Abstract
The transcription factors E2F and Myc participate in the control of cell proliferation and apoptosis, and can act as oncogenes or tumor suppressors depending on their levels of expression. Positive feedback loops in the regulation of these factors are predicted—and recently shown experimentally—to lead to bistability, which is a phenomenon characterized by the existence of low and high protein levels (“off” and “on” levels, respectively), with sharp transitions between levels being inducible by, for example, changes in growth factor concentrations. E2F and Myc are inhibited at the posttranscriptional step by members of a cluster of microRNAs (miRs) called miR-17-92. In return, E2F and Myc induce the transcription of miR-17-92, thus forming a negative feedback loop in the interaction network. The consequences of the coupling between the E2F/Myc positive feedback loops and the E2F/Myc/miR-17-92 negative feedback loop are analyzed using a mathematical model. The model predicts that miR-17-92 plays a critical role in regulating the position of the off–on switch in E2F/Myc protein levels, and in determining the on levels of these proteins. The model also predicts large-amplitude protein oscillations that coexist with the off steady state levels. Using the concept and model prediction of a “cancer zone,” the oncogenic and tumor suppressor properties of miR-17-92 is demonstrated to parallel the same properties of E2F and Myc.
MicroRNAs (miRs) are small noncoding RNAs, 18–24 nt in length, that are predicted to regulate the expression of approximately one-third of all human genes (1, 2). This regulation occurs posttranscriptionally through miR binding to mRNA targets leading to target degradation or inhibition of translation. Current target-prediction computer programs (3, 4) often predict that a miR could target tens to hundreds of genes, and that a gene can be targeted by many miRs—thus, the expectation that miRs play important roles in coordinating many cellular processes, particularly those involved in development and disease (5). Indeed, miRs have been implicated in various cancers, acting either as oncogenes or tumor suppressor genes (6). In this article, we investigate the role of a set of miRs in the important “cancer network” shown in Fig. 1. A cancer network is a molecular or gene interaction network involving oncogenes or tumor suppressor genes. The network shown is associated with the control of the G1-S transition in the mammalian cell cycle (7, 8).
Fig. 1.
A cancer network. This network is part of the mammalian G1-S regulatory network involving oncogenes (black boxes) and tumor suppressor genes (gray boxes). Dashed arrows mean induction of gene expression. For example, the E2F protein induces the expression of its own gene. Solid arrow means activation; for example, the Cdc25A phosphatase activates Cdk2 by catalyzing the removal of an inhibitory phosphate. Hammerheads mean inhibition. Details of the mechanism are discussed in ref. 7. Note that E2F and Myc are labeled as both oncogene and tumor suppressor genes (see text for explanation). Not shown in the figure are pathways downstream of E2F and Myc that lead to caspase-mediated apoptosis (reviewed in ref. 9). Cdk, cyclin-dependent kinase; pRb, retinoblastoma protein.
The extent to which miRs change the levels of their target mRNAs is marginal compared with the effect of other regulators such as transcription factors and posttranslational protein modifiers (10). Thus, it is thought that the primary role of miRs is to modulate or fine-tune the dynamics of regulatory networks (10–13). The significance of this role is now increasingly recognized as there are now many reported cases in which abnormal miR expressions correlate with cancer development (reviewed in ref. 6). Here, we focus on miR-17-92, which behaves as an oncogene or a tumor suppressor in different situations (2, 14).
The miR-17-92 cluster is a polycistronic gene located in human chromosome 13 ORF 25 (C13orf25) located at 13q31-q32. The cluster is composed of 7 mature miRs, namely, miR-17–5p, miR-17–3p, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92–1 (Fig. 2A). The nucleotide sequences and organization of this cluster is highly conserved in vertebrates (reviewed in ref. 2). He et al. (15) reported the first evidence of the oncogenic activity of miR-17-92. Gene expression data show over-expression of miR-17-92 in several tumors, including cancers of the breast, lung, colon, stomach, pancreas, and prostate (16, 17). Although the tumor-suppressor property of miR-17-92 remains to be demonstrated directly in vivo, the following observations are quite suggestive: This cluster is deleted in 16.5% of ovarian cancers, 21.9% of breast cancers, and 20% of melanomas (14, 15).
Fig. 2.
