Abstract
MicroRNA (miRNA)-guided cleavage initiates entry of primary transcripts into the transacting siRNA (tasiRNA) biogenesis pathway involving RNA-DEPENDENT RNA POLYMERASE6, DICER-LIKE4, and SUPPRESSOR OF GENE SILENCING3. Arabidopsis thaliana TAS1 and TAS2 families yield tasiRNA that form through miR173-guided initiation–cleavage of primary transcripts and target several transcripts encoding pentatricopeptide repeat proteins and proteins of unknown function. Here, the TAS1c locus was modified to produce synthetic (syn) tasiRNA to target an endogenous transcript encoding PHYTOENE DESATURASE and used to analyze the role of miR173 in routing of transcripts through the tasiRNA pathway. miR173 was unique from other miRNAs in its ability to initiate TAS1c-based syn-tasiRNA formation. A single miR173 target site was sufficient to route non-TAS transcripts into the pathway to yield phased siRNA. We also show that miR173 functions in association with ARGONAUTE 1 (AGO1) during TAS1 and TAS2 tasiRNA formation, and we provide data indicating that the miR173–AGO1 complex possesses unique functionality that many other miRNA–AGO1 complexes lack.
Keywords: Arabidopsis, ARGONAUTE, microRNA, transacting siRNA
MicroRNA (miRNA) and transacting siRNA (tasiRNA) form through distinct biogenesis pathways, but both function to guide endonucleolytic cleavage or translational modulation of target RNA transcripts (1). For miRNA, self-complementary fold-back structures within primary transcripts are processed into ≈21- to 22-nt miRNA/miRNA* duplexes. For tasiRNA in plants, primary transcripts are first processed by miRNA-guided cleavage. One product of the cleaved transcript is stabilized, possibly by SUPPRESSOR OF GENE SILENCING3 (SGS3), and converted to dsRNA by RNA-DEPENDENT RNA POLYMERASE6 (RDR6) (2–5). The resulting dsRNA is processed sequentially by DICER-LIKE4 (DCL4) into 21-nt siRNA duplexes in register with the miRNA-guided cleavage site (2, 6, 7). One strand of each miRNA or tasiRNA duplex is selectively sorted to one or more ARGONAUTE (AGO) proteins according to the 5′ nucleotide or other sequence/structural elements of the small RNA (8–10). AGO proteins, which contain a 3′ RNA binding domain (PAZ), a mid domain that confers small RNA recognition or binding function, and an RNaseH-like domain (PIWI) (11), provide the effector component for silencing complexes.
Arabidopsis thaliana has eight characterized tasiRNA-generating (TAS) loci belonging to four families. TAS1 and TAS2 tasiRNA target multiple different mRNAs, including several encoding pentatricopeptide repeat (PPR) proteins (3–5, 12–14). TAS4 tasiRNA target mRNA encoding several MYB transcription factors (15). TAS3 tasiRNA target AUXIN RESPONSE FACTORs (ARF3 and ARF4) mRNA, regulation of which is important for proper patterning and developmental timing (4, 16–20). In addition to tasiRNA-based regulation, the RDR6/SGS3/DCL4 silencing pathway contributes to antiviral and transgene silencing (21–25).
It is not clear how transcripts are routed into the RDR6/SGS3/DCL4 silencing pathway, although it is known that transcripts with two or more miRNA or tasiRNA target sites are more likely to spawn secondary siRNA (13, 14) than are singly targeted transcripts. The TAS3a primary transcript is an example of a dual-targeted RNA (13, 14), in which miR390–AGO7 complexes provide distinct functions at two target sites. The miR390–AGO7 complex associates with a target site near the 5′ end of the transcript in a noncleavage mode and with a site near the 3′ end to affect cleavage (9).
The TAS3 family is highly conserved in land plants, whereas the TAS1, TAS2, and TAS4 families are restricted to Arabidopsis or close relatives (3, 4, 13, 15, 26, 27). Unlike TAS3 transcripts, TAS1, TAS2, and TAS4 transcripts each have only a single known miRNA target site. miR173 functions to initiate tasiRNA biogenesis from TAS1 and TAS2 loci, whereas miR828 serves to initiate TAS4 tasiRNA formation. TAS1, TAS2, and TAS4 also differ from TAS3 in that the tasiRNA-generating regions originate from the RNA fragment on the 3′ side of the cleavage site (2, 4, 15). Thus, the features of TAS1, TAS2, and TAS4 tasiRNA formation suggest a distinct route through the biogenesis pathway. Here, the TAS1c locus was manipulated to explore the cis requirements for tasiRNA formation. The results indicate that the miR173–AGO1 complex functioning at a single site possesses unique properties that initiate routing of transcripts through the tasiRNA pathway.
Results
TAS1c-Based Synthetic (syn) tasiRNA.
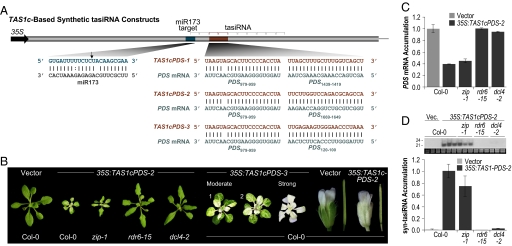
The predictable pattern of tasiRNA formation from the initiation-cleavage site was exploited previously to develop syn-tasiRNA from modified TAS genes (9, 28). TAS3a-based syn-tasiRNA that target the mRNA encoding PHYTOENE DESATURASE (PDS) were effectively used to dissect TAS3 tasiRNA biogenesis and effector requirements (9). We developed a similar system based on the TAS1c locus, in which sequences at DCL4 processing cycles 3 and 4 from the miR173-guided initiation site were substituted for sequences with perfect or near perfect complementarity to two sites within PDS mRNA (Fig. 1A and Fig. S1A). Each of three syn-tasiRNA genes (35S:TAS1cPDS-1, 35S:TAS1cPDS-2, and 35S:TAS1cPDS-3) contained the same syn-tasiRNA at the 3′D3(+) position, but a different syn-tasiRNA at the 3′D4(+) position. Each syn-tasiRNA was designed to possess a 5′ uridine to promote association with AGO1 (8–10). Each construct was expressed by using the 35S promoter and introduced into Arabidopsis thaliana plants. Mapping of both authentic TAS1c and 35S:TAS1cPDS-2 transcripts revealed the dominant poly(A) site at position 810 nt from the transcription start site (Fig. S2A) (4), indicating that the TAS1c-based syn-tasiRNA constructs contained the authentic TAS1c 3′ terminator sequence.
