for autism spectrum disorder, but also exhibited the cognitive–behavioural profile (CBP) associated with Williams–Beuren syndrome (WBS) was examined. The WBS CBP includes impaired visuospatial ability, an overly friendly personality, excessive non‐social anxiety and language delay.
Methods
Using array‐based comparative genomic hybridisation (aCGH), a deletion corresponding to BAC RP11‐89A20 in the distal end of the WBS deletion interval was detected. Hemizygosity was confirmed using fluorescence in situ hybridisation and fine mapping was performed by measuring the copy number of genomic DNA using quantitative polymerase chain reaction.
Results
The proximal breakpoint was mapped to intron 1 of GTF2IRD1 and the distal breakpoint lies 2.4–3.1 Mb towards the telomere. The subject was completely hemizygous for GTF2I, commonly deleted in carriers of the classic ∼1.5 Mb WBS deletion, and GTF2IRD2, deleted in carriers of the rare ∼1.84 Mb WBS deletion.
Conclusion
Hemizygosity of the GTF2 family of transcription factors is sufficient to produce many aspects of the WBS CBP, and particularly implicate the GTF2 transcription factors in the visuospatial construction deficit. Symptoms of autism in this case may be due to deletion of additional genes outside the typical WBS interval or remote effects on gene expression at other loci.
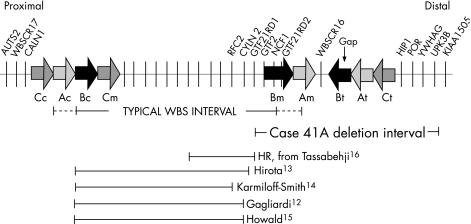
Williams–Beuren syndrome (WBS) results from a ∼1.5 Mb deletion in chromosome 7q11.23 that contains about 20 genes. The disorder occurs at a rate of 1:7500, as estimated in a recent study.1 Three region‐specific low‐copy repeats (LCRs), referred to as centromeric, medial and telomeric blocks A, B and C, flank the WBS deletion site (fig 1). Most WBS deletions are 1.5 Mb in length (95%) and result from unequal crossing over between the centromeric and medial portions of the block B LCR containing GTF2I and NCF1, but sparing GTF2IRD2.2GTF2IRD2 is deleted in the much rarer 1.84 Mb deletion cases mediated by portions of the block A LCR.
Figure 1 Schematic representation of the classic 7q11.23 William–Beuren syndrome (WBS) deletion region and the atypical deletion detected in patient 41A. The thick horizontal arrows represent the three large blocks of low‐copy repeats (LCRs), labelled as blocks A, B and C, with centromeric (c), medial (m) and telomeric (t) sequence indicated. Single‐copy regions flank the LCRs, between Cm and Bm, and between Am and Bt (which contains the gene WBSCR16). The common WBS deletion interval (approximately 1.5 Mb) is depicted as a solid line. The extended boundaries of the rare 1.84 Mb deletion are shown as dotted lines. Roughly 28 genes lie in the WBS deletion interval; only the distal genes that have been the focus of genotype–phenotype correlations are labelled for clarity. Atypical WBS deletions are displayed below the case 41A deletion interval.
The cognitive–behavioural profile (CBP) of WBS includes severely impaired visuospatial ability, mental retardation, an overly friendly personality accompanied by excessive non‐social anxiety, attention deficit, hyperacusis and/or love of music.3 Language delay is usually present, but verbal IQ is generally, although not always, higher than performance IQ in WBS cases.3,4,5,6,7,8 By contrast, although language delay is common to both disorders, subjects with autism tend to have higher performance IQs than verbal IQs and display excessive social versus non‐social anxiety. However, difficulty reading emotions from the face and voice, an inability to get along with peers and poor social judgement are common features of both disorders.9,10,11
Key points
Hemizygosity of GTF2I and GTF2IRD1 results in the WBS cognitive‐behavioural profile including a severe visuospatial construction deficit and an overly friendly personality.
GTF2I and GTF2IRD1 oppositely regulate Goosecoid, a gene involved in craniofacial development.
A gene related to Goosecoid, called Goosecoid‐like resides in the 22q11 DiGeorge/Velocardiofacial syndrome region also associated with autism and a visuospatial construction deficit.
WBS cases with atypical CBPs and nested proximal deletions have helped generate genotype–phenotype correlations. For instance, there have been multiple reports of WBS cases with normal visuospatial ability that carry atypical deletions sparing the GTF2 transcription factors GTF2IRD1 and GTF2I.9,10,11,12,13,14,15 Furthermore, a recent report described a female child carrying a WBS deletion that extended through intron 1 of GTF2IRD1, but spared GTF2I, whose visuospatial abilities were less severely impaired than those of full‐deletion carriers.16 This finding suggests that hemizygosity of both GTF2I and GTF2IRD1 is necessary to produce the severe visuospatial deficits seen in carriers of the classic WBS deletion, a hypothesis supported by our data.
Here, we describe a female child with many aspects of the WBS CBP, who also meets diagnostic criteria for autism spectrum disorder, as assessed by the Autism Diagnostic Interview–Revised17 and the Autism Diagnostic Observation Schedule,18 but lacks the classic medical and physical features of WBS. She carries a novel, de novo deletion between 2.4 and 3.1 Mb in length extending towards the telomere from intron 1 of GTF2IRD1, which results in hemizygosity of the genes at the distal end of the WBS deletion, including GTF2IRD1, GTF2I and GTF2IRD2 (figs 1, 2), as well as roughly 14 other genes which lie downstream of a gap in the genomic sequence that has yet to be resolved. Two of the 14 genes have previously been implicated in neuropsychiatric disorders, Huntingtin‐interacting protein 1 (HIP1)19 and tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein γ (YWHAG).20 Therefore, the symptoms of autism observed in our patient may be caused or exacerbated by hemizygosity of another gene or genes in the deletion interval or result from remote effects on gene expression at other loci.
