Abstract
Individuals with fragile X syndrome (FXS) are cognitively impaired and have marked speech delays and deficits. Our goal was to characterize expression of FMRP, the fragile X mental retardation protein, encoded by the gene FMR1, in an animal model that learns to vocalize, namely the zebra finch Taeniopygia guttata (Tgu). We cloned and sequenced the zebra finch ortholog of FMR1 (TguFmr1) and developed an antibody that recognizes TguFmrp specifically. TguFmrp has structural features similar to its human ortholog FMRP. Because FXS patients exhibit sensorimotor deficits, we examined TguFmrp expression prior to, during, and after sensorimotor song learning in zebra finches. We found that TguFmrp is expressed throughout the brain and in four major song nuclei of the male zebra finch brain, primarily in neurons. Additionally, prior to sensorimotor learning, we observed elevated TguFmrp expression in the RA of post-hatch day 30 males, compared to the surrounding telencephalon, suggesting a preparation for this stage of song learning. Finally, we observed variable TguFmrp expression in the RA of adolescent and adult males: in some males it was elevated and in others it was comparable to the surrounding telencephalon. In summary, we have characterized the zebra finch ortholog of FMRP and found elevated levels in the premotor nucleus RA at a key developmental stage for vocal learning.
Keywords: FMRP, Songbird, Brain Development, Vocal Learning
Fragile X syndrome (FXS) is the most commonly inherited mental retardation, affecting 1:4000 males and 1:6000 females panethnically (O'Donnell and Warren, 2002). The gene encoding the human fragile X mental retardation protein (FMRP in humans; Fmrp in mice and rats; TguFmrp in zebra finch [Taeniopygia guttata]) is FMR1; FXS primarily results from the absence of FMRP (Pieretti et al., 1991). FMRP is an RNA-binding protein involved in localization and translation regulation of its mRNA cargos (Terracciano et al., 2005). The FMR1 gene is expressed ubiquitously in the body, excluding the muscles, and predominantly in the testes and brain (Bächner et al., 1993, Devys et al., 1993, Hergersberg et al., 1995). Within the brain the protein is primarily neuronal (Devys et al., 1993), predominantly in the cytoplasm. Furthermore, it has been observed in both dendrites (Feng et al., 1997, Weiler et al., 1997, Greenough et al., 2001, Ling et al., 2004, Antar et al., 2005) and axons (Antar et al., 2006, Price et al., 2006, Tessier and Broadie, 2008a).
Males with FXS exhibit delays in speech and language development (Ferrando-Lucas et al., 2003, Roberts et al., 2005, Abbeduto et al., 2007), with vocalization deficits from preschool through adolescence (Dykens et al., 1993, Bailey et al., 1998, Kau et al., 2002, Roberts et al., 2007c). FXS vocalization includes perseveration, which is the repetition of a word or phrase, and abnormal prosody such as arrhythmia or inconsistent pronunciation of vowels. Importantly, these vocalization characteristics are not simply due to mental retardation – they are distinct from those of Down Syndrome or idiopathic mental retardation (Wolf-Schein et al., 1987, Sudhalter et al., 1990, Ferrier et al., 1991, Belser and Sudhalter, 2001). Kids with FXS perseverated significantly more than typical, developmentally-age matched (TD) controls and children with Down Syndrome (Roberts et al., 2007a). Further, FXS children have significantly lower expressive language skills than TD controls, but they still performed significantly better than kids with Down Syndrome (Roberts et al., 2007b). It is therefore appropriate to question the role of FMRP in normal vocal learning.
An excellent model organism for the study of speech pathology is the songbird zebra finch (Taeniopygia guttata). Like humans, songbirds are vocal learners – they have a sensitive period during early development in which they must hear adult vocalizations, as well as their own, to learn to properly vocalize (Doupe and Kuhl, 1999, Brainard and Doupe, 2002, Wilbrecht and Nottebohm, 2003). The process by which a zebra finch learns to vocalize is highly similar to the manner in which humans learn to speak (Doupe and Kuhl, 1999, Doupe et al., 2005). Song learning takes place in two overlapping periods – sensory, from approximately post-hatch day 25 or P25 (day of hatch = P1) until ∼P65 (equivalent to human adolescence just before puberty) and sensorimotor, from ∼P35 until sexual maturity at ∼P90 (White, 2001). The song no longer changes after sexual maturity and is said to be crystallized (Doupe and Kuhl, 1999). In humans, babbling begins at about month 7, followed by the first true word spoken at about 1 year, with continued vocal learning that diminishes markedly after sexual maturity (Doupe and Kuhl, 1999). Intriguingly, FXS individuals show impaired sensorimotor development, evident even before completion of the first year (Bailey et al., 2003, Baranek et al., 2005).
The neuronal circuitry for song learning and production (the ‘song circuit’) has been well characterized and includes nuclei in cortical, thalamic, and basal ganglia regions (Fig. 1). The pre-motor output nucleus of the telencephalic control system is the Robust nucleus of the Arcopallium (RA), and its major input develops at ∼P30, just preceding the onset of the sensorimotor phase of song learning (Konishi and Akutagawa, 1985, Mooney and Rao, 1994). Here we identify the zebra finch ortholog of FMRP (TguFmrp) and show its high similarity to human, mouse, and chicken orthologs. Additionally, we describe a novel antibody specific to TguFmrp, with which we show expression throughout neurons with increased expression in the RA nucleus of P30 males and variable expression in adolescent and adult males, indicating a role for TguFmrp in song circuit plasticity and thereby possibly in human vocal learning.
Fig. 1. Map of the zebra finch ‘song circuit’.
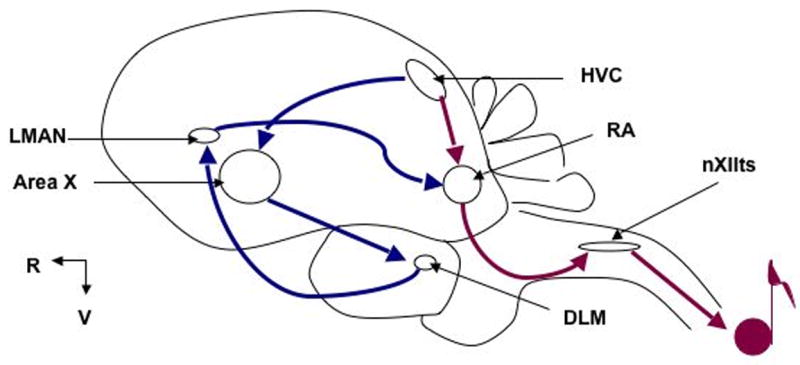
HVC: letter-based name; DLM: DorsoLateral Medial nucleus of the thalamus; LMAN: Lateral Magnocellular nucleus of the Anterior Nidopallium; nXIIts: nucleus trachiosyringealis of cranial nerve XII; RA: Robust nucleus of the Arcopallium. Anterior Forebrain Pathway (AFP) shown in blue; Posterior Pathway (PP) in red. The AFP traditionally has been recognized as important for song learning, while the PP for song production. LMAN is the output nucleus for the AFP. Recent data show that LMAN input into RA induces variability in song in both juveniles and adults, indicating that both pathways contribute to the motor production of song (Kao et al., 2005, Ölveczky et al., 2005, Thompson and Johnson, 2007, Aronov et al., 2008). (Rostral [R] left; Ventral [V] down. Not drawn to scale.)