Reduction of the complex Myc/E2F/miR-17-92 network to an abstract model. (A) Summary of the interactions among the transcription factors Myc, E2F1, E2F2, E2F3, and members of the miR-17-92 cluster (reviewed in ref. 14). All arrows refer to induction of gene expression. The hammerheads from miR-17-92 members to Myc and to the E2Fs refer to inhibition of translation or degradation of mRNAs. (B) First stage in the reduction of the model. (C) The final network model that abstracts the essential structure of the network in A.
Among the experimentally validated targets of some miR-17-92 cluster members are the transcription factors Myc, E2F1, E2F2, and E2F3; interestingly, these same factors have been shown to induce the transcription of miR-17-92 (reviewed in ref. 14). The negative feedback loops thus formed are depicted in Fig. 2A. In the modeling that follows, we will focus on E2F1, which possesses both oncogenic and tumor suppressor properties (18). Earlier, we analyzed a model that views E2F1 and Myc as members of a control node in a regulatory network coordinating cell cycle entry and apoptosis (9, 19). Among the predictions of the model is that increasing levels of E2F or Myc drives the sequence of cellular states, namely, quiescence, cell proliferation, and apoptosis. Here, we postulate that in between levels associated with normal cell cycles and apoptosis, there exists a range of Myc and E2F1 levels with increased probability of inducing cancer. We call this range the cancer zone. We summarize in Fig. 3 our hypothesis on how the oncogenic and tumor suppressive properties of miR-17-92 arise in relation to the cancer zone. Using a mathematical model abstracted from the complex network (Fig. 2), we illustrate the mechanisms and conditions under which these miR-17-92 properties operate.
Fig. 3.
The miR-17-92 cluster as an oncogene or as a tumor suppressor. As the transcriptional activities of E2F or Myc increase, cells transit through quiescence, cell cycle, and apoptosis. It is postulated that there exists a range of E2F or Myc levels—called the cancer zone—that leads to hyperproliferation because cell division is not appropriately balanced by apoptosis (cell death). As shown at the top of the figure, there are two ways that miR-17-92 can keep levels of E2F or Myc in the cancer zone (i.e., acting as an oncogene). (case a) Increasing miR levels drives E2F or Myc levels to enter the cancer zone. (case b) Increasing miR inhibition of E2F or Myc translation suppresses exit from the cancer zone. (cases c and d) The two ways that miR-17-92 can suppress entry into the cancer zone (i.e., acting as a tumor suppressor) are: increasing miR inhibition of E2F or Myc translation suppresses entry into the cancer zone (case c), and increasing miR levels drives E2F or Myc levels to exit the cancer zone and enter apoptosis (case d). It is shown in this article that cases a and c are concomitant, as are cases b and d.
Formulation of the Model
Dimensionless Equations.
Fig. 2 summarizes how the complex regulatory network is reduced to a model with 2 components representing the protein module p (Myc and the E2Fs) and the miR cluster m. Step 1 in Fig. 2C represents the autocatalytic (positive feedback) growth of p, which is inhibited by m. Step 2 in the same figure depicts the p-induced transcription of the miR cluster. The dynamics of the respective concentrations of these modules, p and m, are described by Eqs. 1 and 2.
The rate of step 1 is given by the second term on the right-hand side of Eq. 1 where the Hill exponent of Eq. 2 on p assumes a threshold kinetics; furthermore, the presence of m in the denominator accounts for the miR-dependent down-regulation of protein expression. The aforementioned threshold on the rate of protein expression is determined by the term (Γ1 + Γ2m) in the denominator. The value of the parameter Γ2 is a measure of the efficiency of miR inhibition of protein expression, and lumps all factors that could affect the binding of the members of the miR-17-92 cluster to their targets and the inhibition of protein translation.
The constant term α in Eq. 1 stands for constitutive protein expression due to signal transduction pathways stimulated by growth factors present in the extracellular medium. The parameter α therefore corresponds to an experimentally controllable condition such as the concentration of nutrients in the cell culture medium. The right-most term in Eq. 1 is a first-order protein degradation term with fixed rate coefficient of δ. The constant term β in Eq. 2 represents p-independent constitutive transcription of m. The second term in Eq. 2 is the rate of p-induced transcription of m (assumed to be first order in p for simplicity), and the last term is a degradation term with rate coefficient γ.