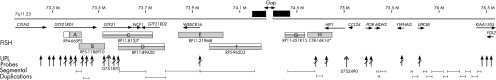
Figure 2 Atypical deletion of the distal William–Beuren syndrome (WBS) deletion interval of approximately 2.4–3.1 Mb. The proximal deletion breakpoint lies in intron 1 of GTF2IRD1, completely deleting GTF2I, NCF1 and GTF2IRD2, the distal genes in the WBS deletion interval. Fluorescence in situ hybridisation (FISH) clones with hatch marks indicate that the subject is hemizygous for that sequence. Clones labelled with half‐hatch marks gave diminished signals owing to cross hybridisation. All Universal Probe Library (UPL) probes listed in table 2 are displayed as upright arrows. A line crossing the arrow signifies that the subject is hemizygous for the sequence corresponding to the UPL probe. The double‐headed upright arrow marks the location of microsatellites D7S1870 and D7S2490 used to determine the deletion that occurred de novo on a paternal chromosome. The location of segmental duplications, indicating highly repetitive sequence, has been approximated from the Database of Genomic Variants.
Materials and methods
This project was approved under the guidelines of the Ministry of Health of Costa Rica, the ethical committee of the National Children's Hospital in San José, California, USA, and the institutional review board at Mount Sinai School of Medicine, New York, USA, in accordance with the Declaration of Helsinki. These approvals remain active. The parents of patient 41A provided written informed consent.
We are conducting an ongoing genetic study of autism in the isolated founder population of the Central Valley of Costa Rica.21 Occasionally, we are referred atypical cases including patient 41A, a female child that smiled often and was overly friendly with strangers. She had a normal karyotype (G banding at 550), was negative for fragile X and lacked a constellation of physical features consistent with a known syndrome. However, we hypothesised that despite the normal karyotype data, mental retardation or atypical behavioural features could be indicative of an underlying submicroscopic chromosomal abnormality, so we decided to screen this patient using array‐based comparative genomic hybridisation (aCGH).
Array‐based comparative genomic hybridisation
Labelling of DNA samples, including a dye swap, and hybridisation to the Spectral Chip 2600 (Spectral Genomics, Houston, Texas, USA) whole‐genome bacterial artificial chromosome (BAC) array, was carried out at Spectral Genomics, according to the manufacturer's protocol. Hybridised microarray slides were analysed with GenePix 4000B scanner (Axon Ins, Union City, California, USA), and data obtained were analysed using the Spectralware V.2.24 software (Spectral Genomics). Normalisation of the Spectral aCGH data was performed using the global linear regression and pin linear options provided in the Spectralware V.2.24 software. Clones were considered to be positive for a duplication or deletion if the log2 fluorescence intensities of the spots were >2 standard deviations (SDs) beyond the mean ratio of intensities for the autosomes.
Fluorescence in situ hybridisation confirmation of aCGH results
BAC DNAs for validation of aCGH results and LB agar stabs of individual BAC clones were obtained directly from Spectral Genomics and BAC PAC Resources Center (San Francisco, California, USA), respectively, and are listed in supplementary table A (details available online at http://jmg.bmj.com/supplemental). BAC DNA was prepared according to the product manual of the Nucleobond Plasmid Maxikit (BD Biosciences Clontech, San Jose, California, USA). Each BAC was labelled using the Nick Translation Reaction Kit (Vysis, Downer's Grove, Illinois, USA) and Spectrum Green‐11‐dUTP (Vysis) or Spectrum Red‐11‐dUTP (Vysis). Single or dual‐coloured fluorescence in situ hybridisation (FISH) was performed on metaphase spreads from the peripheral blood of the participant or established lymphoblastoid lines dropped on to precleaned glass slides using a standard protocol. Images were captured with an ImagePoint cooled CCD video camera (Photometrics, Tuscon, Arizona, USA) through a Labophot‐2A fluorescence microscope (Nikon, Melville, New York, USA) using Cytovision FISH software (Applied Imaging, San Jose, California, USA).
Genomic quantitative polymerase chain reaction for deletion mapping
We adapted the Universal Probe Library (UPL) system from Roche (Roche, Nutley, New Jersey, USA) to perform genomic quantitative polymerase chain reaction (PCR). The Roche system offers a library of eight‐nucleotide fluorescence‐labelled probes containing locked nucleic acids to ensure tight binding with the target sequence. Probes are chosen according to the primer sequences specified using ProbeFinder V.2.04 software (Roche, http://www.universalprobelibrary.com). This system is a precise and much cheaper alternative to traditional TaqMan assays, as the complete UPL covers >99% of the transcriptome using only 90 probes paired with transcript‐specific primers.
Primers to be used with the UPL probes were designed using gene sequences from the UCSC Genome Browser (NCBI Build 35, http://genome.ucsc.edu/) and the ProbeFinder application. Supplementary table B includes all primers and probes used for the genomic deletion analysis (available online at www.jmg.bmjjournals.com/supplemental). Reactions were performed with a volume of 10 ml containing 25 ng DNA, 400 nM of each primer, 100 nM UPL probe (Roche) and 1× Platinum Quantitative PCR SuperMix‐uracil‐N‐glycosylase, with Rox (Invitrogen, Carlsbad, California, USA). All reactions were performed in quadruplicate with an ABI Prism 7900HT sequence detection system (Applied Biosystems, California, USA). In addition to the case and parental DNA (case 41A, 41B father and 41C mother) and the genes to be assayed for copy number, we included a trio (child and two parents) from the CVCR as control genomic DNA samples (C1, C2 and C3) and four reference genes (COBL, GUSB, PPIA and SNCA) on each plate. PCR conditions were as follows: 2 min uracil‐N‐glycosylase activation at 50°C, then 2 min denaturation at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min.