Experimental Procedures
TguFmr1 and TguFmrp sequence generation
The complete sequence (4.2 Kb) was assembled from three shorter overlapping fragments: a PCR amplified open reading frame containing the translation start site through 57 nucleotides upstream of the translation stop site; and two overlapping EST clones containing combined sequence from 185 nucleotides upstream of the translation stop site through the poly-A tail. ESTs #SB03027A1E03.f1 and 0065P0010A12 were generously provided by David F. Clayton and Erich D. Jarvis, respectively. For the PCR, total RNA was harvested from a male zebra finch brain using the QIAGEN RNeasy Mini Kit (QIAGEN Sciences, Valencia, CA). cDNA was made using the BD SMART RACE cDNA Amplification Kit (BD Biosciences, San Jose, CA). PCR amplification was carried out using the following primers, each written 5′ to 3′ (note that the central primer was used in both directions): START: ATGGAGGAGCTGGTGGTGG; EXON6: ATGCTGATTGATATGCACTTTCG; A12-0: GGGGTTCCGCTCCTTGCCCG. Primers START and EXON6 were designed to conserved regions between human, mouse, and chicken Fmr1, that did not overlap with the mammalian autosomal homologs Fxr1 and Fxr2 (Siomi et al., 1995, Zhang et al., 1995). A12-0 was designed from the known sequence of EST #0065P0010A12. The amplified sequences were cloned into the pCRII vector using the TA Cloning Kit Dual Promoter (pCRII) (Invitrogen, Carlsbad, CA) and multiple clones were sequenced in both directions at the UIUC Core Sequencing Facility (https://unicorn.biotec.uiuc.edu/). For sequence from START to EXON6, 10 clones were sequenced; for EXON6 to A12-0, 18 clones were sequenced. Sequences for TguFmr1 and TguFmrp were both deposited into NCBI GenBank as accession number EU555184.
Protein alignment
Zebra finch and chicken sequences (XM_420363) were translated using DNA Strider and aligned to the human (NP_002015) and mouse (NP_03257) protein sequences using EBI ClustalW2 (available at http://www.ebi.ac.uk/Tools/clustalw2/index.html). Alignment at the coding nucleotide level was calculated using NCBI bl2seq (available at http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi).
Northern blot and production of DIG-labeled riboprobe
The northern blot was carried out as described (Zayas et al., 2005), using CSPD (Disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2-(5-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate) (Roche Applied Sciences, Indianapolis, IN) instead of CDP-Star as the chemiluminescent substrate. A CSPD working solution was made according to manufacturer's instructions. The probe was a Digoxigenin (DIG)-labeled RNA probe synthesized using the DIG RNA Labeling Kit (Roche Applied Sciences), off a plasmid template that contained an insert of a PCR-amplified fragment from the 3′-UTR of TguFmr1. Primers used to amplify this fragment were (both written 5′→3′) were FMR1LEFT: CATTGTTACTGAGGGAACAAGTGTA and FMR1RIGHT: TGCCCAGTAAGTTTAACGTGTG. To measure the size of the TguFmr1 transcript, avian rRNA subunits were measured against murine rRNA, Fmr1 mRNA, and an RNA ladder and calculated to measure 4.25 and 2.0Kb (data not shown). We used these values as size markers with which to calculate the length of the TguFmr1 transcript.
ORF construction / Sewing PCR
Two cloned sequences were united into one construct encoding the entire open reading frame (ORF) of TguFmr1 via ‘bridging’ or ‘sewing’ PCR (described in (Liu et al., 2002)). Briefly, in sewing PCR two segments of DNA with short overlapping terminal regions are annealed together; the short overlapping region effectively serves as a primer for the full-length fusion. Here, the two shorter sequences joined together were the two PCR amplified pieces described above under sequence generation; they overlapped at EXON6. An EcoRI site was added to the 5′-end using primer EcoRIFMR1 (GGAGAATTCGATGGAGGAGCTGGTGGTGGAGGTGCGG) and a BamHI site was added 84 nucleotides downstream of the translation stop site using primer A12BAMHI (5′→3′ GCCCGGATCCATCTTGCCTACTATT). This final sequence was cloned into pEGFP-C1 (BD Biosciences) using the EcoRI and BamHI sites. The protein product will be described as TguFmrp-EGFP.
Western blot
The western blots were carried out using the following antibodies: affinity-purified polyclonal anti-TguFmrp produced by Abgent (San Diego, CA) to the TguFmrp-specific peptide sequence KERNPKKEKT; monoclonal anti-EGFP (Clontech, Mountain View, CA); polyclonal anti-eukaryotic translation initiation factor 5 (anti-eIF5, Santa Cruz Biotechnology, Santa Cruz, CA); and monoclonal antibody 1a, derived against human FMRP (Devys et al., 1993). Cos-7 cells were transfected with an EGFP vector containing the TguFmrp ORF (see above) using Lipofectamine 2000 (Invitrogen, as per manufacturer's instructions) or mock-transfected. Protein extracts from zebra finch brain and muscle were prepared as described (George et al., 1995).
Immunoprecipitation (IP)
Cos-7 cells were transfected or mock-transfected with transgene encoding TguFmrp-EGFP as described for the western blot and total protein lysate was immunoprecipitated with anti-TguFmrp coupled to protein A sepharose beads. Immunoprecipitate was used for western blot.
Sizing of TguFmrp
The predicted amino acid sequence for TguFmrp was sized by the Compute pI/Mw tool on the ExPASy Proteomics tools server (available at http://ca.expasy.org/tools/pi_tool.html).
Immunocytochemistry (ICC)
Cos-7 cells were transfected or mock-transfected with transgene encoding TguFmrp-EGFP as described for the western blot, in 4-chamber culture slides (BD Falcon via BD Biosciences). After 24h, cells were fixed using 4% formaldehyde with 4% glucose in PBS and permeabilized with 0.2% Triton X-100 in PBS. Next the cells were blocked in 2% bovine serum albumin, 2% newborn calf serum, 0.2% Tween 20, and 0.02% NaN3 then stained using anti-TguFmrp followed by anti-rabbit rhodamine-red (Jackson ImmunoResearch Laboratories, West Grove, PA), and mounted with fluoro-protectant mounting media (0.2% DABCO) containing 1ug/mL DAPI. Cells were then imaged using a Zeiss AxioVert 200M Microscope (Carl Zeiss, Germany).
Collection and storage of tissue for immunohistochemistry (IHC)
Birds were decapitated and the brains rapidly frozen in TissueTek O.C.T. (VWR, Batavia, IL), in brain molds (Polysciences, Inc., Warrington, PA) submerged in dry ice. The process of dissection to freezing took approximately three minutes. The brains were then sectioned in 20-micrometer sections using a Cryostat. Sections were thaw-mounted onto 3-Triethoxysilylpropylamine (TESPA)-coated microscope slides and stored frozen at -80C until used.
Cresyl Violet stains for anatomy
Every fifth slide was stained with cresyl violet to identify the anatomy. Briefly, sections were fixed in 10% formalin then gradually dehydrated through increasing concentrations of EtOH and allowed to air-dry. Next, sections were rehydratd through decreasing concentrations of EtOH, stained with 0.5% cresyl violet (Acros Organics via Fisher Scientific, Hanover Park, IL) in water, and dehydrated through increasing concentrations of EtOH. Finally, sections were cleared with HistoChoice clearing agent (Sigma-Aldrich, St. Louis, MO) and mounted with DPX Mountant (Fluka via Sigma-Aldrich), and allowed to dry overnight. Sections were imaged as described for IHC.
Fluorescent double-label immunohistochemistry
Sections containing RA were fixed in 10% formalin then dehydrated through increasing concentrations of EtOH. Next the sections were rehydrated in PBS, blocked in 5% normal donkey serum, and incubated with the first primary antibody, anti-TguFmrp. The secondary antibody was donkey anti-rabbit rhodamine-red (Jackson ImmunoResearch Laboratories). Next the sections were incubated with the second primary antibody, anti-NeuN (Chemicon International via Millipore, Charlottesville, VA) followed by donkey anti-mouse Cy2 (Jackson ImmunoResearch Laboratories). Finally, coverslips were mounted and sections imaged as for ICC, above.