Eqs. 1 and 2 can be nondimensionalized as follows
where the dimensionless variables and parameters are
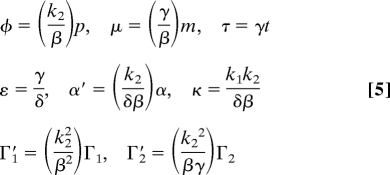 |
Delay Differential Equation.
Step 1 in Fig. 2C is an abstraction of all of the steps involved in protein expression—from transcription factor binding to DNA, gene transcription, to translation in ribosomes. Thus, the rate of synthesis of the protein is not a function of its instantaneous concentration (as assumed in Eq. 3), but rather of its concentration at some time Δ in the past. In a second set of computer simulations presented in the Results Section, this time delay Δ is considered in the second term on the right-hand side of Eq. 3, which is rewritten explicitly in Eq. 6.
Solving for the Steady States.
The steady states of the system of Eqs. 3 and 4 are determined by equating the right-hand sides to zero. After eliminating μ in the steady equations, we obtain the following cubic polynomial whose non-negative roots give the steady states of φ (symbolized by φs):
where
 |
The steady state of μ (symbolized by μs) is given by
We are interested in threshold or switching behavior of the system, and, therefore, the conditions on the parameters for the existence of multiple steady states are relevant. From ref. 20, the set T of parameters that guarantee existence of 3 positive real roots of Eq. 7 is:
 |
where
Thus, the necessary (but not sufficient) conditions for the existence of 3 steady states of the model are:
Parameter Values and Numerical Solution of the Differential Equations.
The parameter ε is expected to be less than unity because miRs are typically more stable than proteins; for example, δ for E2F1 and Myc are ≈0.25 h−1 and ≈0.7 h−1, respectively (21), and γ ≈ 0.02 h−1 (22). The value of ε = 0.02 is used in our computer simulations (noting that δ for Myc is of order unity, and making allowances for the other E2Fs besides E2F1). An estimate of k1 for E2F1 is ≈0.4 μM h−1 and Γ1 ≈0.1 μM2 (21). We arbitrarily set (k2/β) ≈3 μM−1 so that Γ′1 ≈1 and κ ≈5 (the parameter β is assumed to be manipulated experimentally via gene transfection, for example). The dimensionless parameters α′ and Γ′2—whose values can be tuned experimentally—are allowed to vary in the ranges 0–0.4 and 0–2.5, respectively, to explore the effect of increasing rate of growth factor-induced protein synthesis and inhibition efficiency of the miRs. The differential equations of the model are solved using the computer software described in Methods.
Results and Discussion
Steady States of the Model and Significance of the Parameter α′.
According to Eq. 8, the steady states of m and p increase or decrease in the same direction. This model prediction agrees with observations in various tumors that levels of Myc and miR-17-92 are both increased (23, 24). The model also clarifies the interpretation of Hayashita et al. (23) that members of the miR-17-92 cluster promote proliferation—this is because increase in the miR level correlates with increase in the levels of Myc or E2F, which are both proliferative.
The steady state φs as a function of the parameter α′ for different values of Γ′2 is shown in Fig. 4. These diagrams are referred to as a “steady-state bifurcation diagrams,” and α′ is referred to as a bifurcation parameter. Because of physiological constraints, the horizontal axis in Fig. 4 does not extend to infinity but terminates at some maximum value of α′. To generate experimental curves similar to those shown in this figure, one can envisage a laboratory experiment in which cells are grown at different nutrient concentrations and then measuring the long-term protein levels.
Fig. 4.
Steady-state bifurcation diagrams. Steady states of the variable φ (dimensionless protein concentration) as a function of the parameter α′ for different values of the parameter Γ′2 (all parameters are dimensionless, according to Eq. 5). Values of other parameters: κ = 5, Γ′1 = 1 (specific value of ε not required for steady-state calculations). Note that α′ does not extend to infinity, but only to a maximum value dictated by cell physiology (a maximum of α′ = 0.4 is assumed only for illustrative purposes).