Fig. 1.
Syn-tasiRNA. (A) Organization of syn-tasiRNA constructs. Arrow indicates the miR173-guided cleavage site. tasiRNA positions 3′D1(+) through 3′D10(+) are indicated by brackets. (B) Representative images of Col-0 and mutant plants expressing syn-tasiRNA constructs. (C) Mean relative level +/− SEM of PDS mRNA after normalization to ACT2 mRNA, as determined by quantitative RT-PCR (Col-0 vector = 1.0). (D) Mean relative level +/− SEM of syn-tasiRNA as determined by blot assays (Col-0 35S:TAS1cPDS-2 = 1.0). (Inset) Small RNA blot and EtBr-stained 5S rRNA/tRNA.
The majority of WT (Col-0) plants transformed with each TAS1c-based syn-tasiRNA construct displayed a photobleached phenotype characteristic of PDS silencing, although there was variation in phenotype severity among transformants (Fig. 1B and Table S1). Photobleaching was concurrent with accumulation of syn-tasiRNA and significant reduction in PDS mRNA levels (Bonferroni corrected P < 0.05, two-sample t test) (Fig. 1 C and D). In zip-1 mutants, which lack AGO7, the photobleached phenotype was indistinguishable from that in Col-0 plants, whereas photobleaching was undetectable in rdr6–15 and dcl4–2 mutants (Fig. 1B and Table S1). PDS mRNA was reduced to similar levels in 35S:TAS1cPDS-2-transformed Col-0 and zip-1 plants, but unchanged in rdr6–15 and dcl4–2 mutants, relative to vector-transformed Col-0 plants (Fig. 1C). Syn-tasiRNA were detected in Col-0 and zip-1 plants, but not in rdr6–15 and dcl4–2 plants, transformed with each construct (Fig. 1D and Table S1). Relative PDS mRNA and syn-tasiRNA levels were not significantly different between Col-0 and zip-1 35S:TAS1cPDS-2-transformed plants (P = 0.23 and 0.29, respectively, two-sample t tests) (Fig. 1 C and D). These results indicate that TAS1c-based syn-tasiRNA have genetic requirements similar to those of authentic TAS1 tasiRNA and are distinguishable from TAS3 tasiRNA by the lack of a requirement for AGO7 (2–7, 16, 18, 19).
A potential application of syn-tasiRNA is silencing of genes in a wide range of plant species. However, miR173 is only known to exist in Arabidopsis and close relatives (29). Therefore, a dual-gene construct was designed to deliver both miR173 and syn-tasiRNA (35S:TAS1cPDS/MIR173), and it was tested in Nicotiana benthamiana leaves after transient delivery (Fig. S1A) (30). This construct yielded both miR173 and syn-tasiRNA with efficiency similar to coexpression of the independent constructs 35S:TAS1cPDS-4 and 35S:MIR173 (P = 0.43 and 0.50, respectively, two-sample t tests) (Fig. S1 B and C). In transgenic Arabidopsis Col-0 plants, both 35S:TAS1cPDS-4 and 35S:TS1cPDS-4/MIR173 constructs resulted in similar patterns of photobleaching (Fig. S1D and Table S1). Thus, a single-construct, dual-gene system can expand TAS1c-based syn-tasiRNA technology to non-Arabidopsis species.
Role of miR173 in TAS1c-Based syn-tasiRNA Formation.
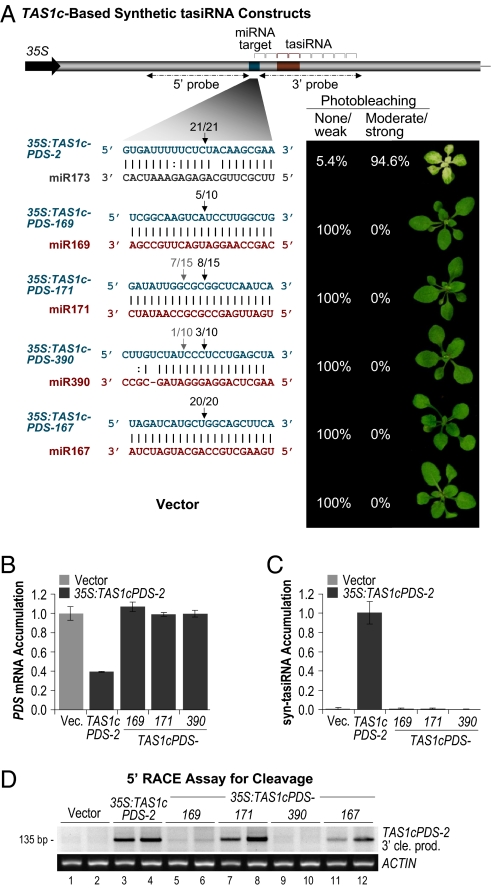
To determine whether miR173 possesses unique properties that facilitate tasiRNA initiation, syn-tasiRNA constructs were developed in which the miR173 target site in 35S:TAS1cPDS-2 was substituted for miR169 (35S:TAS1cPDS-169), miR171 (35S:TAS1cPDS-171), miR390 (35S:TAS1cPDS-390), or miR167 (35S:TAS1cPDS-167) target sites. The substitutions in 35S:TAS1cPDS-390 and 35S:TAS1cPDS-171 resulted in authentic miRNA-target site duplexes with 5 mispairs or no mispairs, respectively. The 35S:TAS1cPDS-167 and 35S:TAS1cPDS-169 substitution resulted in perfect complementarity to their respective miRNA, although authentic miR167- and miR169-target site duplexes contain mispairs (Fig. 2A). Each heterologous miRNA accumulates to relatively high levels in Col-0 inflorescence, seedling, and leaf tissues (31).
Fig. 2.
miR173 specificity in syn-tasiRNA formation. (A) The miR173 target site in 35S:TAS1cPDS-2 was substituted for sites recognized by miR169, miR171, miR390, or miR167. The percentages of transgenic plants (n > 30 primary transformants) with none/weak or modest/strong photobleaching are shown. For plants in the none/weak category, only those containing 35S:TAS1cPDS-167 rarely displayed photobleaching. The proportion of cloned 5′ RACE products corresponding to cleavage at a site is shown above the arrows. Black arrows indicate the canonical miRNA-guided cleavage position. Dashed horizontal arrows indicate the 5′ and 3′ probes used in blot assays in Fig. 3. (B) Mean relative level +/− SEM of PDS mRNA after normalization to ACT2 mRNA, as determined by quantitative RT-PCR (Col-0 vector = 1.0). (C) Mean relative level +/− SEM of syn-tasiRNA, as determined by blot assays (Col-0 35S:TAS1cPDS-2 = 1.0). (D) EtBr-stained 5′ RACE products. 5′ RACE products corresponding to cleavage at the canonical miRNA-guided cleavage site migrate at 135 bp. ACT2 RT-PCR products are shown as a control.