DAB-Immunohistochemistry
Sections containing RA were fixed in 10% formalin then dehydrated through increasing concentrations of EtOH, blocked with 0.3% normal goat serum, incubated with 0.3% H2O2 to quench endogenous peroxidase activity, then stained using the Vectastain ABC System as per manufacturer's instructions (Vector Laboratories, Burlingame, CA). The primary antibody used was anti-TguFmrp. HRP immunoreactivity was visualized using SigmaFast DAB with Metal Enhancer (Sigma-Aldrich).
Semi-quantification of TguFmrp expression (ROD)
Sections were visualized at 10× or 20× magnification using a Zeiss AxioVert 200M Microscope. The images were all exposed under optimal digital conditions, such that the mean pixel grayscale intensity was centered between 0 and 4095 gray values. Condenser aperture and transmitted light intensity were kept constant at 0.55 n.a. and 2.9V, respectively. Images were acquired using a 12bit camera, and converted post-acquisition to 8bit, in which there are 255 gray levels between 0 (black) and (255 white). Tiles (individual images) were merged into a single mosaic using AxioVision software, release 4.6, to give one image. 8bit mosaic images were analyzed using AxioVision release 4.6 or 4.5 for mean pixel grayscale intensity of selected regions. Five regions within and immediately rostral to the RA nucleus were measured per section, and at least 15 sections were examined per age group. A ratio was taken of the average pixel grayscale intensity, to give the relative optical density (ROD) of TguFmrp immunoreactivity in the RA compared to the surrounding brain region. An ROD value greater than 1 indicates greater TguFmrp expression in the RA than in the surrounding telencephalon.
Statistical analysis
To determine if all three finches per treatment were equivalent to each other, the individual average ratios were compared using an ANOVA, allowing a probability of type I error or alpha of 0.05. If the individual ratios were determined to be in the same distribution, a statistical test for the mean ratio and its difference from 1 (“z-test”) was used to determine if there was a significant difference in TguFmrp immunoreactivity in the RA neuropil compared to that in rostral telencephalon neuropil, again allowing a probability of type I error or alpha of 0.05.
Animals
All experiments were performed under protocols approved by the University of Illinois Laboratory Animal Care Advisory Committee. Zebra finches were bred and raised in an aviary of the Beckman Institute animal facility. All birds were kept on a 12 hr light/dark cycle in flight cages approximately 36″ wide × 48″ deep × 72″ high, and sacrificed in the early afternoon.
Results
The zebra finch ortholog is highly homologous to human FMRP but lacks exons 11 and 12
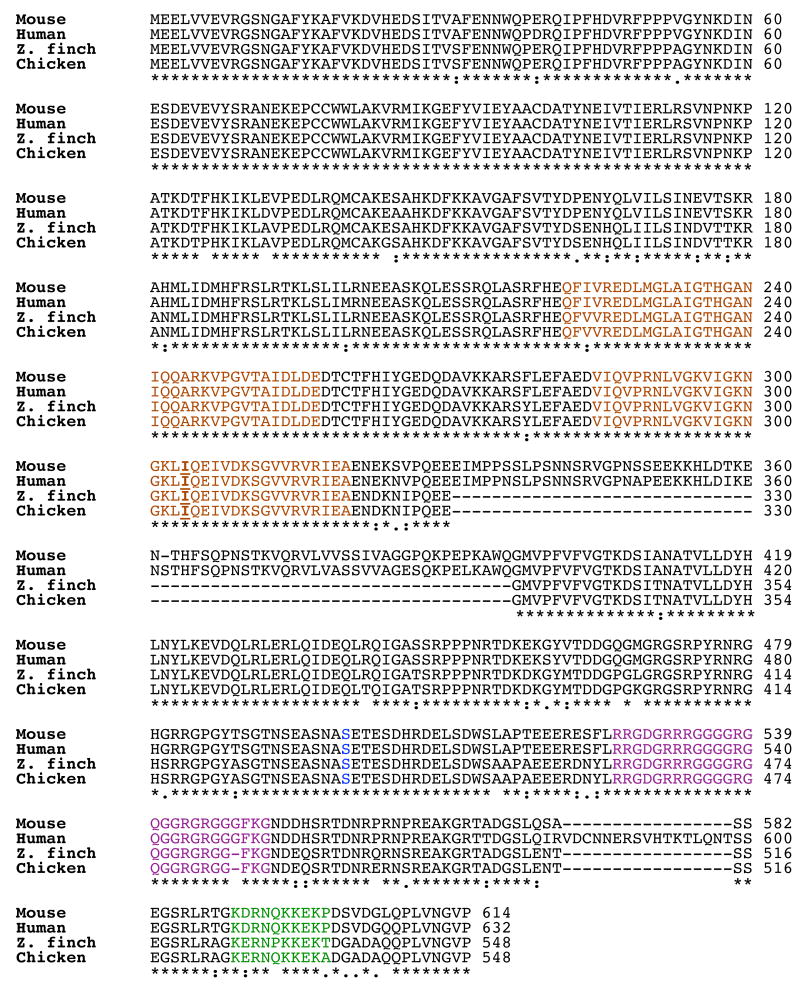
To begin this study, we constructed the TguFmr1 cDNA sequence from the coding start site through the poly-A tail (see Methods). At the nucleotide level, TguFmr1 is most similar to the chicken (Gallus gallus) sequence (90% identity in the coding sequence), which is the only other avian species that has been sequenced for Fmr1 (Price et al., 1996). TguFmr1 is 85% and 84% identical to the coding region of the human and murine orthologs, FMR1 and Fmr1, respectively (data not shown). We next compared the protein sequences and found that all of the orthologs had a strong identity at the amino acid level, indicated by the stars below the alignment (Fig. 2). TguFmrp contains the conserved RNA binding motifs, the KH1 and KH2 domains and the RGG box (Siomi et al., 1993) (Fig.2). Also conserved is the isoleucine in the KH2 domain (at position 304 in the human protein) that when mutated results in a severe FXS phenotype (De Boulle et al., 1993). The serine that is the primary phospho-acceptor (murine serine 499/human serine 500) (Ceman et al., 2003) and flanking serines are also conserved (Figure 2). In contrast, a striking difference was observed between the avian and mammalian orthologs where the TguFmrp and GgFmrp are lacking mammalian sequence residues 330-396, which correspond to exons 11 and 12. FMRP has multiple splice sites from human exons 9-17 (Eichler et al., 1993); to rule out that we had cloned a splice variant, we sequenced at least 10 additional clones and found the same result. Exons 11 and 12 are also absent from the fruit fly Drosophila melanogaster (Wan et al., 2000), the zebra fish Danio rerio (Tucker et al., 2004), and the frog Xenopus tropicalis (Blonden et al., 2005) orthologs, as well as from the mammalian autosomal paralogs Fxr1 and Fxr2 (Siomi et al., 1995). Thus, the zebra finch ortholog of Fmrp is highly similar to the mammalian orthologs but is most similar to the chicken ortholog.
Fig. 2. Alignment of Fmr orthologs of mouse, human, zebra finch, and chicken Fmr proteins.