Quantitative PCR to evaluate cDNA expression
cDNA was generated with SuperScript III First Strand kit according to the manufacturer's instructions. The products were diluted either 1:250 (for reference probes) or 1:25 (for all other probes), and 4.5 μl of the diluted reaction was used in the PCR reactions.
Four genes within the deletion interval and three reference genes were assayed by TaqMan gene expression probes (Applied Biosystems) according to standard protocols. Expression probes are listed in supplementary table C (available online at http://jmg.bmj.com/supplemental).
Quantitative PCR data analysis
The data were first evaluated using the Sequence Detection Software V.2.1 (Applied Biosystems). The baselines and thresholds for each gene were adjusted to measure Ct values at the most linear range possible. Quadruplicates having a SD >0.2 were evaluated and outlying Ct values >2 SDs from the mean of the quadruplicate were excluded from further analysis. The remaining Ct values were then fed into the qBase program22 and analysed using the default options and settings. The reference genes were excluded from the analysis one at a time on the basis of the coefficient of variation, until two remained.23 For genomic quantitative PCR experiments, relative quantification was performed and then calibrated by setting control DNA sample C2 to a value of 1. Single‐copy deletions were detected when relative copy number values were <0.75. In the quantitative PCR expression assays, relative quantification was performed, calibrated by setting 41C to a value of 1, then all values were normalised to the mean of 41B and 41C as 1. (For raw data from both the genomic quantitative PCR mapping assays and quantitative PCR expression assays, refer to supplementary tables D and E, respectively, available online at http://jmg.bmj.com/supplemental.)
Microsatellite genotyping
The microsatellite primer pairs D7S1870 in GTF2I and D7S2490 at the distal end of HIP1 were synthesised by Integrated DNA Technologies (Coralville, Iowa, USA). PCR conditions are available on request. The microsatellites were genotyped using the ABI 31‐30 XL (Applied Biosystems) and genotype calls were performed using Genemapper V.3.7 (Applied Biosystems).
Results
Patient 41A (age 6 years and 6 months at the time of interview)
The mother of patient 41A (41C, 18½ years old at the time of the child's birth) experienced bleeding in both the first and third trimesters and gained <7 kg during the pregnancy. The child was delivered at term without complications, although she was born with an orbital hemangioma that required surgery at 2 years and 8 months of age, resulting in strabismus. Her birth weight, height and head circumference were 2930 g (20th centile), 47 cm (10th centile) and 32 cm (<5th centile), respectively. (These norms are from the National Center for Health Statistics, as none are available for Costa Rica.) She has one younger male sibling who is in good health and had no perinatal complications. She did not have a social smile until she was 1 year old and had delayed acquisition of phrase speech (3½ years of age). She developed sphincter control at 42 months; other motor milestones were generally normal, or only mildly delayed. Her parents placed her in first grade in a regular school, but she had to be removed because of aggression against her peers and other disruptive behaviour. She is very interested in music, and certain sounds, but does not have hyperacusis according to parental reports, and clinical observation, although hyperacusis was not formally tested.
The patient met the Autism Diagnostic Interview–Revised criteria for autism in all three domains. Her most severe symptoms included deficits in the comprehension of simple language, articulation, reciprocal conversation, attention to voice, empathy and socialising with peers. She met the Autism Diagnostic Observation Schedule criteria for autism in the social, play and restricted behaviour domains, but was 2 points below the cut‐off score in the communication domain. She smiles easily and is superficially friendly with strangers, although she exhibits minimal reciprocal social interaction. She also displays excessive non‐social anxiety and moderately severe attention deficits.
Her cognitive profile on the Wechsler Preschool and Primary Scale of Intelligence—third edition24 showed a verbal IQ of 53, a performance IQ of 57 and a Full‐Scale composite IQ of 49, which places her in the extremely low range when compared with her peers. She had an IQ of 67 using the Leiter‐R,25 a non‐verbal IQ test. Her verbal and performance abilities were also both in the extremely low range. We noted no significant differences in performance on the verbal subtests, suggesting that her verbal cognitive abilities are similarly developed. Notably, she showed an extreme weakness in visuospatial construction (block design score of 1; see supplementary tables Fa and Fb for full details of the IQ results, available online at http://jmg.bmj.com/supplemental). Developmental testing showed an adaptive behaviour composite score of 50, a communication skills summary score of 44, a daily living skills domain summary score of 64 and a social skills summary score of 54 (Vineland Adaptive Behavioral Scales).
At the time of the evaluation, the patient weighed 21 kg (50th centile), measured 120 cm in height (50th centile), and had a head circumference of 51 cm (50th centile). The results of our medical evaluation did not show any cardinal medical features of WBS, such as heart defects (negative echocardiogram and no heart murmur), joint laxity, loose skin or genitourinary dysfunction (negative renal ultrasound), although more extensive diagnostic testing might uncover such abnormalities. She also had no endocrine or serum calcium abnormalities and did not have feeding or sleeping difficulties as an infant. Her blood pressure was normal. Her head and facial features were remarkable for asymmetrical, abnormally shaped external ears, most prominent on the left side and iatrogenic strabismus due to the orbital hemangioma (fig 3). The only facial features consistent with WBS that she displayed include widely spaced teeth, a wide mouth and prominent lips. Specifically, she did not display dolichocephaly, bifrontal narrowing, mid‐face hypoplasia, periorbital fullness, an upturned nose, a bulbous nasal tip or a long philtrum.