With parameters satisfying the relationships given in Eq. 10, the model predicts that there is a range of α′ in which the system has 3 coexisting steady states (e.g., those with Γ′2 = 0, 1, 1.5, 1.8, 2). For example, for the curve with Γ′2 = 1.8, values of α′ from 0.05 (corresponding to the left knee of the curve) to 0.18 (right knee) give 3 steady states. We interpret the “right knee” of a steady-state diagram as a “switch-on” point at which a sharp irrevocable increase in protein level occurs until the upper steady state (the “on” position) is attained. In other words, the model predicts that there exists a threshold in growth-factor requirement for cells to “turn on” the protein synthesis. (We interpret the low protein steady states in Fig. 4 as the “off” positions.) Note that one of the hallmarks of cancer is a decreased growth factor requirement for proliferation. As this switch-on value of α′ (growth-factor requirement) decreases as the parameter Γ′2 decreases, the significance of this parameter is discussed next.
Significance of the Parameter Γ′2 and miR Regulation of Protein Levels.
The dimensionless parameter Γ′2 represents the inhibition efficiency of miR-17-92 against its target proteins. The expression of Γ′2 in terms of the 4 parameters Γ2, k2, β, and γ (Eq. 5) suggests the ways to manipulate the miR inhibition efficiency experimentally. For example, Γ′2 can be increased by increasing Γ2 or k2, or by decreasing β or γ. The dependence of Γ′2 on β seems counterintuitive because Eq. 5 states that an increase in the constitutive or p-independent expression of miR-17-92 leads to a decrease in the miR inhibition efficiency.
The case of Γ′2 = 0 represents any of the following situations: deletion of the miR-17-92 cluster; members of the cluster do not bind the transcripts of the target proteins (p) perhaps due to mutations; p does not induce expression of miR-17-92 (case of k2 = 0). Although p is no longer coupled to m, the 1-dimensional model of the autocatalytic variable p is still capable of exhibiting 3 steady states for α′ between 0 and ≈0.05 (see Fig. 4). Standard linear stability analysis shows that the system is bistable in this range of α values—that is, the bottom and upper branches of steady states are stable, whereas the middle branch of steady states are unstable. Very interestingly, bistability involving E2F1 and Myc have recently been demonstrated experimentally by Yao and colleagues (21). In addition to the E2F1 loop, these authors invoked another source of positive feedback loop—specifically, the E2F-Cdk2-pRb-E2F loop shown in Fig. 1—in their model of the system.
Two key observations can be made from Fig. 4 with regards to the role of miR-17-92: (i) as Γ′2 is increased, the switch-on values of α′ (corresponding to the right knees of the curves) increase; (ii) as Γ′2 is increased, the upper branch of steady states (the on states) are lowered. The first observation suggests that the miRs counteract the cancer-associated decreased growth factor requirement for cell proliferation. The second observation agrees with the current thinking about the role of miR-17-92 in preventing a runaway E2F/Myc positive feedback loop that may induce uncontrolled cell proliferation.
The model also predicts 3 qualitatively different types of steady-state bifurcation diagrams as illustrated in Fig. 4: (i) 2 disconnected curves with just the right knee (e.g., Γ′2 = 0, 1, 1.5), (ii) a continuous curve with left and right knees (e.g., Γ′2 = 1.8 and 2), and (iii) a continuous curve with no knees (e.g., Γ′2 = 2.5). Type (i) is an irreversible switch to the upper branch of steady states, whereas type (ii) allows a transition from the upper branch to the lower branch by decreasing α′ below the value corresponding to the left knee of the curve.
Non-Steady-State Behavior and Sensitivity of Protein Levels to miRs.
The functional properties of the E2F/Myc/miR-17-92 network—in particular, the role of the miR cluster—can be further understood by studying its non-steady state kinetics. For example, the dynamics of the system can be very sensitive to the initial levels of the miR cluster. Shown in Fig. 5 are computer simulations from various initial conditions of φ and μ. In Fig. 5A, 5 initial conditions located at the lower left corner of the box are very close to each other, with identical initial φ0 but with 5 close values of μ0. All of the trajectories ultimately approach a stable steady state (shown as empty circle), but the initial conditions where μ0 = 0.340, 0.343, and 0.345 lead to trajectories with wide swings in protein levels that even surpass the upper steady state (see Fig. 4, α′ = 0.1); in contrast, for the initial conditions μ0 = 0.350 and 0.355 the system goes to the steady state directly. (The value of μ0 that delineates these two dynamics is between 0.345 and 0.350, as exhibited in Fig. 5B.) Thus, the model predicts that the system could be prone to large bursts of protein synthesis if the level of mir-17–92 is below a certain threshold.