The alignment was performed using ClustalW. Stars indicate identical residues; dots denote similarity (residues with two dots are more similar than those with one). The human sequence is accession number NP_002015; mouse is NP_03257. Zebra finch cDNA sequence (see methods) was translated using DNA Strider. The chicken cDNA sequence (accession number XM_420363) was translated using DNA Strider. Colors: KH1 and KH2 are shown in brown and the RGG box in purple according to (Siomi et al., 1993); anti-TguFmrp epitope is shown in green; the isoleucine converted to asparagines in one severe case of FXS (De Boulle et al., 1993) is underlined and in bold font within the KH2 domain; the primary phosphorylation site of FMRP and Fmrp (Ceman et al., 2003) is shown in blue.
To determine the size of the TguFmr1 transcript and to confirm the absence of sequence corresponding to mammalian exons 11 and 12 in the TguFmr1 cDNA sequence, we performed a northern blot using total zebra finch mRNA harvested from brain (Fig. 3). FMR1 mRNA measures 4.8 kilobases (Kb) (Verkerk et al., 1991) and we predicted TguFmr1 mRNA to be shorter. Indeed we calculated the mature TguFmr1 transcript to be 3.4kB in size. We suspect the larger, fainter band at 5.0Kb represents prespliced, heterogeneous nuclear RNA for TguFmr1 because no spliceforms were noted in the 28 clones sequenced (see Experimental Procedures).
Fig. 3. TguFmr1 mRNA runs at 3.4Kb.
Total RNA was prepared from songbird brain and 20ug was resolved on a 1.2% agarose/formaldehyde gel and probed with a digoxigenin-labeled riboprobe specific to TguFmr1. Markers indicating sizes in kilobases (Kb) are shown on the left. To measure the transcript length, a standard curve was generated using ribosomal RNA (rRNA) of subunits as size markers. Avian rRNA subunits were measured against murine rRNA and Fmr1 mRNA and calculated to measure 4.25 and 2.0Kb (data not shown). Using these values, we were able to calculate the length of TguFmr1 mRNA as 3.4Kb, with a minor band at 5.0Kb.
Development and characterization of a novel polyclonal antibody specific for TguFmrp
With the protein sequence of TguFmrp determined, we next developed a specific antibody. As mentioned above, there are two autosomal paralogs of FMRP – Fxr1 and Fxr2 (Siomi et al., 1995, Zhang et al., 1995). Thus, a consideration in producing the antibody was to choose an epitope that was not present in the zebra finch paralogs. ESTs corresponding to zebra finch Fxr1 are present in the songbird database (SB02003A2H12.f2 and SB03035B2E05.f1); however the full-length sequence of TguFxr1 is not yet completed. The second paralog Fxr2 has not yet been identified in zebra finch. Interestingly, Xenopus tropicalis is also lacking Fxr2 (Blonden et al., 2005). Thus, in the absence of a complete TguFxr1 sequence, we chose the epitope KERNPKKEKT that was unique to TguFmrp by identifying a stretch of hydrophilic residues in the C-terminal end, where FMRP does not overlap with Fxr1 and Fxr2 or with GgFxr1 (data not shown). By BLAST search of the songbird EST collection, this sequence does not align with any other known or putative proteins in the zebra finch (data not shown).
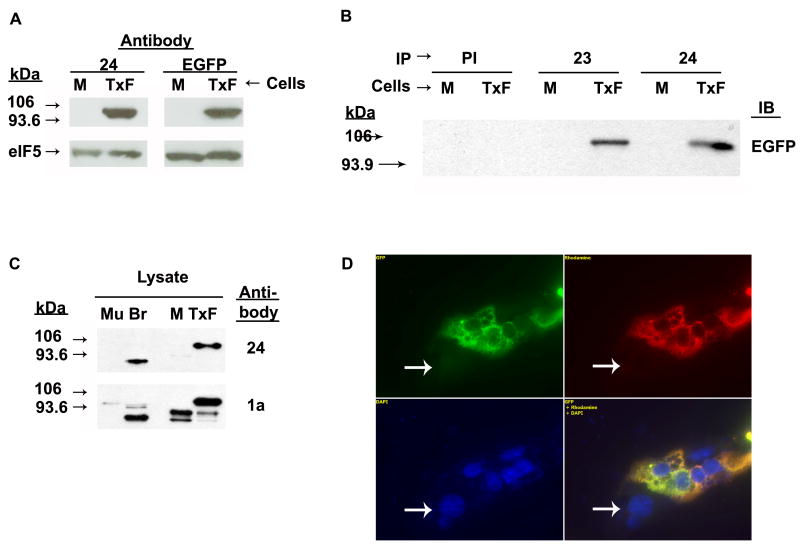
We obtained two affinity-purified rabbit antisera to the KERNPKKEKT peptide (23 and 24; hereafter interchangeably referred to as the anti-TguFmrp antibody). We determined their specificity to TguFmrp in an immunoblot by analyzing lysates prepared from either mock-transfected (M) or transiently transfected TguFmrp-EGFP in Cos-7 cells (Fig. 4A). The EGFP antibody identified the transgene-encoded protein, which was the same size as the protein identified by anti-TguFmrp, suggesting that the antisera recognized TguFmrp encoded by transgene in an immunoblot. To unequivocally demonstrate that it was TguFmrp-EGFP recognized by both antibodies, we immunoprecipitated mock transfected or TguFmrp-EGFP expressing Cos-7 cells with the anti-TguFmrp antibody and found that it specifically immunoprecipitated the Fmr-fusion construct as visualized with the EGFP antibody (Fig. 4B). Taken together, our data show that the anti-TguFmrp antisera can specifically recognize and immunoprecipitate TguFmrp.
Fig. 4. Anti-TguFmrp is specific to TguFmrp.
A. Lysate from Cos-7 cells mock-transfected (M) and transfected (TxF) with TguFmrp-EGFP, immunoblotted with anti-TguFmrp 24 and anti-EGFP. Addition of the 28 kDa EGFP increased the size of the fusion construct to ∼98kDa. eIF5 shown here as a loading control. B. M and TxF lysate was immunoprecipitated with anti-TguFmrp 23 and 24 and the immunoprecipitates were analyzed by western blot using anti-EGFP. C. Protein lysate from zebra finch muscle (Mu) and brain (Br) tissue, as well as lysate from (A) was analyzed by western blot using anti-TguFmrp 24. The blot was followed with the 1a antibody to visualize endogenous Cos-7 Fmrp as well as both zebra finch and Cos-7 Fxr1. Endogenous TguFmrp runs at 75kDa. For A and C, similar results were obtained using anti-TguFmrp 23 (data not shown). D. Cos-7 cells transfected with TguFmr1-EGFP were fixed 24 hours post-transfection and stained with anti-TguFmrp 23. Shown are EGFP expression (green), 23 immunoreactivity (red), and DAPI-labeled nuclei (blue). The overlay of all three exposures is shown in the lower right. Similar results were obtained with anti-TguFmrp 24 (data not shown). Bar = 20μm
Next, it was critical to determine that the anti-TguFmrp antibody recognized endogenous TguFmrp in zebra finch brain lysate (Fig. 4C). Total protein was harvested from samples of male finch male pectoral muscle (Mu), brain (Br) and Cos-7 cells mock-transfected (M) and transgene transfected (TxF), as a negative and positive control, respectively. Brain TguFmrp was recognized by anti-TguFmrp and observed to run at ∼75 kDa (Br in Fig. 4C). The EGFP tag adds approximately 28 kDa to the size of the native protein; thus, the antibody detected a 103kDa fusion protein in the Cos-7 transfected cells (TxF in Fig. 4C). Importantly, there was no reactivity with the muscle extract, which expresses Fxr1 and not Fmrp (Khandjian et al., 1998). To confirm that Fxr1 was indeed present in muscle extract, the membrane was re-probed with anti-Fmrp 1a (Devys et al., 1993), which cross-reacts with Fxr1 (Mazroui et al., 2003) (Fig. 4C, lower). Thus, zebra finch Fxr1 is present in muscle extract but the anti-TguFmrp antibody does not recognize it, leading us to conclude that this antibody is specific for TguFmrp and recognizes it in brain extracts.