In contrast to plants expressing the miR173-targeted 35S:TAS1cPDS-2 construct, no photobleaching was detected in Col-0 plants transformed with 35S:TAS1cPDS-169, 35S:TAS1cPDS-171, or 35S:TAS1cPDS-390 constructs (Fig. 2A). Rarely, very weak photobleaching was observed in plants transformed with the 35S:TAS1cPDS-167 construct (data not shown). PDS mRNA levels were reduced in lines with the miR173-targeted construct, but were not significantly affected in the miRNA-target site substitution lines (P > 0.46, two-sample t tests) (Fig. 2B). Syn-tasiRNA failed to accumulate to detectable levels in leaf tissue of 35S:TAS1cPDS-169-, 35S:TAS1cPDS-171-, and 35S:TAS1cPDS-390-transformed plants (Fig. 2C). Cleavage at each heterologous target site was tested by using RNA ligase-mediated 5′ RACE (30). PCR products with 5′ ends corresponding to positions of canonical miRNA-guided cleavage were detected in plants transformed with each construct, although the levels of 5′RACE products detected were highest in 35S:TAS1cPDS-2- and 35S:TAS1cPDS-171-transformed plants (Fig. 2 A and D).
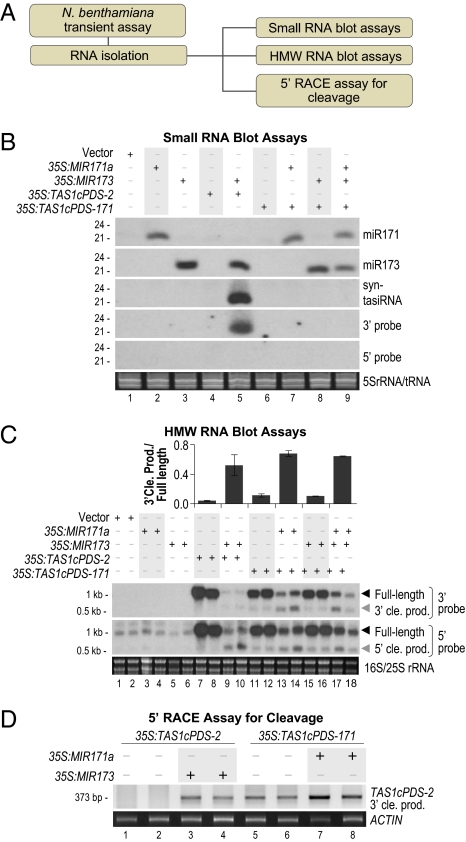
The 35S:TAS1cPDS-2 and 35S:TAS1cPDS-171 constructs were examined in more detail in N. benthamiana leaves in combination with 35S:MIR173 or 35S:MIR171a, which produce active miR173 and miR171, respectively (9) (Fig. 3A). Syn-tasiRNA were efficiently generated from 35S:TAS1cPDS-2 transcripts and depended on coexpression of 35S:MIR173 (Fig. 3B, lanes 4 and 5). In contrast, 35S:TAS1cPDS-171 failed to generate syn-tasiRNA when coexpressed with either 35S:MIR171a alone or a combination of 35S:MIR171a and 35S:MIR173 (Fig. 3B, lanes 7–9). Cleavage of the 35S:TAS1cPDS-2 transcript depended on coexpression of 35S:MIR173 (Fig. 3 C, lanes 7–10 and D, lanes 1–4). With the 35S:TAS1cPDS-171 construct, cleavage was detected in leaves that coexpressed 35S:MIR171a, but also at lower levels in leaves that lacked 35S:MIR171a (Fig. 3 C, lanes 11–14 and D, lanes 5–8). As shown (9, 30), N. benthamiana contains active miR171 that functions on ectopically expressed target transcripts. Therefore, failure to form miR171-initiated syn-tasiRNA in Arabidopsis and N. benthamiana was not caused by a lack of cleavage.
Fig. 3.
Specificity of miR173 for syn-tasiRNA formation in N. benthamiana transient assays. (A) Flow chart. (B) Syn-tasiRNA constructs containing miR173 or miR171 target sites were expressed or coexpressed as indicated above blot panels. One of two biological replicates is shown. Probe sequences for 5′ and 3′ regions of syn-tasiRNA constructs are indicated by horizontal arrows in Fig. 2A. EtBr-stained 5S rRNA/tRNA are shown as a loading control. (C) Blot assays for syn-tasiRNA full-length transcripts and cleavage products (cle. prod.). The ratios of 3′ cleavage product to full-length transcript are shown above the blot images. EtBr-stained 18S/25S rRNA are shown as a loading control. (D) EtBr-stained gel of 5′ RACE products. 5′ RACE products corresponding to cleavage at the canonical miRNA-guided cleavage site migrate at 373 bp. ACTIN RT-PCR products are shown as a control.
TAS1 and TAS2 transcripts normally yield tasiRNA from the 3′ RNA fragment generated by miR173-guided cleavage. To determine whether miR171 initiated aberrant entry of the 5′ fragment into the tasiRNA pathway, RNA blot assays were done by using probes corresponding to the regions directly 5′ or 3′ of the miR173 and miR171 target sites of 35S:TAS1cPDS-2 and 35S:TAS1cPDS-171, respectively, in the N. benthamiana transient system. Using the parental construct, siRNA from the 3′ side (corresponding to the tasiRNA region), but not from the 5′ side, of the miR173 target were detected (Fig. 3B, lane 5). In contrast, targeting by miR171 failed to trigger siRNA formation from either side of the target site (Fig. 3B, lanes 7 and 9). Collectively, these data indicate that miR173 and/or associated factors possess unique properties that facilitate directional routing of cleaved TAS1c RNA precursor into the tasiRNA biogenesis pathway.
miR173 Triggers Phased siRNA Formation from Non-TAS Transcripts.