Fig. 5.
Model dynamics. (A) Phase plane trajectories from different initial conditions: φ (0) = 0.13, μ (0) = 0.340, 0.343, 0.345, 0.350, 0.355. Parameter values: ε = 0.02, α′ = 0.1, κ = 5, Γ′1 = 1, Γ′2 = 1.8. The empty circle represents a steady state of the system. (B) Time courses for 2 very close initial conditions φ (0) = 0.13, μ (0) = 0.345 (black curve) and φ (0) = 0.13, μ (0) = 0.350 (gray curve). (C) Same as A except α′ = 0.2 and μ (0) = 1, 2, 3. The empty circle represents a steady state of the system. (D) Temporal course of φ for parameters and initial conditions identical to C.
At α′ = 0.1 (as in Fig. 5 a and b) the system has 3 coexisting steady states, but only the lowest one is stable (shown as empty circle in Fig. 5A). When α′ is increased beyond the right knee of the curve in Fig. 4 (for Γ′2 = 1.8), only one steady state is available for the system; this steady state is asymptotically stable as shown by the phase plane trajectories plotted in Fig. 5C and the temporal course of φ in Fig. 5D.
The non-steady state behavior of the system as shown in Fig. 5 could explain an experimental observation that seemingly contradicts the prediction of the model at steady state. Several groups (14, 25, 26) have shown that miR-20a and miR-17–5p (members of the miR-17-92 cluster) are antiapoptotic because the down-regulation of these miRs leads to increased cell death and their overexpression decreases cell death. These observations were explained (25, 27) in terms of the down-regulation of E2F1 protein levels by miR-20a and miR-17–5p, and E2F1's induction of apoptosis when overexpressed. However, our model suggests that, at steady state, increased levels of miR-17-92 are associated with increased levels of Myc and the E2Fs (Eq. 8 and Fig. 4) and therefore increased apoptosis. One way to resolve this dilemma is to view the model's transient dynamics instead of steady states, and to illustrate the possibility that reported experimental observations were made under non-steady state conditions. In Fig. 5 A and C, the slow segments of the trajectories (from the maximum φ to the maximum μ) correspond to increasing miR (μ) and decreasing protein (φ). This decrease in the protein, which is associated with increased miR, could then lead to a decreased rate of apoptosis—thus, the reference to the miR as antiapoptotic.
miR-17-92 as an Oncogene and a Tumor Suppressor.
Viewed in terms of miR steady state levels, the oncogenic and tumor suppressor properties of miR-17-92—cases a and d in Fig. 3, respectively—parallel those of E2F1 or Myc because of Eq. 8. This idea is explained more explicitly in Fig. 6. (Note that in this figure, the range of φs that defines the cancer zone is chosen arbitrarily; one would expect that the range of the cancer zone would depend on the specific cellular system and on the perturbations of the system that drives it toward a cancerous state.) In the direction of the arrow in Fig. 6A, the on steady states increase to levels that enter the cancer zone. The corresponding increase in μs is interpreted as promoting cancer and therefore classifies the miR as an oncogene. However, in the direction of the arrow in Fig. 6B, both φs and μs increase—driving exit from the cancer zone and into apoptosis, thereby classifying the miR cluster as a tumor suppressor.
Fig. 6.
In and out of the cancer zone. (A) Decrease in Γ′2 from 2.0 to 1.0 corresponds to a decrease in the miR inhibition efficiency of protein expression (see Fig. 3C) and to an increase in the steady-state level of the miR cluster (Eq. 8; see also Fig. 3A)—leading to entry into the cancer zone. (B) A further decrease in Γ′2 from 1.0 to 0.5 corresponds to a decrease in miR inhibition efficiency (Fig. 3B) and to an increase in miR steady state level (Fig. 3D)—thus driving the exit from the cancer zone and into apoptosis.
An alternative view of the oncogenic and tumor suppressor properties of miR-17-92—cases b and c in Fig. 3, respectively—arises when the focus is on the miR inhibition efficiency parameter, Γ′2, rather than the miR levels as in the preceding paragraph. The inhibition of the exit from the cancer zone (case b in Fig. 3) is carried out by preventing a decrease of Γ′2; in other words, the direction of increasing Γ′2 when miR levels are high would be oncogenic. As a tumor suppressor, the inhibition of the entry into the cancer zone (case c in Fig. 3) is done by preventing a decrease of Γ′2 when miR levels are low.