To determine whether we could use this reagent for immunostains, we transiently transfected Cos-7 cells with TguFmrp-EGFP and performed immunocytochemistry (ICC) with the anti-TguFmrp antibody. We found complete overlap between EGFP expression (green) and reactivity with the antibody, as visualized using a rhodamine-coupled secondary antibody (red, Fig. 4D). Importantly, we saw no staining of nontransfected cells, which were identified with DAPI stain (Fig. 4D, arrows). Our results indicate that the anti-TguFmrp antibody can be used for specific immunohistology, reacting only with cells expressing TguFmrp.
TguFmrp is expressed in male song nuclei and this expression is restricted to neurons
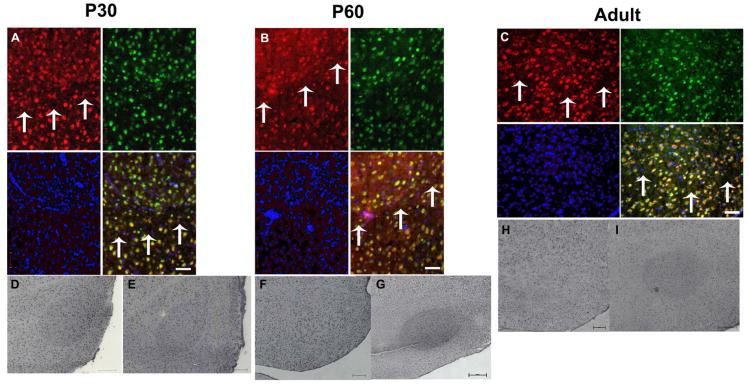
Confident that our antibody could specifically detect TguFmrp, we next examined brain sections for expression of TguFmrp in the song nuclei (Fig. 5). Because FXS patients have impaired sensorimotor function, and in finches the sensorimotor period begins around day P35, we chose to examine finches at the three ages of P30, P60, and Adult (>P90), to address periods prior to, during, and after sensorimotor learning, respectively. We first identified brain sections that contained the song nuclei HVC and RA by cresyl violet staining (Fig. 5A). We then stained subsequent sections with the anti-TguFmrp antibody, visualized with a secondary antibody coupled to rhodamine (red). We identified neurons with an antibody against the neuronal marker NeuN (green) and all cells with the nuclear DAPI stain (blue). Cell bodies expressing TguFmrp colocalized with cells expressing NeuN (Fig. 5 lower right panels). We also identified a number of small cells that stained with DAPI but neither TguFmrp nor NeuN. Since they were not neuronal, we suspect that they were glial cells, which express little or no Fmrp (Feng et al., 1997, Wang et al., 2004, Pacey and Doering, 2007). Using cresyl violet staining, we identified more medial sections containing the song nuclei LMAN and Area X (Fig. 5B). Again, we found results similar to those shown in Fig. 5A where TguFmrp colocalized with the NeuN marker, indicating that TguFmrp is expressed in neurons. Taken together, our results show that TguFmrp is expressed in the song nuclei HVC, RA, LMAN and Area X of male zebra finches in a neuron-specific manner.
Fig. 5. TguFmrp is expressed in neurons in the male zebra finch brain.
Sagital brain sections containing A. HVC (letter-based name) and RA and B. LMAN (Lateral MAgnocellular nucleus of the Nidopallium) and Area X were stained for anatomy with cresyl violet (bar = 1000μm) (upper left). Upper right is an accompanying sketch with the significant anatomical features indicated. Adjacent sections were co-stained with the anti-TguFmrp antibody 24 (red), the neuronal marker NeuN (green), and DAPI (blue). In the overlay, a yellow signal indicates co-fluorescence for red and green. Bar = 20μm. Bst: Brainstem; Cb: Cerebellum; LFM: Lamina frontalis suprema; LFS: Lamina frontalis superior; LH: Lamina hyperstriatica; LMD: Lamina medullaris dorsalis; TeO: Optic Tectum. Shown are images from a P30 brain; P60 and Adult males showed similar results (data not shown).
TguFmrp expression is elevated at P30 and varies in the RA nucleus during and after the sensorimotor learning period
After determining that TguFmrp was expressed in neurons in all brain regions examined and at all ages examined, we evaluated the song nuclei HVC, Area X, LMAN, and RA at a lower magnification. We observed elevated TguFmrp expression in the RA nucleus, compared to the surrounding telencephalon for all three ages (Fig. 6A-C, red, above the arrows). We did not observe disparate TguFmrp expression in the cell bodies (data not shown) nor did we see elevated TguFmrp expression in any of the other song nuclei (data not shown). To visualize the entire RA, we performed immunohistochemistry (IHC) with 3,3′-diaminobenzadine (DAB-IHC). Similar to the results obtained by fluorescent immunostain, we saw TguFmrp staining throughout the telencephalon, with more TguFmrp in the RA in male P30 finches (Figs. 6D and E); however, the strikingly elevated expression of TguFmrp in the RA nucleus was not consistently observed in all of the adolescent (P60) and adult males (Figs. 6F-I). In fact, there was variability in TguFmrp expression in the P60 and Adult males, ranging from no or low elevation (Figs. 6F, H) to highly elevated expression (compare Fig. 6F to Fig. 6G and Fig. 6H to Fig. 6I). The staining is specific since preabsorbing the anti-TguFmrp antiserum with the immunizing peptide completely eliminated reactivity to RA (data not shown). As expected, similar analysis of other song regions showed no difference in TguFmrp expression as compared to the surrounding telencephalon (data not shown).
Fig. 6. TguFmrp is consistently elevated in the RA nucleus of a P30 male zebra finch and variably expressed in P60 and Adult males.
A-C. Representative fluorescent-IHC using anti-TguFmrp 24 on a male P30 (A) P60 (B) and Adult (C) zebra finch RA. Shown are TguFmrp immunoreactivity (red), NeuN stain (green), and DAPI-labeled nuclei (blue), along with the overlay. Arrows denote ventral border of RA. Bar = 100μm. D-F. Representative DAB-IHC using anti-TguFmrp 24 on a male P30 (D) P60 (E) and Adult (F) zebra finch RA. Bar = 200μm.
To semi-quantify the relative amount of TguFmrp in the RA, compared to the flanking telencephalon, relative optical density (ROD) values were determined (see Methods), such that a value greater than 1 indicated more TguFmrp in the RA than in the surrounding telencephalon (Table 1). Analysis of three P30 males showed significantly more TguFmrp expression in the RA of all three birds (analyzed by z-test); these birds also fell into the same distribution (analyzed by ANOVA) and their ROD values could therefore be combined to show a significant increase of TguFmrp expression in the RA nucleus of male P30 zebra finches than in the surrounding telencephalon.
Table 1. Semi-quantification of TguFmrp expression in the RA.