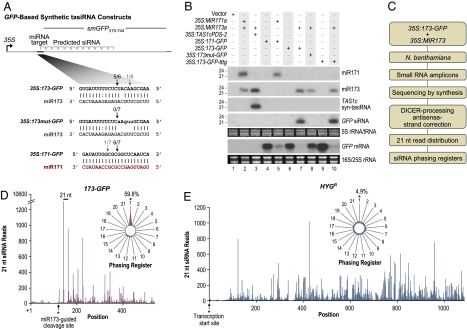
To determine whether a single miR173 target site is sufficient to directionally route a transcript into the tasiRNA pathway, a series of synthetic target sites were fused to a 425-base segment from the 3′ region of the smGFP coding sequence (35S:173-GFP). Also, constructs containing either a defective miR173 target site with four additional target–miR173 mispairs (35S:173mut-GFP) or a functional miR171 target site (35S:171-GFP) were made (Fig. 4A). Strikingly, the 35S:173-GFP construct containing a functional miR173 site, and a 35S:TAS1cPDS-2 control, yielded siRNA specifically when coexpressed with 35S:MIR173 in N. benthamiana leaves (Fig. 4B, lanes 3, 6, and 7). The 35S:173mut-GFP construct failed to yield GFP siRNA when coexpressed with 35S:MIR173, indicating that miR173-guided cleavage is likely necessary (Fig. 4B, lane 8). The 35S:171-GFP construct failed to yield GFP siRNA, even when coexpressed with 35S:MIR171a (Fig. 4B, lanes 4 and 5). Both the 35S:173-GFP and 35S:171-GFP transcripts were efficiently cleaved at the predicted miR173 or miR171 target sites, as determined by 5′ RACE and RNA blot assays (Fig. 4 A and B). No miR173-guided cleavage was detected with 35S:173mut-GFP transcript (Fig. 4A).
Fig. 4.
miR173 triggers phased siRNA formation from modified GFP transcripts. (A) Organization of the GFP-based constructs with engineered miRNA target sites. The proportion of cloned 5′ RACE products corresponding to cleavage at a site is shown above the arrows. Black arrows indicate the canonical miRNA-guided cleavage position. (B) Blot assays. TAS1c- and GFP-based constructs containing miR173 or miR171 target sites were expressed or coexpressed as indicated above the blot panels. One of two biological replicates is shown. EtBr-stained 5S rRNA/tRNA and 18S/25S rRNA are shown as loading controls. (C) Flow chart. (D and E) Twenty-one-nucleotide small RNA read distributions across 173-GFP and HYGR transcripts. (Insets) Radar plots display percentages of reads corresponding to each of the 21 registers, with the 5′ end formed by miR173-guided cleavage or the transcription start site in 173-GFP and HYGR, respectively, defined as register 1.
To determine whether the GFP siRNA from 173-GFP transcript (coexpressed with 35S:MIR173) were in register with the miR173-guided cleavage site, as are authentic TAS1 and TAS2 tasiRNA, small RNA from the N. benthamiana transient assay were analyzed by deep sequencing-by-synthesis (SBS) technology (Illumina 1G) (9) (Fig. 4C). Small RNA formation from the 171-GFP transcript (coexpressed with 35S:MIR171a) was also assessed. Small RNA reads with perfect matches to the 173-GFP or 171-GFP transcripts or to the hygromycin resistance gene (HYGR) transcripts from the base plasmids were quantified. Both 173-GFP- and 171-GFP-derived reads were predominantly 21 nt, whereas HYGR-derived reads were nearly evenly distributed between the 21- and 22-nt or 24-nt size classes (Fig. S3 and Fig. S4). The 21-nt small RNA reads from both strands were consolidated into one set of values by summing sense and antisense reads that were offset by 2 nt (14) to account for the known properties of DICER and DICER-LIKE enzymes, which leave a 2-nt, 3′ overhang at each end of a small RNA duplex (32).
Approximately 98% of 21-nt small RNA reads from 173-GFP transcripts originated from the 3′ side of the miR173 target site. This region yielded a series of abundant siRNA (each with at least 367 reads) that were clustered downstream of the miR173 target site and separated by 21 nts for 5 21-nt cycles (Fig. 4D). Approximately 60% (13,407) of 21-nt reads from 173-GFP were in 21-nt register with the cleavage site, although 78% of those were from the second-phase cycle relative to the target site (Fig. 4D and Table S2). Even with this abundant position subtracted from the analysis, 25% of the remaining reads were in phase with the cleavage site; no other phase exceeded 8% of total reads in the depleted dataset. In contrast, only 5.9% (529) of 21-nt reads that mapped to 171-GFP were in register with the miR171-guided cleavage site (Fig. S4A and Table S3). Small RNA reads from HYGR were evenly distributed across the 21 possible registers, with no more than 9% of the total reads coming from any single register (Fig. 4E, Fig. S4B, Table S4, and Table S5). These results indicate that targeting by miR173 at a single site initiates phased siRNA formation in a directional manner.
Search for Additional cis Elements for TAS1c syn-tasiRNA Biogenesis.
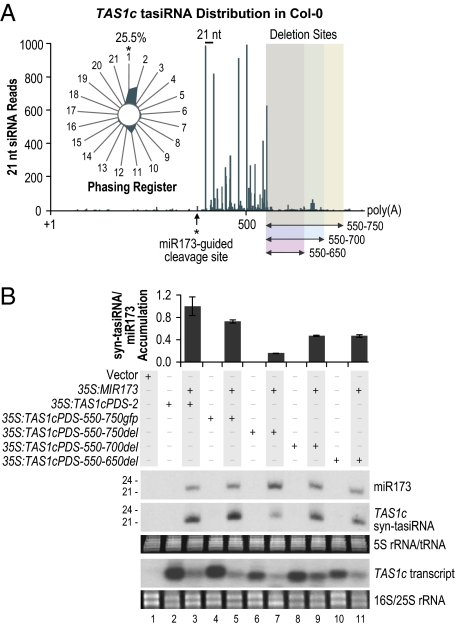
Although the 35S:173-GFP results indicate the sufficiency of a single miR173 target site to trigger tasiRNA biogenesis, authentic TAS1 and TAS2 tasiRNA formation may be enhanced by additional cis regulatory elements. To test for such sequences in the TAS1c locus, a series of 5′, 3′, and internal deletions were introduced into 35S:TAS1cPDS-2 and the resulting constructs were tested in N. benthamiana leaves (Fig. 5A and Fig. S2A). The predominant tasiRNA-yielding region of TAS1c was identified by using Col-0 leaf and whole plant small RNA sequencing data (31). As shown, tasiRNA were abundant from eight processing cycles downstream of the cleavage site, although processing was offset by 1 nt after the fourth cycle (Fig. 5A and Table S6) (14). In an initial series of constructs, the entire TAS1c-derived sequence upstream (5′) of the miR173 target site, and varying amounts at the 3′ end between nucleotide positions 662–837 (relative to the transcription start site), of 35S:TAS1cPDS-2 were removed (Fig. S2A). In each case, the truncated transcripts were susceptible to miR173-guided cleavage (Fig. S2C). Deletion of the 5′ region did not significantly affect syn-tasiRNA levels (P = 0.68, two-sample t test) (Fig. S2B). The 3′ deletion constructs also yielded syn-tasiRNA, although both syn-tasiRNA and transcript levels were reduced proportionally from constructs with deletions upstream of nucleotide position 762 (Fig. S2 B and C). This result suggests that the 3′ region has an effect on transcript accumulation. Considering that the dominant poly(A) site of TAS1c transcripts is located at position 810, this effect could relate to the efficiency of 3′ termination.