Figure 3 Anteroposterior (A) and lateral (B) photographs of patient 41A. Note the abnormal external rotation of the ear on the lateral photograph. Parental consent was obtained for publication of this figure.
Array‐based comparative genomic hybridisation
Analysing the aCGH data, clone RP11‐89A20 showed a signal consistent with a deletion of this region (table 1). The two flanking clones, RP11‐137M14 (proximal) and RP11‐451M14 (distal), located approximately 3.3 Mb apart, were not deleted. Only the clones in the WBS deletion interval were considered for further follow‐up. Refinement of the deletion interval was conducted with a combination of FISH and genomic qualitative PCR with UPL probes.
Table 1 Patient 41A: results of an array‐based comparative genomic hybridisation from Spectral Genomics.
| Clone name | Cyto band | Cy5 test log ratios | Cy3 test log ratios | Clone gain or loss | Known genes in the BAC clones |
|---|---|---|---|---|---|
| CTC‐232B23 | 1pter | 1.542 | 0.803 | G | CENTB5, SCNN1D, B3GALT6, DVL1, PUSL1, TNFRSF18, UBE2J2, TNFRSF4 |
| RP4‐703E10 | 1p36.32–1p36.33 | 1.399 | 0.647 | G | Observed in 14/20 Costa Rican cases |
| RP1‐45P21 | 6p22.2 | 1.36 | 0.733 | G | Cluster of BTN genes in the extended MHC |
| B39H04 | 7q11.23 | 0.799 | 1.236 | L | GTF2IRD1 |
| RP11‐89A20 | 7q11.23–7q11.23 | 0.774 | 1.244 | L | GTF2I |
| RP11‐89A20(E) | 7q11.23 | 0.755 | 1.321 | L | GTF2IRD2 |
| RP11‐65C22 | 17q24–17q24 | 1.487 | 0.797 | G | Solute carrier family 39 (metal ion transporter) |
BAC, bacterial artificial chromosome; G, gain (duplication); L, loss (deletion).
The data were analysed using the global linear and pin lowess options of the Spectral V.2.24 software. The known CNV column indicates whether this particular clone was duplicated or deleted in normal people. We indicate a new CNV because this clone was duplicated in 14/20 Costa Rican cases, with autism hybridised in the original study (results to be published elsewhere). Importantly, we tested many of the putative CNVs indicated by the array data and found them to be false‐positive results, therefore we only followed up the most convincing findings in our patients.
Deletion mapping by FISH and genomic quantitative PCR
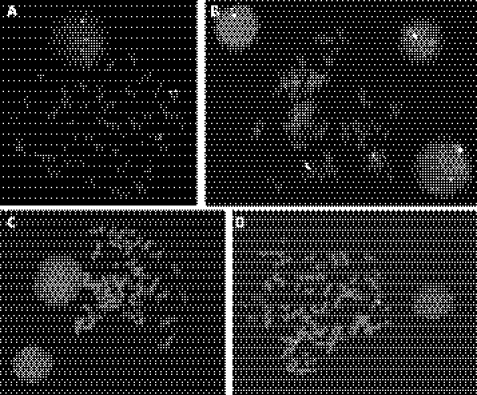
Given the genomic location of the deleted BAC within the classic WBS deletion interval, we first tested the ∼180 kb probe LSI ELN (Vysis) used to screen for the WBS deletion. The LSI ELN probe gave two clear signals on both metaphase chromosomes and interphase nuclei, indicating that patient 41A does not carry a deletion of the ELN gene (fig 4A). We then selected a tiling path of FISH clones across the immediate interval surrounding RP11‐89A20 to confirm the Spectral Genomics array data. The deletion was confirmed using the clones in supplementary table A (available online at http://jmg.bmj.com/supplemental), and the results are depicted schematically in fig 2. Figure 4B shows FISH results for RP11‐89A20 (clone D in fig 2) that gave one normal signal and a second weak signal of varying intensity, although this clone was clearly deleted according to the Spectral Genomics software. A diminished signal could indicate that this segment of DNA was only partially deleted or that the BAC was partially hybridising to non‐allelic homologous sequence elsewhere. The second alternative is most likely, given that others have observed a similar hybridisation pattern using FISH clones in the block B LCR26; the results obtained with the adjacent proximal and distal overlapping clones, the UPL probe data and the original Spectral Genomics array data all indicate a deletion. Hybridisation of RP11‐89A20 (clone D) to paternal and maternal metaphase chromosomes and interphase nuclei showed two clear signals, indicating that this deletion is de novo in patient 41A (fig 5C,D). In addition, parental nuclei often displayed a second weak signal adjacent to a strong signal in many of the nuclei, as did the normal chromosome 7 from 41A. However, this second signal was often too close to resolve from the stronger signal hybridising to its identical sequence in block B and was only clearly visible as a discrete albeit weak signal on the deleted chromosome 7 of 41A.