Table showing ROD values for three P30, three P60, three Adult RAs, one P23, and one P27, where an ROD value >1 indicates more TguFmrp in the RA than in the surrounding brain region. (* p<0.05; ** p<0.01; *** p<0.001; n.s. not significant)
| Bird | ROD | p-value | |
|---|---|---|---|
| P30-1 | 1.111 | ≪0.001 | *** |
| P30-2 | 1.082 | ≪0.001 | *** |
| P30-3 | 1.046 | 0.004 | ** |
|
| |||
| P60-1 | 1.126 | ≪0.001 | *** |
| P60-2 | 1.002 | 0.436 | n.s. |
| P60-3 | 1.043 | 0.004 | ** |
|
| |||
| Adult-1 | 1.099 | ≪0.001 | *** |
| Adult-2 | 1.022 | 0.013 | * |
| Adult-3 | 1.071 | ≪0.001 | *** |
|
| |||
| P23 | 1.007 | 0.327 | n.s. |
|
| |||
| P27 | 1.015 | 0.105 | n.s. |
Out of the three P60 males, two males had significantly increased TguFmrp expression in the RA nucleus; all three Adult males had significantly increased TguFmrp expression in the RA nucleus, although one had significantly less than the other 2 (Table 1). By ANOVA analysis, the three ROD values for each of the older age groups did not fall into the same distribution and therefore could not be averaged to provide a mean ratio of TguFmrp stain intensity in the RA nucleus as compared to the surrounding telencephalon. We saw no birds in which the TguFmrp expression in the RA nucleus was lower than in the surrounding telencephalon. We therefore conclude that TguFmrp expression in the RA nucleus is clearly elevated at P30 with more variable expression at older ages.
To ask when this elevation first occurs, we performed a similar analysis on one male P23 and one male P27 and did not observe elevated TguFmrp levels in the RA or any other brain region (Table 1). Our preliminary data suggest that the elevated expression of TguFmrp in the RA nucleus consistently observed at P30 begins after P27.
Discussion
We found that the zebra finch Taeniopygia guttata Fmr1 cDNA and protein product are remarkably homologous to their human ortholog FMRP with the notable exception that exons 11 and 12 are absent from the zebra finch cDNA. Using a specific antibody, we found that TguFmrp is expressed in neurons throughout the brain and in the song nuclei of male P30 zebra finches, and that this increased expression is variable in RA in adolescent and adult males: it is elevated in some birds and unchanged in others. Our results establish that TguFmrp is present at a key place and time in the developing song system and thus may participate in the cellular and synaptic changes that are occurring in sensorimotor learning. It will be interesting to determine at what age this elevation first presents; preliminary work suggests that it is after P27.
Synaptic plasticity in the RA has been proposed as a primary mechanism for song learning and production (Mooney, 1992, Stark and Perkel, 1999, Fee et al., 2004). RA lies at the convergence of the Anterior Forebrain Pathway and the Posterior Pathway (Figure 1) and is required for song production (Nottebohm et al., 1976). The RA receives input from both the HVC and LMAN, as well as intrinsic interneurons, and therefore is not merely a relay nucleus from the HVC to the motor nucleus nXIIts (Spiro et al., 1999). HVC is a nucleus critical for song production; when lesioned in an adult male, major deficits in song structure are observed (Nottebohm et al., 1976). LMAN is also a nucleus critical for song learning; when lesioned in a juvenile male, the plastic song crystallizes too early, suggesting that input from LMAN is critical for introducing the variability that is required to produce a mature song (Scharff and Nottebohm, 1991, Ölveczky et al., 2005). Additionally, the RA receives input from the Ventral Palleostriatrum (VP – analogous to the mammalian Nucleus Basalis Magnocellularis (NBM) (Li and Sakaguchi, 1997), which does in fact show strong FMR1 expression (Abitbol et al., 1993). The mammalian NBM is critical for distinguishing relevant from irrelevant sensory stimuli (Wenk, 1997), and the finch VP receives auditory input. Together these studies support a role for TguFmrp in sensorimotor learning, as during this stage the adolescent male compares his song to that of his tutor.
At P30 when we detected elevated TguFmrp in RA, the young male is actively listening to his adult male tutor and just beginning the immature vocalizations that will be shaped over the following weeks into a close copy of his tutor's performance. By this age, axons from nucleus HVC have just begun to enter RA (Konishi and Akutagawa, 1985, Mooney and Rao, 1994, Holloway and Clayton, 2001), forming synapses onto the dendrites of neurons that already receive synapses from LMAN (Canady et al., 1988). Sometime during the next three weeks, the terminals from LMAN projections will double in size and decrease in number by half, implying a major change in the organization of RA dendrites (Herrmann and Arnold, 1991).
Since Fmrp is required for normal dendritic spine maturation (Weiler and Greenough, 1999, Greenough et al., 2001, Michel et al., 2004), we propose that the component of the RA nucleus in which TguFmrp is elevated is the reorganizing dendritic compartment. We further hypothesize that TguFmrp is necessary for the synaptic maturation in RA associated with sensorimotor learning. In contrast, TguFmrp may not be necessary for production of mature vocalizations, since in humans, fragile X individuals are capable of vocalization. We also note that Fmrp has been reported in axons (Antar et al., 2006, Price et al., 2006, Tessier and Broadie, 2008b). Thus it is formally possible that the increased TguFmrp in the RA is contributed by axons from HVC, LMAN, or VP instead of or in addition to the dendrites of RA neurons. It should be possible to test all of these hypotheses in zebra finches using viral vector-mediated knockdown of the mRNA, as recently applied in a test of FoxP2 mRNA function in nucleus Area X (Haesler et al., 2007).
Elevated Fmrp in brain neuropil has been previously reported in other experimental models. For example, Fmrp levels transiently increased in the neuropil of visual cortex in rats reared in the dark and then exposed to light (Gabel et al., 2004). Further, neuropil Fmrp levels are increased in response to enriched environment, as well as to whisker stimulation when barrel cortex was examined (Irwin et al., 2000, Todd and Mack, 2000, Irwin et al., 2005). Since Fmrp expression has been shown to change in an experience-dependent fashion, we hypothesized that the variability in TguFmrp expression in RA in P60 and adult males was due to recent experience. Because the RA is a premotor nucleus for singing, finches with elevated TguFmrp in RA may have sung immediately before analysis. Thus, we predicted that P60 males kept in silence overnight would have the same amount of TguFmrp in the neuropil of RA as in the surrounding telencephalon. To test this hypothesis, three P60 males were placed in isolation overnight. The following day, each of the P60 males was analyzed as described for Fig. 6. All three P60 males had an ROD that was not significantly different than 1 (data not shown). Thus, there was no increase in TguFmrp expression in the RA nucleus in the absence of singing. One possible interpretation of this data is that TguFmrp expression in RA is influenced by the motor act of singing. Toward that end, individual P60 males that were isolated and kept in silence over night were either allowed to sing undirected song or were induced to sing by introducing females in separate cages prior to examination of their brains for TguFmrp expression. Preliminary data showed that neither treatment restored increased levels of TguFmrp in RA (data not shown). Thus, although keeping them in silence overnight suppressed elevated TguFmrp in RA, the process of singing after a night in isolation did not induce an increase in TguFmrp expression. Future experiments analyzing more finches and monitoring variables such as the total time singing, the number of motifs sung, and the interval between singing and analysis may reveal a relationship between the motor act of singing and TguFmrp expression in the adolescent male RA. Alternatively, a more socially complex or sustained singing behavior may be required for elevated TguFmrp expression in RA.
The zebra finch model offers a unique opportunity to explore the functional consequences of Fmrp expression on vocal development, an important aspect of the fragile X syndrome. It is our hope that future researchers can utilize this model to develop novel therapeutic techniques that will ameliorate the speech deficit seen in FXS.
Acknowledgments
We would like to thank Dr. Sarah London for advice and technical support, as well as Dr. Ricardo Zayas, Michael Miles and Dr. Mayandi Sivaguru for their technical assistance. DAB-IHC imaging was performed at the Microscopy and Imaging Facility at the Institute for Genomic Biology at UIUC. This work was supported in part by Public Health Service grant HD41591-01 from NICHD, the FRAXA foundation and the Spastic Paralysis Research Foundation of the Illinois-Eastern Iowa District of Kiwanis International to S. Ceman; the NIH Cellular and Molecular Biology Training Grant at UIUC (T32 GM007283) and first-year Neuroscience Program fellowship to C. Winograd; and the Songbird Neurogenomics Initiative (PHS grant RO1 NS045264).