Fig. 5.
The effect of internal deletions on syn-tasiRNA formation. (A) tasiRNA distribution across the endogenous TAS1c transcript in Col-0. 5,888 TAS1c tasiRNA reads from Col-0 leaf and whole plant Illumina 1G sequencing data sets were obtained from the Arabidopsis Small RNA Project database (31). (Inset) The radar plot displays the percentages of reads corresponding to each of the 21 registers, with the 5′ end formed by miR173-guided cleavage defined as register 1. Internal deletion positions are shown by the shaded boxes. (B) Blot assays. Parental 35S:TAS1cPDS-2 and modified constructs containing internal deletions were expressed individually or in combination with 35S:MIR173 as indicated above the blot panels. Mean relative level +/− SEM of syn-tasiRNA relative to miR173 (35S:TAS1cPDS-2 + 35S:MIR173 = 1.0). One of three biological replicates is shown. EtBr-stained 5S rRNA/tRNA and 18S/25S rRNA are shown as loading controls.
In a second series of deletions, internal regions downstream from the main tasiRNA-generating sequences, but upstream from the likely poly(A) signal, were removed (Fig. 5A). For 3 constructs, sequences spanning positions 550–650, 550–700, and 550–750 (35S:TAS1cPDS-550–650del, 35S:TAS1cPDS-550–700del, and 35S:TAS1cPDS-550–750del) were deleted (Fig. 5A). In a fourth construct (35S:TAS1cPDS-550–750gfp), the TAS1c sequence at positions 550–750 was replaced with an equal size segment of smGFP coding sequence, thus maintaining the authentic transcript length. Transcripts from each construct were susceptible to miR173-guided cleavage in the N. benthamiana transient assay (Fig. 5B). Deletions of nucleotide positions 550–650 and 550–700 resulted in a modest reduction in syn-tasiRNA formation, relative to the parental construct (Fig. 5B), whereas deletion of nucleotides 550–750 significantly reduced syn-tasiRNA formation (Bonferroni corrected P < 0.05, two-sample t test). In each case where a reduction in syn-tasiRNA was observed, as with the 3′ deletion constructs, a proportional reduction in transcript level was detected (Fig. 5B). In contrast, 35S:TAS1cPDS-550–750gfp yielded transcript at high levels and syn-tasiRNA in amounts that were indistinguishable from the parental 35S:TAS1cPDS-2 construct (P = 0.19, two-sample t test) (Fig. 5B). These results indicate that regions 5′ of the miR173 target site and 3′ of the predominant tasiRNA-yielding region of TAS1c do not directly affect tasiRNA formation; however, transcript length and presence of the authentic poly(A) signal likely have an indirect effect based on transcript levels.
Consistent with previous studies (2), only weak similarity was identified between the three TAS1 loci and the TAS2 locus outside of the tasiRNA yielding region. However, an 8-nt motif (TTTGTAAT) was identified near the 3′ ends of the tasiRNA generating regions of all TAS1 and TAS2 loci. This motif occurs at least twice at each locus (data not shown). In TAS1c, the motif occurs at nucleotide positions 714 and 744, relative to the transcript 5′ end. Although the results of the deletion-replacement construct suggest that this motif plays no direct role in syn-tasiRNA formation, we nonetheless tested a 61-nt segment of TAS1c, 702–762 nt downstream of the transcription start site and encompassing both of the TTTGTAAT motifs, for siRNA-enhancing activity in the context of the 3′ end of 35S:173-GFP. The resulting construct (35S:173-GFP-tttg) was coexpressed with or without 35S:MIR173 in N. benthamiana leaves. The levels of miR173-dependent siRNA from the GFP sequence were indistinguishable from those measured by using 35S:173-GFP (Fig. 4B), indicating that the TTTGTAAT motifs and other sequences tested do not possess detectable signals for tasiRNA formation.
miR173–AGO1 Complex Functions in TAS1 and TAS2 Initiation Cleavage.
Each of the Arabidopsis TAS1 and TAS2 miR173–target site duplexes has a mispair at position 9, relative to the 5′ end of the miRNA, although the nucleotide at this position differs between the target sites. In TAS1b, there is an additional mispair at position 10 (Fig. S5A). miRNA-guided cleavage occurs between positions 10 and 11, and mispairs at or adjacent to the cleavage site in plant miRNA-target duplexes are rare (4, 33–35). Conceivably, the mispairs could facilitate tasiRNA formation by stabilizing an AGO complex at the target site long enough for RDR6 recruitment to occur, without compromising eventual cleavage. To determine whether the TAS1/TAS2-miR173 mispair pattern is important for tasiRNA formation, the miR173 target site in 35S:TAS1cPDS-2 was mutated to generate a target site with perfect complementarity to miR173, aside from a single G:U pair at position 15 (35S:TAS1cPDS-173fix) (Fig. S5B). Conversely, a mispair base was introduced at position 9 in the miR171–target site of construct 35S:TAS1cPDS-171 (35S:TAS1cPDS-171–2) to mimic the base pair composition of authentic miR173-TAS1 and miR173-TAS2 duplexes. When coexpressed with 35S:MIR173 in N. benthamiana, 35S:TAS1cPDS-173fix yielded syn-tasiRNA at a level similar to 35S:TAS1cPDS-2 (Fig. S5C, lanes 4–7). In contrast, 35S:TAS1cPDS-171–2 failed to yield syn-tasiRNA, even when coexpressed with 35S:MIR171a (Fig. S5C, lanes 8–11). Thus, the ability of miR173 to trigger tasiRNA formation is likely more complicated than can be explained by the base pair–mispair configuration at the TAS1 and TAS2 miR173–target site duplex.