Figure 4 (A) Fluorescence in situ hybridisation (FISH) results for patient 41A performed with the William–Beuren syndrome (WBS) probe LSI ELN (Vysis). The ELN probe (orange signal) is labelled with Spectrum Orange dUTP and LSI D7S486, D7S522 (green signal) is labelled with spectrum green dUTP. (B) FISH results for patient 41A performed with the SGI BAC RP11‐89A20 (green) labelled with spectrum green dUTP, and the control probe for the 7q subtelomere (orange, TelVysion 7q (Vysis)) labelled with spectrum orange dUTP. A normal green signal and a second weaker green signal (indicated with arrowheads) of varying intensity are observed on both metaphase chromosomes and interphase nuclei. Control probe signals appear equal in intensity on both chromosomes 7. (C, D). FISH results for the father and mother, respectively, of patient 41A, performed with the SGI BAC RP11‐89A20 (green) labelled with spectrum green dUTP, and the control probe for the 7q subtelomere (orange, TelVysion 7q (Vysis)) labelled with spectrum orange dUTP. Two normal green signals are observed, and control probe signals are also of equal intensity on both chromosomes 7.
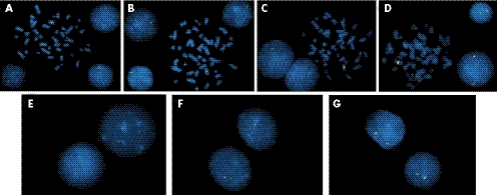
Figure 5 Fluorescence in situ hybridisation (FISH) results for patient 41A performed with BAC and PAC clones that span the distal end of the William–Beuren syndrome (WBS) region (fig 2). (A) BAC RP11‐813J7 labelled with spectrum green dUTP. A normal green signal and a second weaker signal are observed on both metaphase chromosomes and interphase nuclei. (B) BAC RP11‐219M8 labelled with spectrum green dUTP. A normal green signal and a second faint signal are consistently observed on both metaphase chromosomes and interphase nuclei. (C) Dual‐colour FISH results with PAC RP5‐1186P10 (spectrum red dUTP) and BAC RP11‐451K15 (spectrum green dUTP). A single red signal and two green signals of equal intensity are present. (D) Dual‐colour FISH results for PACs RP4‐665P5 (spectrum green dUTP) and RP5‐962D3 (spectrum red dUTP). (E, F, G) Interphase FISH results with BAC CTB‐16K1427 (spectrum green dUTP) for patient 41A, her mother and her father, respectively. Nuclei of patient 41A exhibit a single green hybridisation signal, whereas both parents have normal hybridisation patterns, with two signals of equal intensity.
BACs RP11‐813J7 (clone C) and RP11‐219M8 (clone E) also gave a normal signal and a second weaker signal when hybridised to patient 41A (fig 5A, B, respectively). BAC RP11‐813J7 shares substantial overlap with RP11‐89A20 and therefore displays a similar hybridisation pattern. Dual‐colour FISH with PAC RP5‐1186P10 (clone B) gave one signal indicating that this segment of DNA is deleted. BAC RP11‐451K15 (clone G) gave a strong signal and a slightly diminished signal (fig 5C), consistent with partial cross hybridisation to other repetitive sequences as has been reported by Antonell et al.26 Dual‐colour FISH results for PACs RP4‐665P5 (clone A) and RP5‐962D3 (clone F) also showed two signals for each PAC, but again, the signals appear diminished on one chromosome 7 (fig 5D). Clone A is not completely deleted, as indicated by the UPL probe data (fig 6). Figure 5 (E, F, G) shows the results of interphase FISH with BAC CTB‐16K1427 (clone H) for patient 41A, her mother, and her father, respectively. CTB‐16K14 is an unsequenced BAC from the Caltech BAC library. Nuclei of patient 41A exhibit a single hybridisation signal, indicating a deletion of the sequence containing HIP1, whereas both parents have normal hybridisation patterns, with two signals of equal intensity.
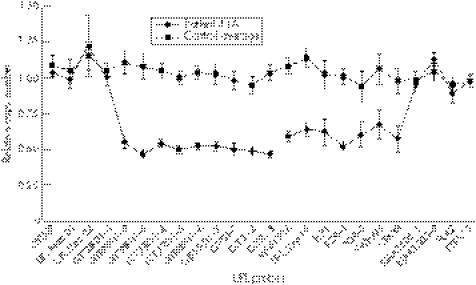
Figure 6 Graph of the Universal Probe Library (UPL) probe data used to fine map the deletion carried by patient 41A, displaying relative genomic copy numbers and standard deviations (SDs) from the qBase program. The control average series is the mean of the values and SDs of the three control DNA samples (C1, C3 and the calibrator C2) as well as the parents of patient 41A (41B and 41C). Single‐copy number deletions were detected in the patient when relative quantification values dropped to <0.75. The deletion spans from probe GTF2IRD1‐2 to UPK3B.
These data are also consistent with the fine‐mapping UPL probe data (fig 6, raw data in supplementary data table D, available online at http://jmg.bmj.com/supplemental) that place the centromeric breakpoint of the deletion within the first intron of GTF2IRD1 in clone A, and the telomeric breakpoint somewhere between gene UPK3B and the putative gene KIAA1505. Note that a gap in the genomic sequence exists between WBSCR16 and HIP1; therefore, placement of additional UPL probes in this region was not possible. Supplementary table B contains a list of the genes in the proximal portion of the deleted interval, upstream of the gap in the genomic sequence and downstream of the gap, respectively (http://jmg.bmj.com/supplemental).