Abbreviations
- EGFP
(enhanced GFP – green fluorescence protein)
- FMR1, Fmr1
(fragile X mental retardation 1 gene or transcript)
- FMRP, Fmrp
(fragile X mental retardation protein)
- Fxr1, Fxr2
(fragile X related protein 1, fragile X related protein 2)
- FXS
(fragile X syndrome)
- Gg
(Gallus gallus)
- ICC, IHC
(immunocytochemistry, immunohistochemistry)
- KH1, 2
(K homology 1, 2)
- LMAN
(lateral magnocellular nucleus of the nidopallium)
- P23, P27, P30, P60, P90
(post-hatch day 23, post-hatch day 27, etc.)
- RA
(robust nucleus of the arcopallium)
- ROD
(relative optical density)
- TD
(typical, developmentally-age matched)
- Tgu
(Taeniopygia guttata)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbeduto L, Brady N, Kover ST. Language development and fragile X syndrome: profiles, syndrome-specificity, and within-syndrome differences. Ment Retard Dev Disabil Res Rev. 2007;13:36–46. doi: 10.1002/mrdd.20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abitbol M, Menini C, Delezoide AL, Rhyner T, Vekemans M, Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Moll Cell Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Aronov D, Andalman AS, Fee MS. A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science. 2008;320:630–634. doi: 10.1126/science.1155140. [DOI] [PubMed] [Google Scholar]
- Bailey DBJ, Hatton DD, Skinner M. Early developmental trajectories of males with fragile X syndrome. Am J Ment Retard. 1998;103:29–39. doi: 10.1352/0895-8017(1998)103<0029:EDTOMW>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bailey DBJ, Skinner D, Sparkman KL. Discovering fragile X syndrome: family experiences and perceptions. Pediatrics. 2003;111:407–416. doi: 10.1542/peds.111.2.407. [DOI] [PubMed] [Google Scholar]
- Baranek GT, Danko CD, Skinner ML, Bailey DB, Jr, Hatton DD, Roberts JE, Mirrett PL. Video analysis of sensory-motor features in infants with fragile X syndrome at 9-12 months of age. J Autism Dev Disord. 2005;35:645–656. doi: 10.1007/s10803-005-0008-7. [DOI] [PubMed] [Google Scholar]
- Belser RC, Sudhalter V. Conversational characteristics of children with fragile X syndrome: repetitive speech. Am J Ment Retard. 2001;106:28–38. doi: 10.1352/0895-8017(2001)106<0028:CCOCWF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Blonden L, van 't Padje S, Severijnen L, Destree O, Oostra BA, Willemsen R. Two members of the Fxr gene family, Fmr1 and Fxr1, are differentially expressed in Xenopus tropicalis. Int J Dev Biol. 2005;49:437–441. doi: 10.1387/ijdb.051974lb. [DOI] [PubMed] [Google Scholar]
- Brainard MS, Doupe AJ. What songbirds teach us about learning. Nature. 2002;417:351–358. doi: 10.1038/417351a. [DOI] [PubMed] [Google Scholar]
- Bächner D, Steinbach P, Wohrle D, Just W, Vogel W, Hameister H, Manca A, Poustka A. Enhanced Fmr-1 expression in testis. Nat Genet. 1993;4:115–116. doi: 10.1038/ng0693-115. [DOI] [PubMed] [Google Scholar]
- Canady RA, Burd GD, DeVoogd TJ, Nottebohm F. Effect of testosterone on input received by an identified neuron type of the canary song system: a Golgi/electron microscopy/degeneration study. J Neurosci. 1988;8:3770–3784. doi: 10.1523/JNEUROSCI.08-10-03770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJMH, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons, and appears normal in carriers of the fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Perkel DJ, Reiner A, Stern EA. Birdbrains could teach basal ganglia research a new song. Trends Neurosci. 2005;28:353–363. doi: 10.1016/j.tins.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Dykens EM, Hodapp RM, Ort SI, Leckman JF. Trajectory of adaptive behavior in males with fragile X syndrome. J Autism Dev Disord. 1993;23:135–145. doi: 10.1007/BF01066423. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Richards S, Gibbs RA, Nelson DL. Fine structure of the human FMR1 gene. Hum Mol Genet. 1993;2:1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- Fee MS, Kozhevnikov A, Hahnloser R. Neural mechanisms of vocal sequence generation in the songbird. Ann N Y Acad Sci. 2004;1016:153–170. doi: 10.1196/annals.1298.022. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando-Lucas MT, Banús-Gómez P, López-Pérez G. Aspects of cognition and language in children with fragile X syndrome. Rev Neurol. 2003;36 Suppl 1:S137–142. [PubMed] [Google Scholar]
- Ferrier LJ, Bashir AS, Meryash DL, Johnston J, Wolff P. Conversational skills of individuals with fragile-X syndrome: a comparison with autism and Down syndrome. Dev Med Child Neurol. 1991;33:776–788. doi: 10.1111/j.1469-8749.1991.tb14961.x. [DOI] [PubMed] [Google Scholar]
- Gabel LA, Won S, Kawai H, McKinney M, Tartakoff AM, Fallon JR. Visual experience regulates transient expression and dendritic localization of fragile X mental retardation protein. J Neurosci. 2004;24:10579–10583. doi: 10.1523/JNEUROSCI.2185-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George JM, Jin H, Woods WS, Clayton DF. Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron. 1995;15:361–372. doi: 10.1016/0896-6273(95)90040-3. [DOI] [PubMed] [Google Scholar]
- Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci U S A. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, Scharff C. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus. Area X PLoS Biology. 2007;5:2885–2897. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergersberg M, Matsuo K, Gassman M, Schaffner W, Lüscher B, Rülicke T, Aguzzi A. Tissue-specific expression of a FMR1/beta-galactosidase fusion gene in transgenic mice. Hum Mol Genet. 1995;4:359–366. doi: 10.1093/hmg/4.3.359. [DOI] [PubMed] [Google Scholar]
- Herrmann K, Arnold AP. The development of afferent projections to the robust archistriatal nucleus in male zebra finches: a quantitative electron microscopic study. J Neurosci. 1991;11:2063–2074. doi: 10.1523/JNEUROSCI.11-07-02063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway CC, Clayton DF. Estrogen synthesis in the male brain triggers development of the avian song control pathway in vitro. Nat Neurosci. 2001;4:170–175. doi: 10.1038/84001. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Christmon CA, Grossman AW, Galvez R, Kim SH, DeGrush BJ, Weiler IJ, Greenough WT. Fragile X mental retardation protein levels increase following complex environment exposure in rat brain regions undergoing active synaptogenesis. Neurobiol Learn Mem. 2005;83:180–187. doi: 10.1016/j.nlm.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Swain RA, Christmon CA, Chakravarti A, Weiler IJ, Greenough WT. Evidence for altered Fragile-X mental retardation protein expression in response to behavioral stimulation. Neurobiol Learn Mem. 2000;74:87–93. [PubMed] [Google Scholar]
- Kao MH, Doupe AJ, Brainard MS. Contributions of an avian basal ganglia-forebrain circuit to real-time modulation of song. Nature. 2005;433:638–643. doi: 10.1038/nature03127. [DOI] [PubMed] [Google Scholar]
- Kau AS, Meyer WA, Kaufmann WE. Early development in males with Fragile X syndrome: a review of the literature. Microsc Res Tech. 2002;57:174–178. doi: 10.1002/jemt.10069. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Bardoni B, Corbin F, Sittler A, Giroux S, Heitz D, Tremblay S, Pinset C, Montarras D, Rousseau F, Mandel J. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum Mol Genet. 1998;7:2121–2128. doi: 10.1093/hmg/7.13.2121. [DOI] [PubMed] [Google Scholar]