The discussion in this section illustrates the confusion that may arise in using the labels “oncogene” and “tumor suppressor” if the attribute of the miR (that is, the miR level, μs, or the inhibition efficiency parameter, Γ′2) used to correlate with entry into or exit from the cancer zone is not clearly specified. However, determining Γ′2 experimentally would be more difficult in practice compared with measuring miR levels, and therefore classification using changes in miR levels is commonly used.
Time Delays and Oscillations.
In this section, Eqs. 6 and 4 are numerically solved. Fig. 7 shows the dynamics of the system as the parameter α′ is increased or decreased. The rate of change of α′ is slow enough so that the system keeps close to the locally stable steady states. As shown by the diagrams on the top row of Fig. 7, for both cases without (Δ = 0) and with time delay (Δ = 0.2), increasing α′ leads to a sharp transition to the upper steady states; this transition occurs at the α′ corresponding to the right knee of the diagram shown in Fig. 4 for Γ′2 = 1.8. Once at the top branch of steady states, oscillations are initially observed with rapidly decreasing amplitudes as α′ is increased further. When the direction of change of α′ is reversed (bottom diagrams of Fig. 7), new interesting dynamics appear. For the case without time delay (Δ = 0), the system initially traces the curve that was made in the direction of increasing α′, but then goes a little bit beyond (to the left) of the right knee before undergoing a sharp drop to the lower branch of steady states. Note that the system does not switch down at the left knee (see Fig. 4 for Γ′2 = 1.8); this is because of the emergence of large-amplitude oscillations (data not shown) that exist in a narrow range of α′ values just to the left of the right knee; these large-amplitude oscillations suddenly disappear and the system gets trapped by the lower branch of stable steady states. In contrast, for the case with time delay of Δ = 0.2, the system exhibits large-amplitude protein oscillations for wider ranges of α′s as indicated by the black region in the bottom right diagram (the accompanying small-amplitude oscillations in μ are also shown in gray). In the same range of α′ values where this oscillatory region is located, the system could get trapped in the lower branch of steady states depending on the initial conditions.
Fig. 7.
Model dynamics with time delay. Eqs. 4 and 6 are numerically integrated, along with slowly changing α′ (with rate dα′/dτ = 0.001) in the increasing (α′ →) and decreasing (← α′) directions, without time delay (Δ = 0, Left) and with time delay (Δ = 0.2, Right). Other parameter values: ε = 0.02, Γ′1 = 1, Γ′2 = 1.8, κ = 5.
Thus, the model with time delay predicts the coexistence between large-amplitude oscillations and low steady-state protein levels. To try to understand the physiological significance of these large-amplitude protein oscillations, we checked the stability of the lower branch of steady states (the off states) and found that these states are quite robust against perturbations—for example, at α′ = 0.1, it takes a perturbation of ≈370% above the value of φs to switch the system to the large-amplitude oscillations (simulations not shown); also, large amounts of perturbation above μs do not induce switching, but, as shown in Fig. 5A (no delay), perturbations that decrease the miR level below a certain threshold induce large-amplitude swings in protein concentrations. The robustness of the off states and of the switch-on values of α′ is required for proteins that control important cellular processes (such as entry into S-phase of the cell cycle, which E2F1 and Myc regulate). However, the model predicts that large bursts in protein levels (the large-amplitude oscillatory states) are possible and can be used by the system to obtain apoptotic levels quickly to avert any danger that may be caused by large perturbations.