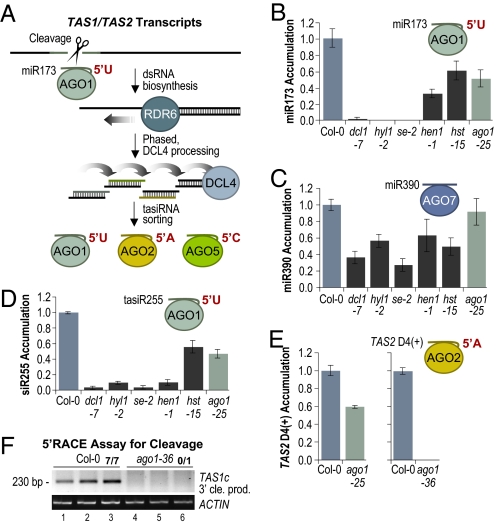
The genetic requirements of miR173, including dependence on DCL1, HYL1, SE, HEN1, and HST, are similar to those of other miRNA (Fig. 6B) (2, 36). The unusual ability of miR173 to trigger tasiRNA formation, therefore, could reflect additional biogenesis or effector factor requirements that distinguish it from other miRNA. Like miR390, it is possible that miR173 associates with a distinct AGO. miR173 was shown to associate with AGO1 (8, 9); however, it is still unclear whether miR173–AGO1 is the functional tasiRNA initiator complex. In ago1–25, a weak allele of AGO1 (37), miR173 levels were reduced by 43% (P = 0.03, two-sample t test) (Fig. 6B), suggesting that at least a portion of the miR173 population is stabilized through association with AGO1. In contrast, miR390, which does not function with AGO1, was unaffected (P = 0.66, two-sample t test) (Fig. 6C). The level of TAS1 tasiR255, an abundant AGO1-associated tasiRNA containing a 5′U (8, 38), was reduced by 52% in ago1–25 (P = 0.0008, two-sample t test) (Fig. 6D) and below detectable levels in the strong ago1–1 mutant (18). In mutants with defects in each of the other 9 AGOs, tasiR255 levels were either unaffected or elevated (Fig. S6).
Fig. 6.
Role of AGO1 in TAS1 and TAS2 tasiRNA formation. (A) Model for TAS1/TAS2 tasiRNA formation and sorting by AGO proteins. (B–E) Mean relative levels +/− SEM of miR173, miR390, and two distinct tasiRNA as determined by blot assays (Col-0 = 1.0). (F) EtBr-stained 5′ RACE products. 5′ RACE products corresponding to cleavage at the canonical miR173-guided cleavage site migrate at 230 bp. ACT2 RT-PCR products are shown as a control. The proportion of cloned 5′ RACE products corresponding to cleavage at the canonical site is shown above lanes 3 and 6.
The loss of TAS1 tasiR255 in ago1 mutants could be caused solely by destabilization of the tasiRNA, rather than lack of initiating cleavage guided by miR173. To determine whether miR173-dependent tasiRNA that do not directly associate with AGO1 are affected by loss of AGO1, and thus indicating that AGO1 functions upstream of tasiRNA sorting into effector complexes (Fig. 6A), TAS2 3′D4(+) levels in the weak ago1–25 and strong ago1–36 (38) mutants were measured. TAS2 3′D4(+) contains a 5′A, which drives association with AGO2 and exclusion from AGO1 (8–10). Misprocessed, 5′U-containing siRNA with part of the TAS2 3′D4(+) sequence (offset by 4 nt), which could conceivably complicate the blot assays, are extremely rare based on deep sequence database searches (31). TAS2 3′D4(+) levels were reduced by ≈40% in ago1–25, relative to Col-0 (P < 0.05, two-sample t test) and absent in ago1–36 (Fig. 6E), supporting the interpretation of AGO1 functionality at an early point during biogenesis.
To test more directly whether the miR173–AGO1 complex is required for cleavage of TAS1c transcripts, 5′ RACE assays were done with RNA extracts from Col-0 and ago1–36 plants. Products corresponding to cleavage at the miR173 target site were efficiently detected in Col-0, but were absent in ago1–36 (Fig. 6F). These results solidify the interpretation that AGO1 functions with miR173 during initiation cleavage of TAS1 and TAS2 transcripts.
Deep Sequencing Analysis of AGO1-Dependent tasiRNA.
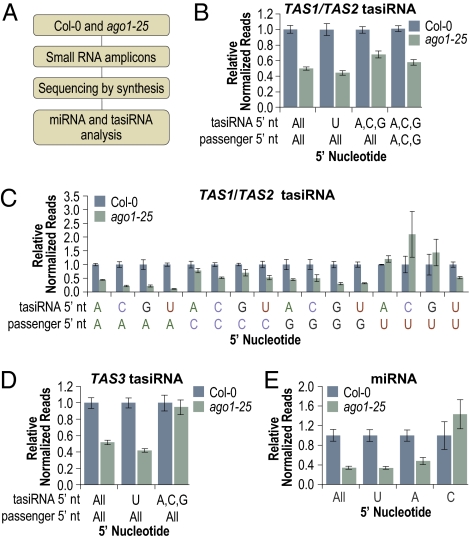
To more exhaustively assess AGO1-dependence across tasiRNA loci, small RNA amplicons were prepared from Col-0 and ago1–25 and sequenced by using SBS technology (Fig. 7A). To facilitate statistical analyses, three biological replicates for both Col-0 and ago1–25 were analyzed. In Col-0, small RNA reads were nearly evenly distributed between the 21- and 24-nt size classes, whereas, in ago1–25, the 21-nt size class was partially depleted (Fig. S7). An average of ≈4.0 million and ≈6.5 million total reads, yielding at least 10,000 tasiRNA reads, were obtained from Col-0 and ago1–25 samples, respectively. Normalized TAS1 and TAS2 tasiRNA reads were reduced by ≈45% in ago1–25, relative to Col-0 (Bonferroni corrected P < 0.05, two sample t test) (Fig. 7B). Subpopulations of TAS1 and TAS2 tasiRNA, however, were differentially affected depending on the 5′ nt of both the sequenced read and its predicted passenger strand (offset by 2 nt) (Fig. 7 B and C, Fig. S8A, and Table S7). 5′U-containing tasiRNA reads were reduced by 56% in ago1–25, relative to Col-0 (Bonferroni corrected P < 0.05, two sample t test) (Fig. 7B). 5′A-, C-, and G-containing sequences were reduced by ≈32% in ago1–25, relative to Col-0 (Bonferroni corrected P < 0.05, two sample t test), consistent with a requirement for AGO1 in TAS1 and TAS2 tasiRNA formation. Although the majority of tasiRNA were reduced in ago1–25, sequences with a 5′A, C, or G and a passenger strand with a 5′U tended to be up or unaffected (Fig. 7C), likely because these passenger sequences normally associate efficiently with AGO1. Removing TAS1 and TAS2 sequences with 5′U-containing passenger strands from the analysis led to a 43% reduction in tasiRNA containing 5′A, C, and G in ago1–25, relative to Col-0 (Bonferroni corrected P < 0.05, two sample t test) (Fig. 7B).