- Konishi M, Akutagawa E. Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch. Nature. 1985;315:145–147. doi: 10.1038/315145a0. [DOI] [PubMed] [Google Scholar]
- Li R, Sakaguchi H. Cholinergic innervation of the song control nuclei by the ventral paleostriatum in the zebra finch: a double-labeling study with retrograde fluorescent tracers and choline acetyltransferase immunohistochemistry. Brain Res. 1997;763:239–246. doi: 10.1016/s0006-8993(97)00417-4. [DOI] [PubMed] [Google Scholar]
- Ling SC, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc Natl Acad Sci U S A. 2004;101:17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Thaler DS, Libchaber A. Signal and noise in bridging PCR. BMC Biotechnology. 2002;2:13. doi: 10.1186/1472-6750-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Huot ME, Tremblay S, Boilard N, Labelle Y, Khandjian EW. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum Mol Genet. 2003;12:3087–3096. doi: 10.1093/hmg/ddg335. [DOI] [PubMed] [Google Scholar]
- Michel CI, Kraft R, Restifo LL. Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J Neurosci. 2004;24:5798–5809. doi: 10.1523/JNEUROSCI.1102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R. Synaptic basis for developmental plasticity in a birdsong nucleus. J Neurosci. 1992;12:2464–2477. doi: 10.1523/JNEUROSCI.12-07-02464.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R, Rao M. Waiting periods versus early innervation: the development of axonal connections in the zebra finch song system. J Neurosci. 1994;14:6532–6543. doi: 10.1523/JNEUROSCI.14-11-06532.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Ann Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Ölveczky BP, Andalman AS, Fee MS. Vocal experimentation in the juvenile songbird requires a basal ganglia circuit. PLoS Biol. 2005;3:e153. doi: 10.1371/journal.pbio.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey LK, Doering LC. Developmental expression of FMRP in the astrocyte lineage: implications for fragile X syndrome. Glia. 2007;55:1601–1609. doi: 10.1002/glia.20573. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang F, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Price DK, Zhang F, Ashley CT, Jr, Warren ST. The chicken FMR1 gene is highly conserved with a CCT 5′-untranslated repeat and encodes an RNA-binding protein. Genomics. 1996;31:3–12. doi: 10.1006/geno.1996.0002. [DOI] [PubMed] [Google Scholar]
- Price TJ, Flores CM, Cervero F, Hargreaves KM. The RNA binding and transport proteins staufen and fragile X mental retardation protein are expressed by rat primary afferent neurons and localize to peripheral and central axons. Neuroscience. 2006;141:2107–2116. doi: 10.1016/j.neuroscience.2006.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J, Martin GE, Moskowitz L, Harris AA, Foreman J, Nelson L. J Speech Lang Hear Res. Vol. 50. 2007a. Discourse skills of boys with fragile X syndrome in comparison to boys with Down syndrome; pp. 475–492. [DOI] [PubMed] [Google Scholar]
- Roberts J, Price J, Barnes E, Nelson L, Burchinal M, Hennon EA, Moskowitz L, Edwards A, Malkin C, Anderson K, Misenheimer J, Hooper SR. Receptive vocabulary, expressive vocabulary, and speech production of boys with fragile X syndrome in comparison to boys with down syndrome. Am J Ment Retard. 2007b;112:177–193. doi: 10.1352/0895-8017(2007)112[177:RVEVAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Roberts JE, Hennon EA, Price JR, Dear E, Anderson K, Vandergrift NA. Expressive language during conversational speech in boys with fragile X syndrome. Am J Ment Retard. 2007c;112:1–17. doi: 10.1352/0895-8017(2007)112[1:ELDCSI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Roberts JE, Long SH, Malkin C, Barnes E, Skinner M, Hennon EA, Anderson K. J Speech Lang Hear Res. Vol. 48. 2005. A comparison of phonological skills of boys with fragile X syndrome and Down syndrome; pp. 980–995. [DOI] [PubMed] [Google Scholar]
- Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Siomi H, Sauer WH, Srinivasan S, Nussbaum RL, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation protein. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro JE, Dalva MB, Mooney R. Long-rage inhibition within the zebra finch song nucleus RA can coordinate the firing of multiple projection neurons. J Neurophysiol. 1999;81:3007–3020. doi: 10.1152/jn.1999.81.6.3007. [DOI] [PubMed] [Google Scholar]
- Stark LL, Perkel DJ. Two-stage, input-specific synaptic maturation in a nucleus essential for vocal production in the zebra finch. J Neurosci. 1999;19:9107–9116. doi: 10.1523/JNEUROSCI.19-20-09107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhalter V, Cohen IL, Silverman W, Wolf-Schein EG. Conversational analyses of males with fragile X, Down syndrome, and autism: comparison of the emergence of deviant language. Am J Ment Retard. 1990;94:431–441. [PubMed] [Google Scholar]
- Terracciano A, Chiurazzi P, Neri G. Fragile X syndrome. Am J Med Genet C Semin Med Genet. 2005;137C:32–37. doi: 10.1002/ajmg.c.30062. [DOI] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008a;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier CR, Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008b doi: 10.1242/dev.015867. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Johnson F. HVC microlesions do not destabilize the vocal patterns of adult male zebra finches with prior ablation of LMAN. Dev Neurobiol. 2007;67:205–218. doi: 10.1002/dneu.20287. [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ. Sensory stimulation increases cortical expression of the fragile X mental retardation protein in vivo. Brain Res Mol Brain Res. 2000;80:17–25. doi: 10.1016/s0169-328x(00)00098-x. [DOI] [PubMed] [Google Scholar]
- Tucker B, Richards R, Lardelli M. Expression of three zebrafish orthologs of human FMR1-related genes and their phylogenetic relationships. Dev Genes Evol. 2004;214:567–574. doi: 10.1007/s00427-004-0438-9. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ku L, Osterhout DJ, Li W, Ahmadian A, Liang Z, Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum Mol Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Greenough WT. Synaptic synthesis of the Fragile X protein: possible involvement in synapse maturation and elimination. Am J Med Genet. 1999;83:248–252. doi: 10.1002/(sici)1096-8628(19990402)83:4<248::aid-ajmg3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci U S A. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk GL. The nucleus basalis magnocellularis cholinergic system: one hundred years of progress. Neurobiol Learn Mem. 1997;67:85–95. doi: 10.1006/nlme.1996.3757. [DOI] [PubMed] [Google Scholar]
- White SA. Learning to communicate. Curr Opin Neurobiol. 2001;11:510–520. doi: 10.1016/s0959-4388(00)00242-7. [DOI] [PubMed] [Google Scholar]
- Wilbrecht L, Nottebohm F. Vocal learning in birds and humans. Ment Retard Dev Disabil Res Rev. 2003;9:135–148. doi: 10.1002/mrdd.10073. [DOI] [PubMed] [Google Scholar]
- Wolf-Schein EG, Sudhalter V, Cohen IL, Fisch GS, Hanson D, Pfadt AG, Hagerman R, Jenkins E, Brown WT. Speech-language and the fragile X syndrome: initial findings. ASHA. 1987;29:35–38. [PubMed] [Google Scholar]
- Zayas RM, Bold TD, Newmark PA. Spliced-Leader trans-Splicing in Freshwater Planarians. Mol Biol Evol. 2005;22:2048–2054. doi: 10.1093/molbev/msi200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, O'Connor JP, Siomi MC, Srinivasan S, Dutra A, Nussbaum RL, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]