Analyses of gene expression
Quantitative reverse transcriptase‐PCR analyses of GTF2IRD1, GTF2I, HIP1 and YWHAG were performed on cDNA generated from RNA extracted from transformed lymphocytes derived from patient 41A, her father (41B) and her mother (41C). Expression of GTF2I was decreased to about 60%, and expression of YWHAG was decreased to about 50%, in patient 41A compared with both parents (supplementary data fig A, supplementary table C, raw data in supplementary data table E, available online at http://jmg.bmj.com/supplemental). Expression of HIP1 was unchanged in either patient 41A or in the WBS control. However, expression of GTF2IRD1, using probes targeting exons 2 and 11, deleted in patient 41A, was increased in both patient 41A and the WBS control (NIGMS cell line GM13460, Coriell) and was at least 2.5 times the expression observed in 41C. The father, 41B, exhibited an intermediate increase in expression for both GTF2IRD1 probes that fell between the levels of the deletion carriers and 41C. The abnormal expression values for GTF2IRD1 in the father may be indicative of some kind of genomic rearrangement; however, we were unable to detect the parental inversion that has been described previously in the father.28
Microsatellite data
Data from microsatellites upstream, D7S1870 and downstream of the gap, D7S2490 (fig 2), indicate that the deletion in our patient occurred de novo on a paternal chromosome (data not shown).
Discussion
Our results show that hemizygosity of the GTF2 transcription factors is sufficient to produce multiple aspects of the WBS CBP, including impaired visuospatial construction abilities, an overly friendly personality accompanied by excessive non‐social anxiety and language delay. Although mental retardation and autistic traits in patient 41A might well be a consequence of a reduced dosage of the GTF2 transcription factors, these symptoms might also be caused or exacerbated by other genes deleted in patient 41A, the prime candidates being HIP1 and YWHAG. Expression of GTF2I and YWHAG in transformed lymphocytes from patient 41A was reduced to 60% and 50% of the parental values, respectively. Expression of GTF2IRD2 was not evaluated, as this gene has an extra copy in humans that is transcribed, although it is not clear if it is functional. The expression data for GTF2I in patient 41A falls within the range described in a previous analysis of gene expression of transformed lymphocytes from 10 full WBS deletion carriers and their parents using semiquantitative reverse transcriptase‐PCR (range 42–70%).29 We found increased expression of GTF2IRD1 in both our cases and in the WBS control cell line. These data are consistent with a recent report documenting decreased expression of GTF2IRD1 in cultured skin fibroblasts from cases with typical WBS deletions, but normal or even increased expression of GTF2IRD1 in transformed lymphocytes.30 Only one group has reported decreased expression of GTF2IRD1 in a typical WBS deletion carrier16; the same group also found decreased expression of GTF2IRD1 in the atypical deletion carrier HR using semiquantitative reverse transcriptase‐PCR. The reason for this discrepancy is unclear; however, expression of GTF2IRD1 in transformed lymphocytes is not representative of gene expression in fibroblasts, and different regulatory elements are probably important for region‐specific expression. Finally, expression of HIP1 was reduced in both transformed lymphocytes and skin fibroblasts from typical WBS deletion carriers who have two copies of HIP130; however, we did not observe decreased expression of HIP1 in our patient or in the WBS typical deletion carrier used as a control. This discrepancy might have arisen because our Taqman probe was designed to the 5′ untranslated region of HIP1, whereas the probe used by Merla et al30 targeted exon 31 of HIP1. Thus, it is possible that we have measured different splice forms and that expression of only one of the splice forms is altered.
GTF2 transcription factors, visuospatial ability and symptoms of autism
In addition to evidence from atypical WBS deletion carriers, other data also implicate GTF2I in visuospatial ability. Holinger et al31 examined neurons in the brain of five patients with WBS and five age‐matched controls and observed a lack of staining for GTF2I in neurons from the posterior parietal lobe, Brodmann area 7. These neuropathological data are consistent with a functional MRI study showing isolated hypoactivation of the parietal portion of the dorsal visual stream in 13 patients with WBS (and normal IQ) on a task designed to test the integrity of both the dorsal and ventral visual streams.32 Additionally, a recent study showed down regulation of GTF2I in association with age‐dependent deficits of hippocampal‐dependent spatial memory in mice.33 Furthermore, defects in the anterior hippocampus have been noted in a structural and functional imaging study of patients with WBS.34 Therefore, reduced expression of GTF2I in the hippocampus may contribute to impaired spatial cognition and navigation observed in patients with WBS.35
Other evidence suggests that the GTF2 family of transcription factors might contribute to abnormalities in speech and language observed in patients with autism and WBS. For example, Laws and Bishop5 studied language impairment and social deficits in traditional WBS deletion carriers and found many similarities between autism spectrum disorders and WBS, especially in the realm of pragmatic language. In fact, patients with WBS resemble patients with Asperger's disorder in that they often fail to discriminate relevant from irrelevant information in conversation, and their conversational content is often incongruent with the conversational intention of others. They may also convey a paucity of information despite apparent volubility and an extensive vocabulary. Another group found that children and adults with WBS did poorly on a basic test of emotional recognition from facial and vocal cues, especially negative emotions, despite the fact that people with WBS are drawn to faces and seek eye contact.8 Finally, a duplication of the WBS deletion interval has been associated with abnormalities in language,29 and delayed acquisition of language is characteristic of both WBS and autism; therefore the GTF2 transcription factors could also have a role in normal speech and language development.
Other candidate genes for autism outside the WBS deletion interval
Of the roughly 14 genes deleted downstream of the WBS deletion interval, HIP1 and YWHAG are the most compelling candidates for susceptibility to autistic traits. HIP1 is a multifunctional nucleocytoplasmic protein that was first identified in screens for proteins that interact with Huntingtin.19 It is a proapoptotic mediator involved in the differentiation and growth of hippocampal neurons36 and also mediates the endocytosis of GluR1‐containing AMPA receptors in primary hippocampal neurons.37Hip1‐null mice show no gross changes in brain morphology, or hippocampal or striatal cell densities and appear normal during embryogenesis, yet develop a neurological phenotype by 3 months of age, characterised by wasting, tremor and gait ataxia secondary to a rigid thoracolumbar kyphosis. (Patient 41A does not have a history of spinal abnormalities.) HIP1 exhibits dosage sensitivity in mice.