Fig. 7.
Sequencing analysis of AGO1-dependent small RNA. (A) Flow chart. (B) Mean relative level +/− SEM of TAS1 and TAS2 tasiRNA (Col-0 = 1.0). (C) Mean relative level +/− SEM of TAS1 and TAS2 tasiRNA for each possible sequenced siRNA/predicted passenger strand 5′ nucleotide combination (Col-0 = 1.0). (D) Mean relative level +/− SEM of TAS3 tasiRNA (Col-0 = 1.0). (E) Mean relative level +/− SEM of miRNA (Col-0 = 1.0).
Unlike TAS1 and TAS2 tasiRNA formation, TAS3 tasiRNA formation does not require AGO1, although TAS3 tasiRNA containing 5′U likely function through AGO1 (8, 9). Thus, whereas TAS3 tasiRNA with 5′U should be destabilized by the reduced activity of AGO1 in ago1–25, TAS3 tasiRNA containing 5′A, C, and G should be unaffected. Indeed, TAS3 tasiRNA containing 5′U were reduced by ≈58% in ago1–25 relative to Col-0 (Bonferroni corrected P < 0.05, two-sample t test), whereas the level of tasiRNA containing 5′A, C, and G was unchanged (P = 0.72, two-sample t test) (Fig. 7D and Fig. S8B). Some specific TAS3 tasiRNA containing a 5′A, C, or G, such as tasiR1778 (containing a 5′C) were reduced in ago1–25 (Table S7). These tasiRNA were typically in phase with cleavage guided by a 5′U-containing secondary tasiRNA derived from the complementary strand of TAS3 (TAS3a 5′D2[−]) that was shown to associate with AGO1 (4, 8).
Total 5′U-containing miRNA were reduced by ≈66% in ago1–25 (Bonferroni corrected P < 0.05, two sample t test) (Fig. 7E), although the majority of miRNA reads (≈83–90%) were the abundant miR159 (Table S8). When miR159 was subtracted from the analysis, the remaining 5′U-containing miRNA were reduced by ≈56% in ago1–25, relative to Col-0 (Bonferroni corrected P < 0.05, two sample t test). In contrast, miRNA containing 5′C were unaffected (P = 0.24, two-sample t test). Although the majority of individual 5′A-containing miRNA were unaffected in ago1–25, the level of the 5′A-containing miR172a/miR172b was reduced by ≈55% (P = 0.006, two-sample t test), resulting in an overall reduction in 5′A-containing miRNA (Fig. 7E and Table S8). For each of the two known 5′G-contaning miRNA (miR172e and miR861a), too few reads were obtained to assess AGO1 dependence (Table S8). Based on these results and those of the previous section, we conclude that AGO1 is required for initiation cleavage of TAS1 and TAS2 transcripts and for stabilization of most 5′U-containing tasiRNA. They also indicate that 5′ nucleotide identity plays a significant role in strand selection and sorting of tasiRNA, irrespective of the TAS family.
Discussion
miR173 guides cleavage of TAS1 and TAS2 transcripts at a single site, defining a discrete 5′ end of the pre-tasiRNA transcript and an initiation point for phased siRNA formation by DCL4. Heterologous miRNA target sites in TAS1c-based syn-tasiRNA constructs were cleaved, but failed to initiate syn-tasiRNA formation. Introduction of a single miR173 target site, but not a miR171 site, into a segment of the GFP coding sequence was sufficient to direct phased, unidirectional, miR173-dependent siRNA biogenesis, suggesting that miR173 has unique properties or associated cofactors for routing transcripts to the RDR6/SGS3/DCL4 pathway. Based on current and previous results of functional and association assays, miR173 interacts with, and functions through, AGO1 for TAS1 and TAS2 tasiRNA biogenesis and activity (8, 9, 38–41). Interestingly, TAS1 and TAS2 transcripts stand in contrast to the high proportion of RDR6/DCL4-dependent siRNA-generating transcripts, including those from TAS3 loci, that are targeted by two or more miRNA and/or tasiRNA in Arabidopsis (13, 14).
There are several events, including transcript cleavage and RDR6 recruitment, necessary for efficient TAS1 and TAS2 tasiRNA biogenesis. Cleavage may be necessary to separate the siRNA-generating fragment from the 5′ cap and cap-associated factors, such as the cap-binding complex in the nucleus (13, 42, 43). Most miRNA targets that undergo cleavage, however, do not spawn siRNA, as they may be degraded by other mechanisms, such as through the XRN4/EIN5-dependent pathway (43–47). Recruitment of RDR6 and subsequent DCL4-mediated siRNA duplex formation distinguish TAS1 and TAS2 transcripts from the vast majority of other AGO1–miRNA-directed cleavage products.
Introduction of a single miR173 target site, but not other target sites, into a foreign sequence is sufficient to trigger phased siRNA formation, indicating that miR173 possess unique information for RDR6 recruitment. Similar to authentic TAS1 and TAS2, the 35S:173-GFP construct containing an ectopic miR173 target site lacked a functional coding sequence, and it is possible that nontranslatability enhances secondary siRNA formation. Conceivably, miR173–AGO1 complexes associate with a unique cofactor that recruits RDR6. An attractive candidate for such a cofactor is SGS3, which was shown to stabilize the cleavage fragments of TAS1 and TAS2 transcripts (2). One possibility is that SGS3 associates with AGO1 during loading of miR173 and is subsequently directed to TAS1 and TAS2 transcripts by the miR173–AGO1 complex. SGS3, in turn, may recruit RDR6 to the transcript. Alternatively, miR173–AGO1 may interact directly with RDR6 for recruitment. Exactly how miR173–AGO1 transmits a signal to recruit RDR6 is a key problem to solve, as is understanding the differences between the single-site functionality of miR173–AGO1 and the dual-site functionality of miR390–AGO7 during TAS3 tasiRNA formation. miR828, miR168, and miR393 also trigger siRNA formation through interaction at a single site on target transcripts, although at low levels relative to miR173-targeted TAS1 and TAS2 transcripts (13, 15, 48). It will be important to learn whether other miRNA possess routing properties similar to miR173.