Conclusions
We proposed and analyzed a simple model of the interactions between miR-17-92 and the transcription factors E2F and Myc. Our goal is to explore the broad consequences of the structure of the network on the levels, steady states and dynamics of the miR and the group of proteins that the miR targets. The simplicity of the 2-variable model precludes it from capturing the different properties of Myc, E2F1, E2F2, and E2F3 with respect to their proliferative or apoptotic effects or nature of repression by miR-17-92 members. The model couples the positive feedback loops involving E2F and Myc—generating bistability and the concomitant off-on switching behavior of the system—and the negative feedback loop between these proteins and members of the miR-17-92 cluster. The model predicts that the steady states of these proteins and the miRs change in the same direction, although slow non-steady state or transient dynamics are possible where the changes could be in opposite directions. We have illustrated how changes in the miR inhibition efficiency—the parameter Γ′2 in Eq. 3—controls the value of the off-on switch in growth-factor requirement and how it attenuates the on levels of the proteins. An important prediction of the model is that decreasing Γ′2 leads to decreasing growth-factor requirement for switching the protein on, and that the on levels increase with decreasing Γ′2. Possible experimental means of manipulating the value of Γ′2 are discussed. Due to the negative feedback loop in the network, large-amplitude protein oscillations are predicted to coexist with the off steady state levels, allowing the system to respond through apoptosis to dangerously large perturbations. Finally, using the postulate of a cancer zone, we have shown that the oncogenic and tumor suppressor properties of miR-17-92 parallel those of E2F and Myc.
Methods
For the first set of computer simulations, the system composed of Eqs. 3 and 4 is solved using the routine ode23 in MATLAB (The Mathworks). For the second set of simulations involving time delay, the system composed of Eqs. 6 and 4 is solved using the routine dde23 in MATLAB.
Acknowledgments.
This work was supported by U.S. National Science Foundation Agreement 0112050 and National Institutes of Health Grant RO-1HL67176.
Footnotes
The authors declare no conflict of interest.
References
- 1.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT. myRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. Methods Enzymol. 2007;427:65–86. doi: 10.1016/S0076-6879(07)27004-1. [DOI] [PubMed] [Google Scholar]
- 4.Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today. 2007;12:452–458. doi: 10.1016/j.drudis.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Gusev Y. Computational methods for analysis of cellular functions and pathways collectively targeted by differentially expressed microRNA. Methods. 2008;44:61–72. doi: 10.1016/j.ymeth.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Cho WC. OncomiRs: The discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476–4598-6–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguda BD, Tang Y. The kinetic origins of the restriction point in the mammalian cell cycle. Cell Proliferation. 1999;32:321–335. doi: 10.1046/j.1365-2184.1999.3250321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aguda BD, Goryachev AB. From pathways databases to network models of switching behavior. PLoS Comput Biol. 2007;3:e152. doi: 10.1371/journal.pcbi.0030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguda BD, Algar CK. Structural analysis of the qualitative networks regulating the cell cycle and apoptosis. Cell Cycle. 2003;2:538–544. doi: 10.4161/cc.2.6.550. [DOI] [PubMed] [Google Scholar]
- 10.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen SM, Brennecke J, Stark A. Denoising feedback loops by thresholding—a new role for microRNAs. Genes Dev. 2006;20:2769–2772. doi: 10.1101/gad.1484606. [DOI] [PubMed] [Google Scholar]
- 13.Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci USA. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coller HA, Forman JJ, Legesse-Miller A. Myc'ed Messages: Myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrocca F, et al. E2F1-regulated microRNAs impair TGFβ-dependent cell cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Johnson DG. The paradox of E2F1: Oncogene and tumor suppressor gene. Mol Carcinog. 2000;27:151–157. doi: 10.1002/(sici)1098-2744(200003)27:3<151::aid-mc1>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 19.Craciun G, Aguda BD, Friedman A. A detailed mathematical analysis of a modular network coordinating the cell cycle and apoptosis. Math Biosci Engineering. 2005;2:473–485. doi: 10.3934/mbe.2005.2.473. [DOI] [PubMed] [Google Scholar]
- 20.Aguda BD, Clarke BL. Bistability in chemical reaction networks: Theory and application to the peroxidase–oxidase reaction. J Chem Phys. 1987;87:3461–3470. [Google Scholar]
- 21.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 22.Khanin R, Vinciotti V. Computational modeling of post-transcriptional gene regulation by microRNAs. J Comput Biol. 2008;15:305–316. doi: 10.1089/cmb.2007.0184. [DOI] [PubMed] [Google Scholar]
- 23.Hayashita Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 24.Rinaldi A, et al. Concomitant MYC and microRNA cluster miR-17-92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48:410–412. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 25.Sylvestre Y, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 26.Matsubara H, et al. Apoptosis induction by antisense oligonucleotides against miR-17–5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 27.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic microRNA cluster by E2F transcription factors. J Biol Chem. 2006;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]