The other gene in the deletion interval that might confer aspects of the autism phenotype is YHWAG, a member of a conserved multigene family of phosphopeptide‐binding proteins involved in signal transduction, protein localisation, cell‐cycle checkpoint control and apoptosis.
Conclusion
In summary, our most important finding is that, in addition to GTF2IRD1, hemizygosity of GTF2I is necessary for full manifestation of the visuospatial construction deficit that is a core component of the WBS CBP. GTF2I may also contribute to impaired hippocampal‐dependent spatial navigation, and other roles for this gene related to language or the overly friendly personality are also possible. Finally, several genes both flanking and within the WBS deletion interval could be candidate genes for autism susceptibility owing to their role in neurodevelopment. Association studies and mutation screens targeting the genes discussed in this paper are warranted
The supplementary tables are available at http://jmg.bmj.com/supplemental
Acknowledgements
We thank Kathryn Castelle for her help in editing and formatting this manuscript and Safiana Katz for her careful processing of immortalised cell lines. We also thank Dr Olga Arguedas from the Bioethical Board and Dr Abdon Castro, Presidente de la Fundación Pro Hospital Nacional de Ninos “Dr Carlos Sáenz Herrera.”
Above all, we thank the family of patient 41A, as well as all the other families who have participated in our study, and the Autism Parents' Association of San José, Costa Rica.
Abbreviations
aCGH - array‐based comparative genomic hybridisation
BAC - bacterial artificial chromosome
CBP - cognitive–behavioural profile
FISH - fluorescence in situ hybridisation
HIP1 - Huntingtin‐interacting protein 1
LCR - low‐copy repeat
PCR - polymerase chain reaction
UPL - Universal Probe Library
WBS - Williams–Beuren Syndrome
YWHAG - tyrosine 3‐monooxygenase/tryptophan 5‐monooxygenase activation protein γ
Footnotes
Funding: LAM is supported by NINDS grant number R01 043540, a Young Investigator Award from the Seaver Center for Excellence in Autism Research and a grant from the General Clinical Research Center at the Mount Sinai School of Medicine, New York, USA.
Competing interests: None.
The supplementary tables are available at http://jmg.bmj.com/supplemental
References
- 1.Stromme P, Bjornstad P G, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol 200217269–271. [DOI] [PubMed] [Google Scholar]
- 2.Bayes M, Magano L F, Rivera N, Flores R, Perez Jurado L A. Mutational mechanisms of Williams‐Beuren syndrome deletions. Am J Hum Genet 200373131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellugi U, Lichtenberger L, Mills D, Galaburda A, Korenberg J R. Bridging cognition, the brain and molecular genetics: evidence from Williams syndrome. Trends Neurosci 199922197–207. [DOI] [PubMed] [Google Scholar]
- 4.Porter M A, Coltheart M. Cognitive heterogeneity in Williams syndrome. Dev Neuropsychol 200527275–306. [DOI] [PubMed] [Google Scholar]
- 5.Laws G, Bishop D. Pragmatic language impairment and social deficits in Williams syndrome: a comparison with Down's syndrome and specific language impairment. Int J Lang Commun Disord 20043945–64. [DOI] [PubMed] [Google Scholar]
- 6.Karmiloff‐Smith A, Grant J, Berthoud I, Davies M, Howlin P, Udwin O. Language and Williams syndrome: how intact is “intact”? Child Dev 199768246–262. [PubMed] [Google Scholar]
- 7.Mervis C B. Williams syndrome: 15 years of psychological research. Dev Neuropsychol 2003231–12. [DOI] [PubMed] [Google Scholar]
- 8.Plesa‐Skwerer D, Faja S, Schofield C, Verbalis A, Tager‐Flusberg H. Perceiving facial and vocal expressions of emotion in individuals with Williams syndrome. Am J Ment Retard 200611115–26. [DOI] [PubMed] [Google Scholar]
- 9.Einfeld S L, Tonge B J, Florio T. Behavioral and emotional disturbance in individuals with Williams syndrome. Am J Ment Retard 199710245–53. [DOI] [PubMed] [Google Scholar]
- 10.Baron‐Cohen S, Leslie A M, Frith U. Does the autistic child have a “theory of mind”? Cognition 19852137–46. [DOI] [PubMed] [Google Scholar]
- 11.Tassabehji M, Metcalfe K, Karmiloff‐Smith A, Carette M J, Grant J, Dennis N, Reardon W, Splitt M, Read A P, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet 199964118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagliardi C, Bonaglia M C, Selicorni A, Borgatti R, Giorda R. Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet 200340526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirota H, Matsuoka R, Chen X N, Salandanan L S, Lincoln A, Rose F E, Sunahara M, Osawa M, Bellugi U, Korenberg J R. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med 20035311–321. [DOI] [PubMed] [Google Scholar]
- 14.Karmiloff‐Smith A, Grant J, Ewing S, Carette M J, Metcalfe K, Donnai D, Read A P, Tassabehji M. Using case study comparisons to explore genotype‐phenotype correlations in Williams‐Beuren syndrome. J Med Genet 200340136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howald C, Merla G, Digilio M C, Amenta S, Lyle R, Deutsch S, Choudhury U, Bottani A, Antonarakis S E, Fryssira H, Dallapiccola B, Reymond A. Two high‐throughput technologies to detect segmental aneuploidies identify new Williams‐Beuren Syndrome patients with atypical deletions. J Med Genet 200543266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tassabehji M, Hammond P, Karmiloff‐Smith A, Thompson P, Thorgeirsson S S, Durkin M E, Popescu N C, Hutton T, Metcalfe K, Rucka A, Stewart H, Read A P, Maconochie M, Donnai D. GTF2IRD1 in craniofacial development of humans and mice. Science 20053101184–1187. [DOI] [PubMed] [Google Scholar]