Although deletions near the 3′ end dampened accumulation of TAS1c-based syn-tasiRNA, the low tasiRNA levels could be accounted for by indirect effects on transcript accumulation or stability. The fact that the deletion effect could be suppressed by insertion of a comparable-size heterologous sequence suggests that the 3′ region, or the entire transcript, is sensitive to length. Thus, although it is likely that miR173 and associated factors provide specific information for routing transcripts through the tasiRNA pathway, more general factors involving RNA metabolism clearly influence tasiRNA biogenesis.
TAS1c-based syn-tasiRNA effectively silenced PDS expression. As a tool for gene silencing, syn-tasiRNA offer several advantages over other RNAi methods. First, because tasiRNA formation occurs in phase with the miRNA-guided cleavage site, syn-tasiRNA formation is highly predictable. In contrast, small RNA formation from hairpin RNAi constructs is unpredictable and prone to off-target effects (49). Second, although amiRNA constructs are also very predictable (4, 33–35), the syn-tasiRNA approach is more amenable to stacking multiple functional small RNA sequences into a single construct. In fact, TAS1c gives rise to at least 7 relatively abundant tasiRNA (14), suggesting that many more syn-tasiRNA could be generated from a single construct compared to what we demonstrated here. The ability to generate multiple syn-tasiRNA enables silencing of multiple transcripts or transcript families from a single construct. Additionally, varying the number of syn-tasiRNA that target a specific gene might be useful for fine-tuning the level of suppression of a particular gene. TAS1c-based syn-tasiRNA have some disadvantages as well, most notably their dependence on miR173, a miRNA known only to exist in Arabidopsis and close relatives (29). But this limitation can be overcome by using the dual-gene construct that expresses both syn-tasiRNA transcripts and miR173 precursor, thus expanding functionality to non-Arabidopsis species.
Methods
The mutant alleles rdr6–15, dcl4–2, zip-1, dcl1–7, hyl1–2, se-2, hen1–1, hst-15, ago1–25, and ago1–36 have been described (4, 7, 37, 38, 50–55). RNA blot assays, quantitative RT-PCR, RNA ligase-mediated 5′ RACE, and transient expression assays in N. benthamiana were done as described (30). Small RNA amplicons for sequencing with the Illumina 1G system were prepared as described (9). Phasing analysis and graphical display were done as described (14). Small RNA library normalization and subsequent statistical analysis procedures using S-PLUS (Insightful) and Excel (Microsoft) have been described (9). Bonferroni adjustments were made to significance level cutoffs when doing multiple comparisons. Additional methods and details about syn-tasiRNA vectors are in SI Text.
Supplementary Material
Acknowledgments.
We thank Amy Shatswell, Jesse Hansen, Molly Blatz, Sarah Dvorak, Josh Cuperus, and Scott Givan for technical and computational assistance; Mark Dasenko and Kristin Kasschau for DNA sequencing assistance; Detlef Weigel and Felipe Felippes for helpful discussions and suggestions; and Jian-Kang Zhu (University of California, Riverside, CA) for ago6–1 and ago6–2 seed. This work was supported by the Monsanto Corporation, National Science Foundation Grant MCB-0618433, National Institutes of Health Grant AI43288, Department of Agriculture Grant 2006-35301-17420, Crop Functional Genomics Center Grant CG1211, and Plant Signaling Network Research Center Grant R11-2003-008-03001-0.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited with the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI), www.ncbi.nlm.nih.gov/geo (accession no. GSE13605).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810241105/DCSupplemental.
References
- 1.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 2.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Peragine A, et al. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montgomery TA, et al. Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 10.Takeda A, et al. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008;49:493–500. doi: 10.1093/pcp/pcn043. [DOI] [PubMed] [Google Scholar]
- 11.Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 12.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Howell MD, et al. Genomewide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahlgren N, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 tasiRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 17.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 18.Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 19.Hunter C, et al. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 22.Muangsan N, Beclin C, Vaucheret H, Robertson D. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 2004;38:1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x. [DOI] [PubMed] [Google Scholar]
- 23.Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- 24.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 25.Dalmay T, et al. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101:543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 26.Talmor-Neiman M, et al. Identification of trans-acting siRNAs in moss and an RNA-dependent RNA polymerase required for their biogenesis. Plant J. 2006;48:511–521. doi: 10.1111/j.1365-313X.2006.02895.x. [DOI] [PubMed] [Google Scholar]
- 27.Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Luz Gutierrez-Nava M, et al. Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol. 2008;147:543–551. doi: 10.1104/pp.108.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 30.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 31.Backman TW, et al. Update of ASRP: The Arabidopsis Small RNA Project database. Nucleic Acids Res. 2007;36:D982–D985. doi: 10.1093/nar/gkm997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 34.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab R, et al. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, et al. Expression of Arabidopsis miRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morel JB, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in posttranscriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Y, et al. Distinct catalytic and noncatalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 42.Herr AJ, Molnar A, Jones A, Baulcombe DC. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc Natl Acad Sci USA. 2006;103:14994–15001. doi: 10.1073/pnas.0606536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gazzani S, et al. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306:1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 44.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Olmedo G, et al. ETHYLENE-INSENSITIVE5 encodes a 5′ → 3′ exoribonuclease required for regulation of the EIN3-targeting F-box proteins EBF1/2. Proc Natl Acad Sci USA. 2006;103:13286–132893. doi: 10.1073/pnas.0605528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.German MA, et al. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 48.Ronemus M, Vaughn MW, Martienssen RA. MicroRNA-targeted and small interfering RNA-mediated mRNA degradation is regulated by argonaute, dicer, and RNA-dependent RNA polymerase in Arabidopsis. Plant Cell. 2006;18:1559–1574. doi: 10.1105/tpc.106.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu P, et al. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol. 2006;142:429–440. doi: 10.1104/pp.106.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 51.Chen X, Liu J, Cheng Y, Jia D. HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development. 2002;129:1085–1094. doi: 10.1242/dev.129.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 53.Golden TA, et al. Short integuments1/suspensor1/carpel factory, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.