- 17.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview–Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 199424659–685. [DOI] [PubMed] [Google Scholar]
- 18.Lord C, Risi S, Lambrecht L, Cook E H, Jr, Leventhal B L, DiLavore P C, Pickles A, Rutter M. The autism diagnostic observation schedule‐generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord 200030205–223. [PubMed] [Google Scholar]
- 19.Kalchman M A, Koide H B, McCutcheon K, Graham R K, Nichol K, Nishiyama K, Kazemi‐Esfarjani P, Lynn F C, Wellington C, Metzler M, Goldberg Y P, Kanazawa I, Gietz R D, Hayden M R. HIP1, a human homologue of S. cerevisiae Sla2p, interacts with membrane‐associated huntingtin in the brain. Nat Genet 19971644–53. [DOI] [PubMed] [Google Scholar]
- 20.Peyrl A, Weitzdoerfer R, Gulesserian T, Fountoulakis M, Lubec G. Aberrant expression of signaling‐related proteins 14‐3‐3 gamma and RACK1 in fetal Down syndrome brain (trisomy 21). Electrophoresis 200223152–157. [DOI] [PubMed] [Google Scholar]
- 21.McInnes L A, Gonzalez P J, Manghi E R, Esquivel M, Monge S, Delgado M F, Fournier E, Bondy P, Castelle K. A genetic study of autism in Costa Rica: multiple variables affecting IQ scores observed in a preliminary sample of autistic cases. BMC Psychiatry 2005515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellemanset al qBase, http://medgen.ugent.be/qbase/ (accessed 24 Oct 2006)
- 23.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 20023RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wechsler D.Wechsler Preschool and Primary Scale of Intelligence—third edition. San Antonio, TX: Harcourt Assessment, 2002
- 25.Roid G, Miller L.Leiter International Performance Scale–Revised. Wood Dale, IL: Stoelting, 1997
- 26.Antonell A, de Luis O, Domingo‐Roura X, Perez‐Jurado L A. Evolutionary mechanisms shaping the genomic structure of the Williams‐Beuren syndrome chromosomal region at human 7q11.23. Genome Res 200591179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peoples R, Franke Y, Wang Y K, Perez‐Jurado L, Paperna T, Cisco M, Francke U. A physical map, including a BAC/PAC clone contig, of the Williams‐Beuren syndrome—deletion region at 7q11.23. Am J Hum Genet 20006647–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne L R, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui L C, Scherer S W. A 1.5 million‐base pair inversion polymorphism in families with Williams‐Beuren syndrome. Nat Genet 200129321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somerville M J, Mervis C B, Young E J, Seo E J, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez‐Jurado L A, Morris C A, Scherer S W, Osborne L R. Severe expressive‐language delay related to duplication of the Williams‐Beuren locus. N Engl J Med 20053531694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merla G, Howald C, Henrichsen C N, Lyle R, Wyss C, Zabot M T, Antonarakis S E, Reymond A. Submicroscopic deletion in patients with Williams‐Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet 200679332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holinger D P, Bellugi U, Korenberg J R, Mills D L, Reiss A L, Galaburda A M. Genes, neural systems, and cognition: neuronal measures in area 39‐40 of inferior parietal lobule in Williams Syndrome support dorsal‐ventral hypothesis. Paper presented at the Society for Neuroscience. Program number 666.12 Abstract Viewer/Itinerary Planner 2004. Washington, DC 2004
- 32.Meyer‐Lindenberg A, Kohn P, Mervis C B, Kippenhan J S, Olsen R K, Morris C A, Berman K F. Neural basis of genetically determined visuospatial construction deficit in Williams syndrome. Neuron 200443623–631. [DOI] [PubMed] [Google Scholar]
- 33.Verbitsky M, Yonan A L, Malleret G, Kandel E R, Gilliam T C, Pavlidis P. Altered hippocampal transcript profile accompanies an age‐related spatial memory deficit in mice. Learn Mem 200411253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer‐Lindenberg A, Mervis C B, Sarpal D, Koch P, Steele S, Kohn P, Marenco S, Morris C A, Das S, Kippenhan S, Mattay V S, Weinberger D R, Berman K F. Functional, structural, and metabolic abnormalities of the hippocampal formation in Williams syndrome. J Clin Invest 20051151888–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Hearn K, Landau B, Hoffman J E. Multiple object tracking in people with Williams syndrome and in normally developing children. Psychol Sci 200516905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang J E, Choi S A, Park J B, Chung K C. Regulation of the proapoptotic activity of huntingtin interacting protein 1 by Dyrk1 and caspase‐3 in hippocampal neuroprogenitor cells. J Neurosci Res 20058162–72. [DOI] [PubMed] [Google Scholar]
- 37.Metzler M, Li B, Gan L, Georgiou J, Gutekunst C A, Wang Y, Torre E, Devon R S, Oh R, Legendre‐Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson P S, Nagy A, Wang Y T, Roder J C, Raymond L A, Hayden M R. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J 2003223254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]